Abstract
Background
2,3-Butanediol is a chemical compound of increasing interest due to its wide applications. It can be synthesized via mixed acid fermentation of pathogenic bacteria such as Enterobacter aerogenes and Klebsiella oxytoca. The non-pathogenic Saccharomyces cerevisiae possesses three different 2,3-butanediol biosynthetic pathways, but produces minute amount of 2,3-butanediol. Hence, we attempted to engineer S. cerevisiae strain to enhance 2,3-butanediol production.
Results
We first identified gene deletion strategy by performing in silico genome-scale metabolic analysis. Based on the best in silico strategy, in which disruption of alcohol dehydrogenase (ADH) pathway is required, we then constructed gene deletion mutant strains and performed batch cultivation of the strains. Deletion of three ADH genes, ADH1, ADH3 and ADH5, increased 2,3-butanediol production by 55-fold under microaerobic condition. However, overproduction of glycerol was observed in this triple deletion strain. Additional rational design to reduce glycerol production by GPD2 deletion altered the carbon fluxes back to ethanol and significantly reduced 2,3-butanediol production. Deletion of ALD6 reduced acetate production in strains lacking major ADH isozymes, but it did not favor 2,3-butanediol production. Finally, we introduced 2,3-butanediol biosynthetic pathway from Bacillus subtilis and E. aerogenes to the engineered strain and successfully increased titer and yield. Highest 2,3-butanediol titer (2.29 g·l-1) and yield (0.113 g·g-1) were achieved by Δadh1 Δadh3 Δadh5 strain under anaerobic condition.
Conclusions
With the aid of in silico metabolic engineering, we have successfully designed and constructed S. cerevisiae strains with improved 2,3-butanediol production.
Keywords: 2,3-Butanediol; Saccharomyces cerevisiae; Metabolic engineering; Flux balance analysis; Alcohol dehydrogenase; OptKnock
Background
With soaring oil price but indefinitely high demand for petroleum, various sustainable forms of alternative energy and chemicals have been sought after. Microorganisms are able to utilize a wide range of substrate such as plant biomass or agricultural waste and convert them into valuable chemicals and biofuel. With rapid development in microbial engineering technology, this bio-based refinery will be more feasible in terms of cost in the future and eventually reduce the dependency on fossil fuel.
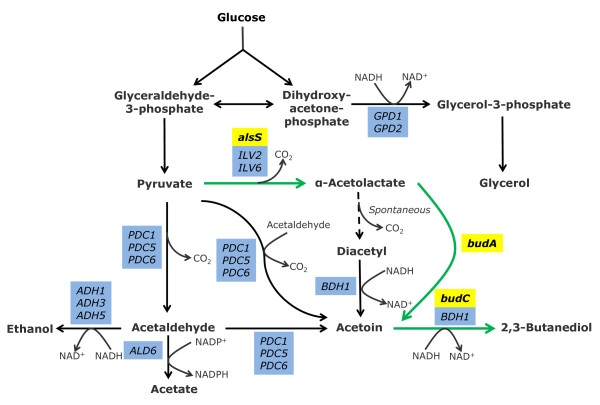
2,3-Butanediol is an interesting metabolic product as its derivatives can be used in wide arrays of industries ranging from synthetic rubber, solvents and drugs. 2,3-Butanediol can be produced efficiently via mixed acid fermentation with prokaryotes such as Klebsiella pneumoniaKlebsiella oxytocaEnterobacter aerogenesSerratia, and Bacillus polymyxa[1]. In these bacteria, pyruvate is first converted into α-acetolactate by acetolactate synthase. In anoxic state, α-acetolactate decarboxylase catalyzes the conversion of α-acetolactate into acetoin (Figure 1, green arrow). In the presence of oxygen, spontaneous decarboxylation of α-acetolactate produces diacetyl. Diacetyl reductase then converts diacetyl into acetoin. 2,3-Butanediol is resulted from the reduction of acetoin by butanediol dehydrogenase.
Figure 1.
Metabolic pathways for 2,3-butanediol biosynthesis and pyruvate metabolism in S. cerevisiae. Green arrow shows the bacterial 2,3-butanediol synthesis pathway through α-acetolactate decarboxylase. Blue boxes contain the S. cerevisiae innate genes, while yellow boxes indicate the foreign genes introduced to the engineered strains.
Most of these bacteria, however, belong to class 2 microorganisms, which are not desirable in industrial-scale fermentation in terms of safety regulations [2]. The need for safe 2,3-butanediol producers are undeniably important when 2,3-butanediol are used as precursors for food additives and cosmetics. Yang et al. has successfully screened a GRAS (generally regarded as safe) microorganism Bacillus amyloliquefaciens B10-127 that has high yield (0.42 g·g-1glucose) and could produce up to 92.3 g·l-1 of 2,3-butanediol during fed-batch fermentation [3]. The model eukaryotic microorganism Saccharomyces cerevisiae which is widely used in the industry is also a promising host microorganism.
In S. cerevisiae, acetaldehyde, pyruvate and α-acetolactate are the precursors of 2,3-butanediol (Figure 1). The biosynthetic pathway with diacetyl as intermediate is similar with that of the bacteria’s. However, α-acetolactate decarboxylase is not found in most of the S. cerevisiae strain. Instead, S. cerevisiae can synthesize acetoin via the condensation of active acetaldehyde with acetaldehyde by pyruvate decarboxylase [4]. In addition, the carboligase mechanism of pyruvate decarboxylase for the synthesis of acetoin from the reaction between pyruvate and acetaldehyde has been elucidated previously [5]. Acetoin is then converted into 2,3-butanediol by butanediol dehydrogenase.
Hitherto, there are few reports regarding 2,3-butanediol production by engineered S. cerevisiae strain. 2,3-Butanediol production has been favored over acetoin and diacetyl in beer and wine fermentation due to its neutral organoleptic characteristics. Diacetyl production in brewing process is undesirable due to its unpleasant flavor and its low taste threshold. Previous attempts successfully reduced the diacetyl production in brewer’s yeast by expressing heterologous α-acetolactate decarboxylase from Klebsiella terrigena and Enterobacter aerogenes[6,7]. 2,3-Butanediol which has a higher flavor threshold than diacetyl is subsequently produced as the final product. Other than that, overexpression of GPD1 in wine yeast strains also led to increased 2,3-butanediol production [8]. Recently, Ehsani et al. [9] have shown that by the overproduction of native Bdh1p or by changing the coenzyme specificity of BDH1 of a strain that overproduces glycerol and lacks ALD6, they manage to yield 0.0307 - 0.033 g·g-1 of 2,3-butanediol in synthetic medium that simulated standard grape juice. Nevertheless, the focus of previous attempts did not target 2,3-butanediol as the main product.
Despite possessing more than two innate pathways that led from pyruvate to 2,3-butanediol [10], the productivity is extremely poor in wild-type S. cerevisiae strain when compared with bacteria[2]. We believed that the capability of S. cerevisiae in producing 2,3-butanediol can be further improved. For this purpose, computational strain design and optimization procedures can guide researcher in seeking for the best metabolic engineering strategy that coupled product formation with cellular objective such as growth [11]. Several genome-scale reconstruction models of yeast have been published previously [12-15]. Using these constraint-based stoichiometric models of yeast, we can predict the metabolic flux redistribution in a strain caused by a genetic or an environmental perturbation by flux balance analysis (FBA) methods [16,17].
OptKnock framework[18] was one of the earliest FBA-based algorithms being developed to predict gene deletion strategies for the overproduction of a target chemical while maximizing the cellular objective such as growth. Since then, this algorithm has been successfully applied to design Escherichia coli strain for the overproduction of lactate [19], malonyl-CoA [20] and 1,4-butanediol [21]. In comparison with E. coli stoichiometric models, the eukaryote S. cerevisiae models are compartmentalized and more complex in terms of its metabolic network. Constraint-based modeling approach for chemical overproduction in S. cerevisiae has been received with mixed opinions [22-24]. Kennedy et al. demonstrated a successful FBA-based design approach for the production of formic acid, a non-fermentative metabolite in yeast [25]. Meanwhile, Bro et al. has applied the genome-scale cell model of S. cerevisiae to construct a strain with 40% reduced glycerol yield and increased ethanol yield [26].
In this paper, we attempted to engineer S. cerevisiae strain for the production of 2,3-butanediol. To achieve this objective, in silico strain design was performed with a recently published S. cerevisiae genome-scale metabolic model of yeast, iMM904 [13]. OptKnock algorithm identified genes that have to be deleted for the overproduction of 2,3-butanediol. Based on the in silico prediction, we constructed a few strains with different combinations of gene deletions and quantitatively analyzed the extracellular metabolites in batch fermentations. We also evaluated whether two other strategies that would reduce by-products and increase pre-cursors of 2,3-butanediol will favor its production: 1. deletion of a NADP + dependent aldehyde dehydrogenase encoded by ALD6 and 2. deletion of NADH dependent glycerol-3-phosphate (G3P) dehydrogenase encoded by GPD2. Finally, we compared the effect of the amplification of S. cerevisiae innate 2,3-butanediol biosynthesis pathway with the introduction of bacterial pathway to different engineered host strains.
Results and discussion
In silico design of 2,3-butanediol producing strain using OptKnock
The flux through butanediol dehydrogenase leading to the biosynthesis of R,R-2,3-butanediol was being considered as our bioengineering objective and biomass production as the cellular objective in OptKnock. All simulations were performed under aerobic condition with glucose as the substrate. We could not identify single reaction deletion or double reaction deletion strategy that will yield significant amount of 2,3-butanediol in iMM904 model.
As we proceeded with higher numbers of reactions for deletion, we initially obtained model C and model D with four and five target reactions respectively (Table 1). Both models include the deletion of pyruvate dehydrogenase (PDH) complex and glutamate dehydrogenase. The former is a mitochondrial multi-protein complex that catalyzes the conversion of pyruvate to acetyl-coA with concomitant generation of NADH [27]. With the deletion of this reaction, more flux from the pyruvate node could be redistributed towards the bioengineering objective. However, the deletion of PDH reaction reduces NADH formation that would favor 2,3-butanediol production. Other than that, the deletion of PDH complexes required deletion of up to five genes encoding different subunits that formed the complex. The NADPH dependent glutamate dehydrogenase encoded by GDH1and GDH3 genes is responsible for glutamate biosynthesis and has a vital role in ammonium metabolism in yeast. It has been reported that deletion of GDH1 increased ethanol formation and reduced glycerol formation in wild-type strain but reduced growth [28]. We could not clearly interpret the other reactions predicted in model C and D as they are non-intuitive. Aspartate transaminase is an important enzyme in amino acid metabolism that catalyzes the reversible conversion of aspartate and α-ketoglutarate to glutamate and oxaloacetate. NAD kinase is responsible for the phosphorylation of NAD+ to NADP+; but the combination of POS5 and UTR1 genes deletion is synthetically lethal [29]. Meanwhile, cytidylate kinase and nucleoside diphosphate kinase are reactions in nucleotide salvage pathway, whereas thymidine phosphorylase is an enzyme in purine and pyrimidine biosynthesis pathway (Additional file 1(a)).
Table 1.
Deletion strategies suggested by OptKnock algorithm for the production of 2,3-butanediol
| ID | Target reaction for knockouts* | Corresponding gene(s) for knockouts | Maximum specific growth rate (1/hr) | R,R-2,3-Butanediol yield (g/g glucose) |
|---|---|---|---|---|
| WT |
- |
- |
0.288 |
0 |
| A |
1. ALCD2ir: |
ADH1, ADH4, ADH5 |
0.231 |
0.313 |
| |
2. ALCD2irm |
ADH3 |
|
|
| |
3. ALCD2x |
SFA1 |
|
|
| B |
1. ALCD2ir |
ADH1, ADH4, ADH5 |
0.190 |
0.362 |
| |
2. ALCD2x |
SFA1 |
|
|
| |
3. GTPCI |
FOL2 |
|
|
| |
4. MDH |
MDH2 |
|
|
| C |
1. GLUDyi |
GDH1, GDH3 |
0.266 |
0.151 |
| |
2. NADK |
UTR1, UTR2, POS5 |
|
|
| |
3. NDPK3 |
YNK1 |
|
|
| |
4. PDHm |
PDB1, PDA1, LAT1, LPD1, PDX1 |
|
|
| D |
1. ASPTA |
AAT2 |
0.264 |
0.151 |
| |
2. CYTK1 |
- |
|
|
| |
3. GLUDyi |
GDH1, GDH3 |
|
|
| |
4. PDHm |
PDB1, PDA1, LAT1, LPD1, PDX1 |
|
|
| 5. TMDPP | PNP1 |
* Additional descriptions are provided in Additional file 1.
The maximum specific growth rate and R,R-2,3-butanediol yield were calculated based on 10 mmol·gDCW-1.hr-1 of glucose uptake. WT stands for the complete network of iMM904 model.
By removing these reactions from the set of target reactions for knockout, we managed to obtain model A. In this model, elimination of all alcohol dehydrogenase (ADH) reactions was proposed. 2,3-Butanediol and ethanol not only share the same precursor molecules, which are pyruvate and acetaldehyde; the conversion of these precursor molecules to 2,3-butanediol and ethanol also require the input of NADH. Thus, removal of the highly active alcoholic fermentation pathway of S. cerevisiae may favor the production of 2,3-butanediol by increasing the supply of pyruvate, acetaldehyde and NADH. Even though this strain has a growth rate 20% lower than that of the reference strain, it is predicted to yield 0.313 g·g-1 glucose of 2,3-butanediol (Additional file 1(b)).
Another quadruple deletion mutant strain with 15% higher yield of 2,3-butanediol was also proposed as a solution. In addition to the cytosolic ADH reaction of model A, cytosolic malate dehydrogenase (MDH2), which converts malate to oxaloacetate with a concomitant reduction of NAD+, was suggested as a target reaction. This gluconeogenic reaction is subject to glucose repression. However, during simulation with iMM904 model, this flux showed negative value for glucose grown wild-type strain, suggesting that the opposite reaction is active. OptKnock might have suggested the deletion of this reaction, which competes for NADH with 2,3-butanediol. The reason for GTP cyclohydrolase deletion is unknown. Furthermore, this quadruple deletion mutant has a growth rate equal to 66% of the reference strain.
Due to the large number of reactions in the genome scale metabolic model, computational strain design with OptKnock produced some non-intuitive deletion strategies that are difficult for logical interpretation. Though, OptKnock has predicted the deletion of PDH and ADH reactions, which are highly competitive for the carbon flux to 2,3-butanediol, as our strain design strategies. Since model A and model B have higher 2,3-butanediol yields than model C and model D (Table 1), we decided to evaluate the deletion of the ADH reaction that is required in both model A and model B.
Strain construction and microaerobic fermentations
To test this hypothesis provided by OptKnock, we would have to delete all the genes that are responsible for the conversion of acetaldehyde to ethanol. A total of four ADH genes, ADH1ADH3ADH4 and ADH5, along with SFA1 gene have to be disrupted. Previous report [30] showed that ADH4 gene product does not contribute to ethanol production. Meanwhile, SFA1 is known as a long-chain ADH [31] and formaldehyde dehydrogenase [32]. Therefore, our target genes for deletion were reduced to three – ADH1ADH3 and ADH5.
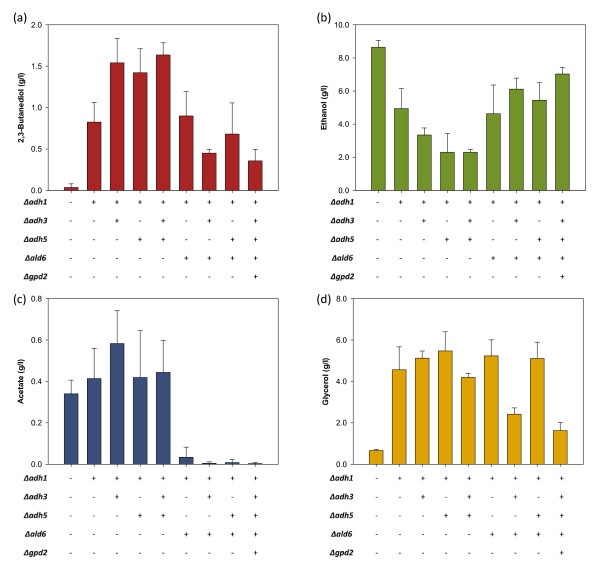
To identify the contribution of each gene deletion to 2,3-butanediol yield, we have constructed deletion strains with different combination of the three ADH genes (Table 2). Shake-flask cultivations were then performed with these strains at microaerobic condition. Engineered strains took longer time (~ 60 h) to complete fermentation than the wild-type strain (~ 40 h). Only up to 0.04 g·l-1 of 2,3-butanediol (0.002 g·g-1) was detected in the culture of BY4742 wild-type strain. In comparison with the wild-type strain, single deletion of the major ADH gene ADH1 reduced ethanol yield by almost half and increased 2,3-butanediol yield by up to 20.5-fold, with a 2,3-butanediol maximum titers and product yields of 0.825 g·l-1 and 0.041 g·g-1, respectively (Figure 2 and Table 3). The double deletion strains B2C-a1a3 (Δadh1 Δadh3) and B2C-a1a5 (Δadh1 Δadh5) produced up to 1.54 g·l-1 and 1.42 g·l-1 of 2,3-butanediol, respectively, with a 2,3-butanediol yield of 85% - 88% more than the Δadh1 single deletion strain. Strain B2C-a1a3a5, of which three ADH isozymes encoded by ADH1ADH3 and ADH5 genes were deleted, achieved the highest 2,3-butanediol titer and yield among the engineered strains, up to 1.64 g·l-1 and 0.093 g·g-1. This triple deletion strain produced 73.3% lesser ethanol than wild-type strain. Consistent with previous reports [33,34], this strain still retains its ability to produce ethanol due to residual ADH isozymes. It has higher 2,3-butanediol yield per substrate than strain B2C-a1a3 and strain B2C-a1a5 owing to the fact that up to 2.41 g·l-1 of residual glucose was detected at the end of fermentation.
Table 2.
S. cerevisiaestrains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| BY4742 |
MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
EUROSCARF |
| B2C-a1 |
BY4742 Δadh1::loxP |
This study |
| B2C-a1a3 |
BY4742 Δadh1::loxP Δadh3::loxP |
This study |
| B2C-a1a5 |
BY4742 Δadh1::loxP Δadh5::loxP |
This study |
| B2C-a1a3a5 |
BY4742 Δadh1::loxP Δadh5::loxP Δadh3::loxP |
This study |
| B2C-a1a6 |
BY4742 Δadh1::loxP Δald6::loxP |
This study |
| B2C-a1a3a6 |
BY4742 Δadh1::loxP Δald6::loxP Δadh3::loxP |
This study |
| B2C-a1a5a6 |
BY4742 Δadh1::loxP Δald6::loxP Δadh5::loxP |
This study |
| B2C-a1a6g2 |
BY4742 Δadh1::loxP Δald6::loxP Δgpd2::loxP |
This study |
| B2C-a1a3a5a6g2 |
BY4742 Δadh1::loxP Δald6::loxP Δgpd2::loxP Δadh3::loxP Δadh5::loxP |
This study |
| B2C-SB |
BY4742 p423GPD-BDH1 |
This study |
| B2C-ABC |
BY4742 p423GPD-budC pCTP-URA::TEF1p-alsS + PGK1p-budA |
This study |
| B2C-a1a5-SB |
BY4742 Δadh1::loxP Δadh5::loxP p423TEF-BDH1 |
This study |
| B2C-a1a5-ABC |
BY4742 Δadh1::loxP Δadh5::loxP p423GPD-budC pCTP-URA::TEF1p-alsS + PGK1p-budA |
This study |
| B2C-a1a6-SB |
BY4742 Δadh1::loxP Δald6::loxP p423GPD-BDH1 |
This study |
| B2C-a1a6-ABC | BY4742 Δadh1::loxP Δald6::loxP p423GPD-budC pCTP-URA::TEF1p-alsS + PGK1p-budA | This study |
*Genotype is described in the order of the corresponding genetic manipulations.
Figure 2.
Extracellular metabolite concentrations for batch cultivation. Concentration of (a) 2,3-butanediol, (b) ethanol, (c) acetate and (d) glycerol at the end of batch cultivation under microaerobic conditions. Plus or minus signs indicate presence or absence of the corresponding gene deletion. The first column corresponds to the wild-type strain. Error bars represent standard deviations associated with four independent experiments (except for strains B2C-a1, B2C-a1a5a6, B2C-a1a3a5a6g2, where n = 3; and for strains B2C-a1a3a5 and B2C-a1a3a6, where n = 2).
Table 3.
Fermentation time, extracellular metabolite concentrations and yields for shake-flask culture under microaerobic condition
| Strain | Fermentation time, hr | Dry cell weight, g·l-1 | Residual glucose, g·l-1 | Acetaldehyde†, g·l-1 |
Yield, g·g-1glucose |
||||
|---|---|---|---|---|---|---|---|---|---|
| 2,3-Butanediol | Glycerol | Ethanol | Acetoin | Acetate | |||||
| BY4742 |
40 |
1.349 |
0 |
0.118 |
0.002 |
0.032 |
0.417 |
0 |
0.017 |
| B2C-a1‡ |
56 |
0.787 |
0 |
0.663 |
0.041 |
0.225 |
0.243 |
0.016 |
0.02 |
| B2C-a1a3 |
54 |
0.876 |
0 |
1.194 |
0.076 |
0.254 |
0.166 |
0.012 |
0.029 |
| B2C-a1a5 |
72 |
0.634 |
1.80 |
1.106 |
0.079 |
0.295 |
0.123 |
0.010 |
0.024 |
| B2C-a1a3a5† |
54 |
0.900 |
2.41 |
1.316 |
0.093 |
0.238 |
0.131 |
0.008 |
0.025 |
| B2C-a1a6 |
57 |
0.740 |
0.09 |
0.493 |
0.046 |
0.265 |
0.231 |
0.010 |
0.002 |
| B2C-a1a3a6† |
63 |
0.619 |
0.78 |
0.312 |
0.023 |
0.125 |
0.318 |
0.011 |
0 |
| B2C-a1a5a6‡ |
54 |
0.842 |
0 |
0.263 |
0.033 |
0.247 |
0.259 |
0.011 |
0 |
| B2C-a1a3a5a6g2‡ | 64 | 0.806 | 0 | 0.220 | 0.018 | 0.081 | 0.347 | 0.007 | 0 |
*All cultures were performed with an initial concentration of 20 g·l-1 of glucose under microaerobic condition. The extracellular metabolite concentrations and yields (g metabolite produced/g glucose consumed) were determined at the time of glucose exhaustion or when the strains ceased to consume glucose. Fermentation time indicates the mean time of the end of fermentation. Residual glucose indicates the amount of glucose detected in the supernatant at the end of fermentation.
** All data correspond to the mean of four independent experiments (n = 4) unless indicated. For all data, standard error of the mean is lower than 10%.
‡ Data correspond to the mean of three independent experiments (n = 3).
† Data correspond to the mean of two independent experiments (n = 2).
Previous works stated that acetaldehyde accumulation in Δadh1 strain led to poor growth in glucose [34,35]. Acetaldehyde at a level higher than 0.3 g·l-1 inhibits cell growth [36]. We found that the concentrations of acetaldehyde in the supernatant at the end of fermentation were significantly higher in deletion strains (B2C-a1, B2C-a1a3, B2C-a1a5 and B2C-a1a3a5) than the wild-type strain (Table 3). Accordingly, the final biomass yields for these deletion strains were highly reduced. We also thought that perhaps the high acetaldehyde level in the supernatant, which is toxic to the cells, causes the cessation in glucose uptake of some of the deletion strains. In addition, deletion strains also exhibited 22- 71% higher level of acetate and 6 - 8-fold increase in glycerol level over wild-type strain. Analysis of strain lacking ADH1 or ADH1-4 isozymes previously has shown that glycerol is the main fermentation product for these strains, while acetaldehyde and acetate are produced in significant amounts [30]. This is because reducing the flux towards ethanol led to the accumulation of NADH and acetaldehyde. Generation of glycerol allows the strain to reoxidize excess NADH in the cytosols, while acetate formation reduces the acetaldehyde accumulated. Nevertheless, 2,3-butanediol production in these strains has not been reported. 2,3-Butanediol can be synthesized directly from pyruvate via yeast innate pathway. Production of 2,3-butanediol requires the oxidation of NADH, whereas oxidation of acetaldehyde to acetate reduced the cofactor NAD(P)+. Thus, accumulation of acetaldehyde in strains B2C-a1, B2C-a1a3 and B2C-a1a5 strains greatly increased the production of 2,3-butanediol. The results of additional gene deletion strategies to reduce by-products (acetate and glycerol) and their effect on 2,3-butanediol production are discussed in the next sections.
Production of 2,3-butanediol in the engineered strain mostly occurs through the pathways that are catalyzed by pyruvate decarboxylase. Deletion of PDC1 from Δadh1 background was reported to reduce acetaldehyde production [37]. In our study on Δadh1 Δpdc1 strain, less than 0.60 g·l-1 of 2,3-butanediol was detected (unpublished observations). Deletion of PDC1 from Δadh1 strain reduced the flux towards acetaldehyde and consequently decreased acetoin production. This attested that pyruvate decarboxylase enzyme is important in the production of acetaldehyde and the production of 2,3-butanediol. Nevertheless, the acetoin synthase activity of pyruvate decarboxylase enzyme has been reported to reach saturation level [5] at a very low concentration and increase only by 10% even with larger pool of acetaldehyde. So, further deletion of more ADH genes did not resulted in a more drastic increase in 2,3-butanediol production.
The effect of ALD6 gene deletion on 2,3-butanediol production
ALD6 gene encodes the main cytosolic aldehyde dehydrogenase that catalyzes the conversion of acetaldehyde to acetate. Eglinton et al. has shown that acetic acid production can be reduced in glycerol overproducing strain of S. cerevisiae by deleting the ALD6 gene [38]. As discussed in the previous section, strains B2C-a1, B2C-a1a3, B2C-a1a5 and B2C-a1a3a5 produced more acetate when compared to the wild-type strain. High production of acetic acid lowers the pH of the culture broth and can induce programmed cell death in S. cerevisiae cells [39]. So, we investigated the effect of ALD6 deletion on the production of 2,3-butanediol in the engineered strains with deletion in one or more ADH genes.
Deletion of ALD6 significantly reduced the production of acetate in all the engineered strains, from an average of 0.47 g·l-1 to undetectable level (Figure 2). However, this decrease in acetate did not correspond to the increase in 2,3-butanediol production in most of the engineered strains. Only strain B2C-a1a6 showed a slight increase in 2,3-butanediol yield when compared to its parent strain (B2C-a1), from 0.041 g·g-1 to 0.046 g·g-1. On the contrary, the 2,3-butanediol yield of B2C-a1a3a6 was equal to only 30% of strain B2C-a1a3. Similarly, strain B2C-a1a5a6 had a 58% lower 2,3-butanediol yield than strain B2C-a1a5 (Table 3).
We had initially anticipated that deletion of the aldehyde dehydrogenase gene, ALD6, in strains that contained one or more deleted ADH genes would lead to the accumulation of acetaldehyde. This acetaldehyde could then be converted to acetoin through pyruvate decarboxylase reaction and consequently improve 2,3-butanediol yield. In contrast to our initial hypothesis, strains B2C-a1a5a6 and B2C-a1a3a6 yielded higher levels of ethanol and lower levels of acetaldehyde than strains B2C-a1a5 and B2C-a1a3 which harbor the native ALD6 gene. This suggests an increase in the activity of the remaining ADH isozymes of these mutant strains as a survival mechanism of the cells due to the toxicity of acetaldehyde. These residual ADH isozymes compete more efficiently for acetaldehyde than the acetoin-synthesizing pyruvate decarboxylase, which is another pathway for acetaldehyde detoxification. Similarly, Roustan et al. [40] have reported that the ADH activity of wine yeast increased when acetaldehyde was added to the stationary phase medium of wine fermentation.
The effect of GPD2 gene deletion on extracellular metabolite yields
In strains carrying deletion of ADH1, excess G3P is produced by G3P dehydrogenase from dihydroxyacetone phosphate while regenerating NAD+. Glycerol is then formed by the dephosphorylation reaction of glycerol-3-phosphatase. Two strains (B2C-a1a6 and B2C-a1a3a5) that produced glycerol as the major fermentation product under microaerobic condition were evaluated for the effect of the disruption of G3P dehydrogenase reaction.
GPD1 and GPD2 encode the isozymes of G3P dehydrogenase. It has been shown that the deletion of both genes cease glycerol production in S. cerevisiae cells [41]. Nevertheless, deletion of both genes was lethal in strain B2C-a1a6 as the cell lost the ability to re-oxidize excess NADH and produce ATP. Single deletion of the osmoregulated GPD1 in Δadh1 Δald6 background has minor effect in the reduction of glycerol production (data not shown). On the contrary, the deletion of GPD2 in strain B2C-a1a6 reduced glycerol production when compared to the double deletion strain (Figure 3). GPD2 activity is affected by redox control. The limitation in NADH re-oxidation ability due to the deletion of ADH1 in strain B2C-a1a6, as well as the low oxygen condition during microaerobic cultivation, contributes to the stressful condition where GPD2 expression is highly induced. The increase in pyruvate and NADH due to GPD2 deletion, however, did not favor 2,3-butanediol production in strain B2C-a1a6g2 as ethanol became the major fermentation product. Similarly, deletion of both ALD6 and GPD2 from strain B2C-a1a3a5 also completely altered the final product composition of the medium. The quintuple deletion strain displayed lower yield of 2,3-butanediol and glycerol, but higher yield of ethanol (Table 3).
Figure 3.
The effect ofGPD2deletion on 2,3-butanediol, glycerol and ethanol yields. Metabolite yields for strains (a) BY4742, (b) B2C-a1a6, (c) B2C-a1a6g2, (d) B2C-a1a3a5 and (e) B2C-a1a3a5a6g2 at the end of batch cultivation under microaerobic condition. Columns and error bars correspond to the mean value and standard deviations of four independent experiments for (a) and (b), duplicate experiments for (c) and (d) and triplicate experiments for (e).
In strains with ADH1 deletion, glycerol is greatly produced to regenerate the reducing equivalents [34]. By reducing the ability of the cells to oxidize excess NADH through glycerol production, ethanol was again produced as major fermentation product in strains B2C-a1a6g2 and B2C-a1a3a5a6g2. It is possible that the expressions of the remaining ADH genes were induced to cope with the NADH surplus condition. This is because the synthesis of two moles of ethanol from one mole of pyruvate and one mole of acetaldehyde consumes twice the amount of NADH that is required for the formation of one mole of 2,3-butanediol from the same amount of precursor molecules [40].
Anaerobic batch fermentation
In order to test whether 2,3-butanediol production can be improved in a highly fermentative condition, strains B2C-a1a5, B2C-a1a6, B2C-a1a3a5 and wild-type strain BY4742 were selected for anaerobic batch cultivation. In the absence of oxygen, S. cerevisiae cells are known to produce significant amount of ethanol and glycerol as a mean to maintain redox balance and generate ATP in the cells. Due to slower growth in anaerobic state, the biomass yield of wild-type and all engineered strains were greatly reduced while the fermentation time increased significantly. All strains exhibited higher glycerol yield due to anaerobiosis. Meanwhile, a decline in the production of the more oxidized acetate was observed in wild-type and the engineered strains (Figure 4).
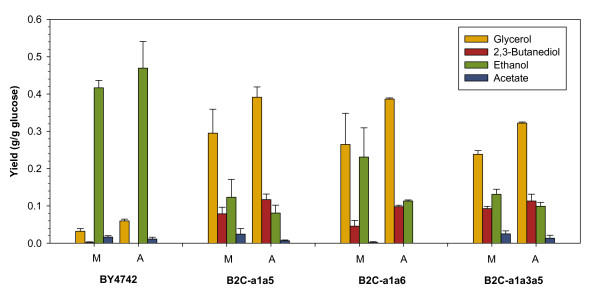
Figure 4.
Metabolite profile of cultivation under microaerobic (M) and anaerobic (A) conditions. The bar chart shows glycerol, 2,3-butanediol, ethanol and acetate yields for batch fermentation of (a) BY4742, (b) B2C-a1a5, (c) B2C-a1a6 and (d) B2C-a1a3a5 under microaerobic condition and anaerobic condition with 20 g·l-1 of glucose as substrate. Error bars represent standard deviations.
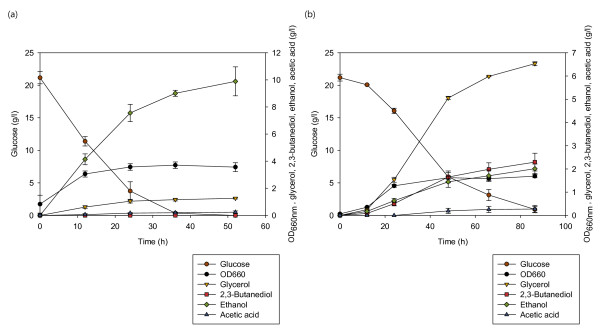
With deficiency in its ADH genes, the engineered strains B2C-a1a5, B2C-a1a6 and B2C-a1a3a5 produced glycerol as the major fermentation product. Under microaerobic condition, the second major by-product was ethanol, followed by 2,3-butanediol. Interestingly, growth in anoxic state led to the opposite case for strains B2C-a1a5 and B2C-a1a3a5, where 2,3-butanediol yield was higher than ethanol yield (Figure 4). In comparison to the microaerobic state, the yield of ethanol in anaerobic state was lowered by more than 34% and 24% for strains B2C-a1a5 and B2C-a1a3a5, respectively. Up to 2.29 g·l-1 of 2,3-butanediol was detected in the fermentation broth for the triple deletion strain, with a yield of 0.113 g·g-1 (Figure 5). 2,3-Butanediol was not detected in wild-type strain culture broth under anaerobic condition.
Figure 5.
Changes in metabolite concentrations for strains (a) BY4742 (wild-type) and (b) B2C-a1a3a5 under anaerobic condition. Glucose (brown circles), OD660nm (black circles), glycerol (inversed triangles), 2,3-butanediol (squares), ethanol (diamonds) and acetate (triangles) concentrations are shown. Symbols and error bars represent the mean and standard deviation for (a) triplicate and (b) duplicate experiments.
An intriguing relationship between the level of oxygen with the production concentration of 2,3-butanediol and ethanol can be deduced from our observation. 2,3-Butanediol yield correlates inversely with the level of oxygen and thus the biomass yield. As low oxygen is known to led to higher fermentative energy metabolism in S. cerevisiae, it is not surprising to obtain more fermentation products under anaerobic condition. It is harder to interpret the connection between 2,3-butanediol and ethanol when combined with oxygen level. Since 2,3-butanediol and ethanol shares the same precursor molecule, their concentrations should correlate in an inverse manner. We suspected that the rate of acetaldehyde formation determines the amount of ethanol and 2,3-butanediol produced. However, we could not track the dynamic changes in the concentration of intracellular acetaldehyde in this study.
Improved 2,3-butanediol yield by gene overexpression strategies
In this section, we compared the single overexpression of innate BDH1 and introduction of additional 2,3-butanediol synthesis pathway from bacteria in three different host: wild-type strain, strain B2C-a1a5 and strain B2C-a1a6. The heterologous pathway was introduced by the co-expression of B. subtilis alsS gene, E. aerogenes budA gene and E. aerogenes budC gene using two multicopy plasmids (Table 4). We used the acetolactate synthase of B. subtilis encoded by alsS gene because it has stronger preference for pyruvate over 2-ketobutyrate [42]. On the contrary, S. cerevisiae native acetolactate synthase encoded by ILV2 has higher affinity for 2-ketobutyrate. E. aerogenes budA encodes α-acetolactate decarboxylase, which is absent in S. cerevisiae strain. The E. aerogenes 2,3-butanediol dehydrogenase, encoded by budC, displays higher affinity for both NADH and acetoin (Km,NADH = 5–7 μM and Km,acetoin = 0.4 mM) than the S. cerevisiae native Bdh1p (Km,NADH = 45 μM and Km,acetoin = 3 mM) [43,44]. The mRNA expression levels of all the genes in the strains we constructed have been confirmed by real-time PCR (data not shown).
Table 4.
Plasmid used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pFA6a-KanMX6 |
E. coli kanr gene under the control of Ashbya gossypii TEF promoter-driven |
[45] |
| pCNKanMX6 |
loxP flanking KanMX6 at SalI/BamHI and SacI/EcoRV sites |
This study |
| pTKURA3 |
loxP flanking S. cerevisiae URA3 gene |
This study |
| p423GPD |
2 μ, HIS3, PGPD1 |
[46] |
| p423TEF |
2 μ, HIS3, PTEF1 |
[46] |
| p423GPD-BDH1 |
S. cerevisiae BDH1 gene under the control of GPD1 promoter in p423GPD |
This study |
| p423TEF-BDH1 |
S. cerevisiae BDH1 gene under the control of TEF1 promoter in p423GPD |
This study |
| p423GPD-E.a.budC |
E. aerogenes KCTC 2190 budC gene under the control of GPD1 promoter in p423GPD |
This study |
| pESC-URA |
2 μ, URA3, PGAL10 PGAL1 |
Stratagene |
| pESC-LEU |
2 μ, LEU2, PGAL10 PGAL1 |
Stratagene |
| pESC-LEU-Cre |
P1 bacteriophage Cre recombinase under the control of GAL10 promoter |
This study |
| pCTP-URA |
2 μ, URA3, PTEF1 PPGK1 |
This study |
| pCTP-URA::TEF1p-alsS + PGK1p-budA | Bacillus subtilis alsS gene under the control of TEF1 promoter and E. aerogenes KCTC 2190 budA gene under the control of PGK1 promoter in pCTP-URA | This study |
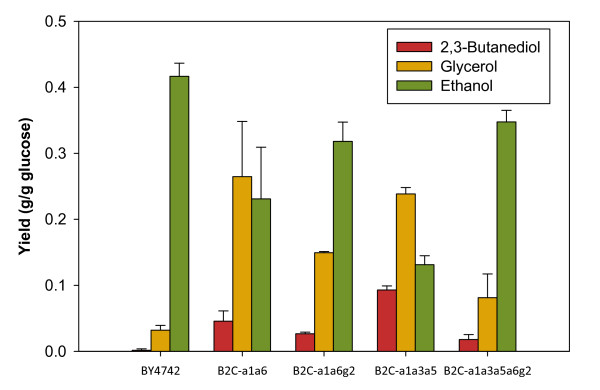
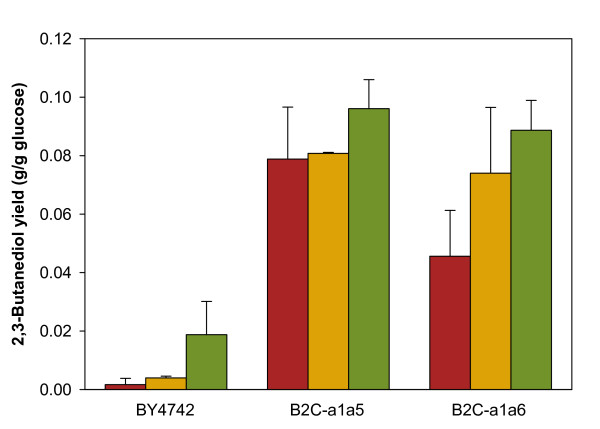
In the wild-type strain, overexpression of BDH1 and the bacterial pathway genes increased 2,3-butanediol yield by 2.4-fold and 11.1-fold, respectively (Figure 6). However, the yield of these strains is very low when compared with the deletion strains. The introduction of acetolactate decarboxylase pathway managed to double the yield of 2,3-butanediol in strain B2C-a1a6 to 0.089 g·g-1. In the engineered strain B2C-a1a5, 2,3-butanediol yield was only slightly affected by the overexpression of endogenous butanediol dehydrogenase gene. The introduction of the heterologous pathway, however, successfully increased 2,3-butanediol yield by 22% when compared with its parent strain B2C-a1a5. Despite the small enhancement of 2,3-butanediol yield, strain B2C-a1a5-ABC produced the highest titer of 2,3-butanediol, with up to 1.67 g·l-1, corresponding to 2,3-butanediol yield of 0.096 g·g-1 during microaerobic shake-flask culture.
Figure 6.
Comparison of 2,3-butanediol yield with gene overexpression strategies. Each bar represents the 2,3-butanediol yield for shake-flask cultivation under microaerobic condition for three different host strains: (a) BY4742, (b) B2C-a1a5 and (c) B2C-a1a6 expressing . Red bars, host strain (reference); yellow bars, overexpression of S. cerevisiae innate BDH1; green bars, bacterial alsS-budA-budC co-expression. Values and error bars correspond to the mean and standard deviation for triplicate experiments (except for all the red bars, where four independent experiments were conducted)
In wild-type strain and strain B2C-a1a5, overexpression of BDH1 rendered minimal effect on 2,3-butanediol yield. It seems likely that butanediol dehydrogenase reaction is not the bottleneck in 2,3-butanediol biosynthetic pathways in these strains. This is because the flux towards acetoin was probably not adequate in the wild-type strain expressing BDH1; whereas in the case of strain B2C-a1a5, it is possible that the conversion of acetaldehyde to acetoin is the rate-limiting step as significant amount of acetaldehyde accumulated in the medium (Table 3). Strain B2C-a1a6 is the only host strain that demonstrated a greater increase in 2,3-butanediol yield due the amplification of BDH1. It is also surprising that this strain yielded more 2,3-butanediol than ethanol (0.0887 g·g-1 and 0.0713 g·g-1, respectively) when the bacterial pathway genes are introduced. However, this strain (B2C-a1a6-ABC) only managed to consume half of the amount of glucose supplied in the fermentation medium under microaerobic condition implicating that the cell growth was greatly inhibited by certain metabolites or its cellular redox state is highly imbalanced (Data not shown). To enhance the overall flux from pyruvate to 2,3-butanediol in S. cerevisiae, our data in this section suggests that the overexpression of budC or BDH1 together with alsS and budA can be an alternative to forcing carbon fluxes through the rate-limiting pyruvate decarboxylase pathway.
Conclusions
In silico S. cerevisiae strain design for the production of 2,3-butanediol suggested the deletion of mitochondrial and cytosolic ADH genes. We thus constructed deletion strains with low ADH activity by the deletion of ADH1, ADH3 and ADH5 genes. Single deletion of major cytosolic ADH gene ADH1 increased the production titer by around 21-fold. Detoxification mechanism of cells as a response to increased acetaldehyde pool [8], along with the failure to regenerate excess NADH efficiently through ethanol production stimulate 2,3-butanediol production in the deletion strains. Further reduction of ADH activity by additional disruption of ADH3 or ADH5 or both genes increased production by ~2-fold when compared to single deletion strain. But distinctively higher production was not observed, showing that the maximal conversion of acetaldehyde to acetoin and subsequently to 2,3-butanediol has been achieved by reducing ADH activity. High accumulation of acetaldehyde in these deletion strains also implies that the conversion of acetaldehyde to acetoin is rate-limiting. We have thus shown that 2,3-butanediol production can be enhanced in the engineered strains through the introduction of foreign biosynthetic pathway. Further improvement of the conversion efficiency of precursor molecules to 2,3-butanediol have to be considered in future study.
OptKnock framework has suggested a deletion strategy that led us to the hypothesis that increase in NADH, acetaldehyde and pyruvate could improve 2,3-butanediol production. This is supported by our data with strains lacking major ADH genes. Nevertheless, this hypothesis is true only to a certain extent as rational deletion strategies based on this hypothesis, including ALD6 deletion to accumulate acetaldehyde and GPD2 deletion to further increase NADH and pyruvate in strains with low ADH activity, did not enhance 2,3-butanediol production but increased ethanol formation instead. Problems remained to be solved to further engineer the industrial workhorse S. cerevisiae for 2,3-butanediol production.
In conclusion, we have demonstrated a successful implementation of in silico guided metabolic engineering approach for the production of a fermentative product in S. cerevisiae. 2,3-Butanediol producing S. cerevisiae strains were constructed by referring to OptKnock’s prediction. Currently, among all the engineered strains, strain B2C-a1a3a5 (Δadh1 Δadh3 Δadh5) achieved the highest 2,3-butanediol titer and yield under anaerobic condition (2.29 g·l-1 and 0.113 g·g-1). To our knowledge, this is the first ever report about 2,3-butanediol production in S. cerevisiae strain specifically engineered for its production.
Methods
Computational procedure
FBA-based simulation and strain design using OptKnock algorithm [18] were performed in Matlab R2010a (The MathWorks, Inc., Massachusetts, USA) with the COBRA Toolbox v2.0 [47]. Gurobi Optimizer 4.5 (Gurobi Optimization, Houston, USA) was used to performed linear programming calculations. The recently reported yeast genome-scale model iMM904 [13], consisting of 904 genes, 1577 reactions and 1228 metabolites, was used. All simulations were performed with the following constraints: oxygen uptake rate, 2mmol·gDCW-1·hr-1; glucose uptake, 10mmol·gDCW-1·hr-1; ATP required for maintenance, 1mmol·gDCW-1·hr-1. We excluded all exchange and transports reactions from the target knockouts.
Strains, plasmid and growth conditions
All engineered strains used in this study are derived from S. cerevisiae BY4742 strain family (BY4742 MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). During constructions, strains were grown and maintained on complex (YPD) medium consisting of 1% (w/v) bacto yeast extract and 2% (w/v) yeast bacto peptone supplemented with 20 g·l-1 glucose as carbon source. Transformants and strains harboring recombinant plasmid(s) were grown on synthetic complete dropout (SC) medium consisted of 20 g·l-1 glucose, 6.7 g·l-1 yeast nitrogen base without amino acid (Difco, BD Biosciences) and the appropriate amino acid supplement mixture (either CSM, CSM-URA, CSM-HIS, CSM-LEU or CSM-HIS-URA, MP Biomedicals). In all cases, glucose was autoclaved separately. Escherichia coli DH5α strain was used for amplification of plasmids. Transformed E. coli DH5α cells were grown at 37°C in Luria-Bertani (LB) medium containing 100ug·ml-1 of ampicillin.
Construction of S. cerevisiae deletion strains
All the strains used in this work are listed in Table 2. Gene deletion of S. cerevisiae was performed by short flanking homology PCR method with Cre/loxP system for recycling of selection marker gene. pCNKanMX6, a plasmid containing loxP-kanMX-loxP module, were constructed from pFA6a-KanMX6 according to [48]. pTKURA3 were constructed by replacing the KanMX6 cassette with S. cerevisiae URA3 gene including 200 nucleotides upstream and +77 nucleotides downstream of URA3 gene. ADH1 gene was deleted by introduction of linearized gene deletion cassette consisting of loxP-URA3-loxP sequence flanked by the 45 nucleotides upstream and downstream of ADH1 gene to BY4742 wild-type strain. ADH3ADH5 or ALD6 genes were then deleted in Δadh1 background using the same method to construct double gene deletion strains B2C-a1a3, B2C-a1a5 and B2C-a1a6. To construct the triple deletion strains B2C-a1a3a6 and B2C-a1a5a6, ADH3 and ADH5 were again deleted separately from Δadh1 Δald6 background with the previously described method. Meanwhile, strain B2C-a1a3a5 was constructed by the deletion of ADH3 from strain B2C-a1a5. Strain B2C-a1a6g2 was constructed by the deletion of GPD2 gene from strain B2C-a1a6. Further deletion of ADH3 and ADH5 genes from strain B2C-a1a6g2 resulted in the quintuple deletion strain B2C-a1a6g2a3a5. All strains were verified by colony polymerase chain reaction (PCR) with appropriate confirmation primers (Additional file 2). The aragose gel images of all gene deletion strains are provided (Additional file 3).
Plasmid construction and transformation
The plasmids used in this study are listed in Table 4. S. cerevisiae BDH1 gene was amplified from the genomic DNA of BY4742 strain with the primers BDH1-F-BamHI and BDH1-R-XhoI. The fragment amplified was ligated to p423GPD and p423TEF that were digested with BamHI and XhoI. The resulting construct is p423GPD-BDH1 and p423TEF-BDH1. E. aerogenes budC gene was amplified from the genomic DNA of E. aerogenes KCTC 2190 strain by PCR using the primers E.a.budC-F-BamHI and E.a.budC-R-EcoRV. budC gene fragment was then inserted between the BamHI-EcoRV sites of p423GPD, resulting in p423GPD-E.a.budC.
The GAL1/GAL10 promoter regions of pESC-URA(Stratagene) were removed and replaced with the PGK1/TEF1 divergent promoters by two-step cloning. PGK1 promoter sequence was amplified from the genomic DNA of S. cerevisiae BY4742 wild-type strain with primers PGK1p-F-AatII-NotI and PGK1p-R-BamHI. It was then digested with BamHI and NotI and cloned into pESC-URA to produce pCP-URA. TEF1 promoter region was amplified from p425TEF with primers TEF1p-R-NotI and TEF1p-F-AatII. The TEF1 promoter fragment and pCP-URA were digested by AatII and NotI and then ligated to each other. The new plasmid generated is pCTP-URA. Bacillus subtilis alsS gene was amplified from the genomic DNA of B. subtilis using primers B.s.alsS-F-NotI and B.s.alsS-R-BglII. Meanwhile, E. aerogenes budA was amplified from the genomic DNA of E. aerogenes KCTC 2190 strain with primers E.a.budA-F-BamHI and E.a.budA-R-XhoI. B. subtilis alsS and E. aerogenes budA fragment were cloned into pCTP-URA under the control of TEF1 promoter and PGK1 promoter, respectively.
In all cases, PCR was performed using TaKaRa LA Taq Polymerase with GC Buffer (Takara Bio Inc, Shiga, Japan). Yeast strains were transformed using the lithium acetate/PEG/SS-DNA method [49]. After transformation, successful transformants were selected on SC media without the appropriate amino acids.
Batch fermentations
For all fermentations, pre-cultures were prepared by inoculating 5 ml of SC medium in a 50 ml screw-capped tube with a fresh colony grown on solid medium. Batch fermentations were performed in 250 ml Erlenmeyer flasks containing 50 ml of synthetic complete dropout medium. Microaerobic condition was maintained in the flasks by sealing with rubber stoppers. For anaerobic fermentations, 50 ml of SC medium supplemented with 0.42 g·l-1 Tween 80 and 0.01 g·l-1 cholesterol in a 150 ml serum bottle was purged with pure nitrogen gas for 15 minutes to establish anaerobic condition. Initial OD660nm was adjusted at 0.05. Fermentations were performed in shaking incubator at 30°C, with agitation at 250 rpm.
Analytical methods
Cell growth was determined by measuring the absorbance at 660 nm using UV–VIS spectrophotometer (Shimazu UV mini 1240, Tokyo, Japan). In addition, we determined the dry cell mass of yeast at the end of shake-flask cultivation by filtering 30 ml of culture through pre-weighed cellulose acetate membrane filters (Chmlab Group, Barcelona, Spain) with a pore size of 0.45 μm. Filters were dried at 65°C for 48 hours and weighed again. Then, we calculated the dry cell mass and estimated the cell biomass by using the dry cell mass/OD660 ratio at the end of cultivation.
For extracellular metabolite analysis, 1 ml of culture samples were collected at appropriate time point. Isopropyl alcohol was added to the 1 ml-sample as internal standard and vortexed for 1 min. The samples were then centrifuged (13500 rpm, 5 min) and 200 μl of supernatants were transferred to HPLC autosampler vials. Extracellular concentrations of glucose, glycerol, ethanol, acetate, succinate, acetoin and 2,3-butanediol were determined with a high performance liquid chromatography (HPLC) system (ACME-9000, Younglin Instrument, Seoul, South Korea) equipped with a refractive index detector (RID). Analytes were separated using Sugar SH1011 column (Shodex, Tokyo, Japan) with 10 mM sulfuric acid as mobile phase pumped at 0.5ml·min-1. The column and the detector were set to 75°C and 45°C, respectively. The order of elution was glucose, succinic acid, glycerol, acetate, acetoin, 2,3-butanediol, ethanol and isopropyl alcohol. For quantitative analysis, standards for all metabolite at five different concentration levels were prepared. A calibration curve for each component was made and from these calibration curves concentrations of metabolites were calculated. The standard for 2,3-butanediol (≥99.0% (GC), Fluka Analytical, Sigma-Aldrich) and the extracellular 2,3-butanediol from the fermentation samples are a mixture of racemic and meso forms. Extracellular acetaldehyde concentrations were determined with Megazyme Acetaldehyde Assay Kit (Megazyme International Ireland, Wicklow, Ireland) according to the manufacturer’s instructions.
Abbrevation list
FBA: flux balance analysis; PDH: pyruvate dehydrogenase; ADH: alcohol dehydrogenase; G3P: glycerol-3-phosphate.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MKO and JL conceived the study. MKO designed and supervised the study, and contributed to manuscript writing. CYN participated in the design of the study, performed the experiments and drafted the manuscript. MYJ helped to conduct HPLC analysis. All authors read and approved the final manuscript.
Supplementary Material
Additional description for Table 1. (a) A list of the corresponding enzymes, reaction equations and subsystems for target reactions listed in Table 1 and (b) a figure showing the 2,3-butanediol production envelope of OptKnock strain A.
Oligonucleotides used in this study. A list of oligonucleotides that were used in this study.
Verification of gene deletion strains by colony PCR. The PCR profiles of wild-type and gene deletion strains.
Contributor Information
Chiam Yu Ng, Email: ngchiamyu@gmail.com.
Moo-young Jung, Email: artsora@naver.com.
Jinwon Lee, Email: jinwonlee1@gmail.com.
Min-Kyu Oh, Email: mkoh@korea.ac.kr.
Acknowledgements
This research was financed by the Research & Development Program of Ministry of Knowledge Economy (MKE) and Korea Evaluation Institute of Industrial Technology (KEIT) (No. 10035578, Development of 2,3-butanediol and derivative production technology for C-Zero bio-platform industry), Republic of Korea. We would also like to thank Tae-yeon Kim for constructing pTKURA3 plasmid.
References
- Ji X-J, Huang H, Ouyang P-K. Microbial 2,3-butanediol production: A state-of-the-art review. Biotechnol Adv. 2011;29:351–364. doi: 10.1016/j.biotechadv.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Celinska E, Grajek W. Biotechnological production of 2,3-butanediol–current state and prospects. Biotechnol Adv. 2009;27:715–725. doi: 10.1016/j.biotechadv.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Yang T, Rao Z, Zhang X, Lin Q, Xia H, Xu Z, Yang S. Production of 2,3-butanediol from glucose by GRAS microorganism Bacillus amyloliquefaciens. J Basic Microbiol. 2011;51:650–658. doi: 10.1002/jobm.201100033. [DOI] [PubMed] [Google Scholar]
- Chen GC, Jordan F. Brewers' yeast pyruvate decarboxylase produces acetoin from acetaldehyde: a novel tool to study the mechanism of steps subsequent to carbon dioxide loss. Biochemistry. 1984;23:3576–3582. doi: 10.1021/bi00311a002. [DOI] [PubMed] [Google Scholar]
- Sergienko EA, Jordan F. Catalytic acid–base groups in yeast pyruvate decarboxylase. 2. Insights into the specific roles of D28 and E477 from the rates and stereospecificity of formation of carboligase side products. Biochemistry. 2001;40:7369–7381. doi: 10.1021/bi002856m. [DOI] [PubMed] [Google Scholar]
- Blomqvist K, Suihko ML, Knowles J, Penttila M. Chromosomal Integration and Expression of Two Bacterial alpha-Acetolactate Decarboxylase Genes in Brewer's Yeast. Appl Environ Microbiol. 1991;57:2796–2803. doi: 10.1128/aem.57.10.2796-2803.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suihko ML, Blomqvist K, Penttila M, Gisler R, Knowles J. Recombinant brewer's yeast strains suitable for accelerated brewing. J Biotechnol. 1990;14:285–300. doi: 10.1016/0168-1656(90)90113-P. [DOI] [PubMed] [Google Scholar]
- Remize F, Roustan J, Sablayrolles J, Barre P, Dequin S. Glycerol overproduction by engineered Saccharomyces cerevisiae wine yeast strains leads to substantial changes in by-product formation and to a stimulation of fermentation rate in stationary phase. Appl Environ Microbiol. 1999;65:143. doi: 10.1128/aem.65.1.143-149.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehsani M, Fernandez MR, Biosca JA, Julien A, Dequin S. Engineering of 2,3-butanediol dehydrogenase to reduce acetoin formation by glycerol-overproducing, low-alcohol Saccharomyces cerevisiae. Appl Environ Microbiol. 2009;75:3196–3205. doi: 10.1128/AEM.02157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano P, Suzzi G. Origin and production of acetoin during wine yeast fermentation. Appl Environ Microbiol. 1996;62:309. doi: 10.1128/aem.62.2.309-315.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NE, Nagarajan H, Palsson BO. Constraining the metabolic genotype–phenotype relationship using a phylogeny of in silico methods. Nat Rev Microbiol. 2012;10:291–305. doi: 10.1038/nrmicro2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo ML, Palsson BO, Herrgard MJ. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol. 2009;3:37. doi: 10.1186/1752-0509-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrgard MJ, Swainston N, Dobson P, Dunn WB, Arga KY, Arvas M, Bluthgen N, Borger S, Costenoble R, Heinemann M. et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol. 2008;26:1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Herrgard MJ, Palsson BO. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 2004;14:1298–1309. doi: 10.1101/gr.2250904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JS, Covert M, Palsson B. Metabolic modelling of microbes: the flux-balance approach. Environ Microbiol. 2002;4:133–140. doi: 10.1046/j.1462-2920.2002.00282.x. [DOI] [PubMed] [Google Scholar]
- Burgard AP, Pharkya P, Maranas CD. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- Fong SS, Burgard AP, Herring CD, Knight EM, Blattner FR, Maranas CD, Palsson BO. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol Bioeng. 2005;91:643–648. doi: 10.1002/bit.20542. [DOI] [PubMed] [Google Scholar]
- Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MA. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng. 2011;13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R. et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- Matsuda F, Furusawa C, Kondo T, Ishii J, Shimizu H, Kondo A. Engineering strategy of yeast metabolism for higher alcohol production. Microb Cell Fact. 2011;10:70. doi: 10.1186/1475-2859-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadollahi MA, Maury J, Patil KR, Schalk M, Clark A, Nielsen J. Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab Eng. 2009;11:328–334. doi: 10.1016/j.ymben.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Schuster S, Pfeiffer T, Fell DA. Is maximization of molar yield in metabolic networks favoured by evolution? J Theor Biol. 2008;252:497–504. doi: 10.1016/j.jtbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Kennedy CJ, Boyle PM, Waks Z, Silver PA. Systems-level engineering of nonfermentative metabolism in yeast. Genetics. 2009;183:385. doi: 10.1534/genetics.109.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bro C, Regenberg B, Forster J, Nielsen J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng. 2006;8:102–111. doi: 10.1016/j.ymben.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Pronk JT, Steensma HY, Van Dijken JP. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(SICI)1097-0061(199612)12:16<1607::AID-YEA70>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Nissen TL, Kielland-Brandt MC, Nielsen J, Villadsen J. Optimization of Ethanol Production in Saccharomyces cerevisiae by Metabolic Engineering of the Ammonium Assimilation. Metab Eng. 2000;2:69–77. doi: 10.1006/mben.1999.0140. [DOI] [PubMed] [Google Scholar]
- Bieganowski P, Seidle HF, Wojcik M, Brenner C. Synthetic lethal and biochemical analyses of NAD and NADH kinases in Saccharomyces cerevisiae establish separation of cellular functions. J Biol Chem. 2006;281:22439–22445. doi: 10.1074/jbc.M513919200. [DOI] [PubMed] [Google Scholar]
- Drewke C, Thielen J, Ciriacy M. Ethanol formation in adh0 mutants reveals the existence of a novel acetaldehyde-reducing activity in Saccharomyces cerevisiae. J Bacteriol. 1990;172:3909. doi: 10.1128/jb.172.7.3909-3917.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson JR, Salgado LE, Hewlins MJ. The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J Biol Chem. 2003;278:8028–8034. doi: 10.1074/jbc.M211914200. [DOI] [PubMed] [Google Scholar]
- Weimer EP, Rao E, Brendel M. Molecular structure and genetic regulation of SFA, a gene responsible for resistance to formaldehyde in Saccharomyces cerevisiae, and characterization of its protein product. Mol Gen Genet MGG. 1993;237:351–358. doi: 10.1007/BF00279438. [DOI] [PubMed] [Google Scholar]
- Smith MG, Des Etages SG, Snyder M. Microbial Synergy via an Ethanol-Triggered Pathway. Mol Cell Biol. 2004;24:3874–3884. doi: 10.1128/MCB.24.9.3874-3884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smidt O, du Preez JC, Albertyn J. Molecular and physiological aspects of alcohol dehydrogenases in the ethanol metabolism of Saccharomyces cerevisiae. FEMS Yeast Res. 2012;12:33–47. doi: 10.1111/j.1567-1364.2011.00760.x. [DOI] [PubMed] [Google Scholar]
- Skory CD. Lactic acid production by Saccharomyces cerevisiae expressing a Rhizopus oryzae lactate dehydrogenase gene. J Ind Microbiol Biotechnol. 2003;30:22–27. doi: 10.1007/s10295-002-0004-2. [DOI] [PubMed] [Google Scholar]
- Stanley GA, Douglas NG, Every EJ, Tzanatos T, Pamment NB. Inhibition and stimulation of yeast growth by acetaldehyde. Biotechnol Lett. 1993;15:1199–1204. doi: 10.1007/BF00130297. [DOI] [Google Scholar]
- Tokuhiro K, Ishida N, Nagamori E, Saitoh S, Onishi T, Kondo A, Takahashi H. Double mutation of the PDC1 and ADH1 genes improves lactate production in the yeast Saccharomyces cerevisiae expressing the bovine lactate dehydrogenase gene. Appl Microbiol Biotechnol. 2009;82:883–890. doi: 10.1007/s00253-008-1831-5. [DOI] [PubMed] [Google Scholar]
- Eglinton JM, Heinrich AJ, Pollnitz AP, Langridge P, Henschke PA, de Barros Lopes M. Decreasing acetic acid accumulation by a glycerol overproducing strain of Saccharomyces cerevisiae by deleting the ALD6 aldehyde dehydrogenase gene. Yeast. 2002;19:295–301. doi: 10.1002/yea.834. [DOI] [PubMed] [Google Scholar]
- Ludovico P, Sousa MJ, Silva MT, Leão C, Côrte-Real M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology. 2001;147:2409–2415. doi: 10.1099/00221287-147-9-2409. [DOI] [PubMed] [Google Scholar]
- Roustan J, Sablayrolles J. Impact of the addition of electron acceptors on the by-products of alcoholic fermentation. Enzym Microb Technol. 2002;31:142–152. doi: 10.1016/S0141-0229(02)00086-8. [DOI] [Google Scholar]
- Ansell R, Granath K, Hohmann S, Thevelein JM, Adler L. The two isoenzymes for yeast NAD + −dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J. 1997;16:2179–2187. doi: 10.1093/emboj/16.9.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollop N, Damri B, Chipman D, Barak Z. Physiological implications of the substrate specificities of acetohydroxy acid synthases from varied organisms. J Bacteriol. 1990;172:3444–3449. doi: 10.1128/jb.172.6.3444-3449.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo J, Martin R, Bernardo A, Gonzalez J. Purification, characterization and some properties of diacetyl(acetoin) reductase from Enterobacter aerogenes. Eur J Biochem. 1991;198:327–332. doi: 10.1111/j.1432-1033.1991.tb16019.x. [DOI] [PubMed] [Google Scholar]
- Ehsani M, Fernandez MR, Biosca JA, Dequin S. Reversal of coenzyme specificity of 2,3-butanediol dehydrogenase from Saccharomyces cerevisae and in vivo functional analysis. Biotechnol Bioeng. 2009;104:381–389. doi: 10.1002/bit.22391. [DOI] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3<259::AID-YEA901>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S. et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS‒DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional description for Table 1. (a) A list of the corresponding enzymes, reaction equations and subsystems for target reactions listed in Table 1 and (b) a figure showing the 2,3-butanediol production envelope of OptKnock strain A.
Oligonucleotides used in this study. A list of oligonucleotides that were used in this study.
Verification of gene deletion strains by colony PCR. The PCR profiles of wild-type and gene deletion strains.