Abstract
The spatial coordination of growth is of central importance for the regulation of plant tissue architecture. Individual layers, such as the epidermis, are clonally propagated and structurally maintained by symmetric cell divisions that are oriented along the plane of the layer. The developmental control of this process is poorly understood. The simple cellular basis and sheet-like structure of Arabidopsis integuments make them an attractive model system to address planar growth. Here we report on the characterization of the Arabidopsis UNICORN (UCN) gene. Analysis of ucn integuments reveals localized distortion of planar growth, eventually resulting in an ectopic multicellular protrusion. In addition, ucn mutants exhibit ectopic growth in filaments and petals, as well as aberrant embryogenesis. We further show that UCN encodes an active AGC VIII kinase. Genetic, biochemical, and cell biological data suggest that UCN suppresses ectopic growth in integuments by directly repressing the KANADI transcription factor ABERRANT TESTA SHAPE. Our findings indicate that UCN represents a unique plant growth regulator that maintains planar growth of integuments by repressing a developmental regulator involved in the control of early integument growth and polarity.
Keywords: AGC protein kinase, growth suppression, floral development, ovule, signal transduction
Plant tissue morphogenesis depends on the coordination of cellular behavior within tissue layers. The formation of distinct layers depends on asymmetric cell division, the developmental control of which is under intense investigation (1, 2). After initiation, individual cell layers are propagated by symmetric cell divisions (3) with division planes often oriented along the plane of the layer (planar growth). For example, the layered structure of the epidermis is maintained by anticlinal cell divisions, whereas periclinal cell divisions are usually suppressed (4). Division planes of symmetrically dividing cells can be accurately predicted by a mathematical rule linking cell geometry and cytoskeletal dynamics (5), and much progress has been made in the elucidation of the cellular machinery controlling the division plane of dividing plant cells. However, developmental controls that maintain the planar orientation of symmetrical cell divisions within a tissue layer remain poorly understood (3).
Arabidopsis integuments represent an attractive model to study tissue morphogenesis. They are lateral determinate tissues of the ovule, the progenitors of the seed coat, and undergo a regular mode of development resulting in simple tissue architecture. An inner and an outer integument originate from the epidermis of the central chalaza (6, 7). They grow into laminar extensions with each integument consisting of a bilayered sheet of anticlinally dividing cells. The outer integument eventually develops into a hood-like structure that envelops the distal nucellus carrying the developing embryo sac and the inner integument.
Coordination of symmetrical cell divisions along the plane of the extending integument represents an intriguing feature of integument development. However, with the exception of the two receptor-like kinase genes, ACR4 and ALE2 (8–10), none of the many genes affecting integument morphogenesis (11, 12) have been implied to play a primary role in the maintenance of the sheet-like organization of the integument cell layers. Thus, the signaling mechanisms regulating a central aspect of integument growth are poorly understood.
Here we show that UNICORN (UCN) regulates planar integument development by suppressing aberrantly oriented growth. In addition, UCN restricts growth in stamen filaments, petals, and cotyledons, and it affects the division planes in the proembryo. Our molecular characterization revealed that UCN encodes a functional AGC VIII kinase. Furthermore, we provide evidence that UCN regulates growth patterns in ovules by repressing the KANADI transcription factor ABERRANT TESTA SHAPE (ATS).
Results
Wild-Type Integument Development in Arabidopsis.
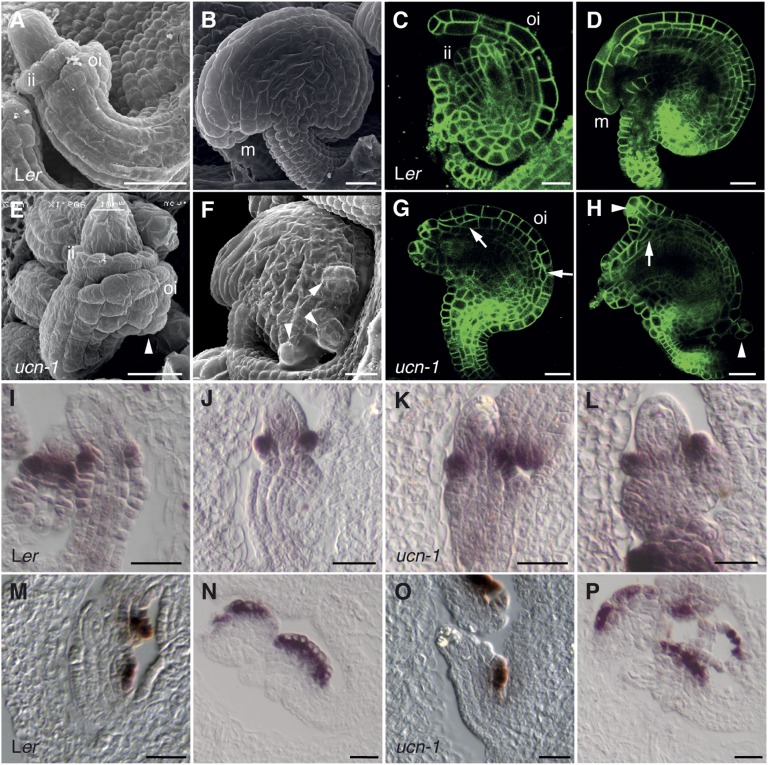
Both integuments were initiated, in cross-section view, in single epidermal cells that underwent oblique or periclinal cell divisions (7). Further development resulted in bilayered sheets of cells that expanded within the plane of the integument and underwent anticlinal cell divisions (Fig. 1 A–D) (7, 13). The early oblique or periclinal cell divisions likely include asymmetric cell divisions because early differences in gene expression and morphology indicate that cells of the two cell layers rapidly adopt different fates (Fig. 1 I and M) (7, 14–17). Thus, each integument exhibits tissue polarity and consists of an upper, or adaxial, and lower, or abaxial, cell layer.
Fig. 1.
Defective planar growth in ucn-1 integuments. (A and B) Scanning electron micrographs (SEMs) of wild-type ovules. (A) Stage 2-IV. ii, inner integument; oi, outer integument. (B) Stage 4. m, micropyle. (C and D) Confocal micrographs of wild-type ovules expressing the plasma membrane marker pUBQ::EGFP:LTI6B (51). (C) Stage 3-I. (D) Early stage 4. (E–H) Ovules of ucn-1. A similar series to that in A–D is shown. (E) Early protrusion formation (arrowhead). (F) Protrusions (arrowheads). (G and H) Irregular cell divisions (arrows) and protrusions (arrowheads). (I–L) In situ hybridization on sectioned ovules using a PHB probe (16). (I) Stage 2-III wild-type ovule. Signal is detectable in the adaxial layer of the inner integument. (J) Stage 2-IV wild-type ovule. (K and L) ucn-1. Ovules of comparable stages as in I and J are depicted. Note the normal PHB expression. (M–P) In situ hybridization on sectioned ovules using an INO probe (16). Early stage 2-III and late stage 2-IV ovules, respectively, are depicted. (M and N) Wild-type. INO expression is restricted to the abaxial cell layer in the outer integument. (O and P) ucn-1. Normal spatial expression domain of INO is shown. (Scale bars: 20 μm.)
UCN Restrains Growth in Various Lateral Organs and the Embryo.
Integuments of the recessive ucn-1 mutant (18) showed spontaneous local ectopic growths caused by aberrant changes in cell size, shape, and proliferation (Fig. 1 E–H). The earliest irregularities could be observed shortly after integument initiation (stage 2-IV/V; stages according to ref. 7) (Fig. 1E). In most ovules, however, the phenotype became apparent at slightly later stages (early stage 3 onward). At various positions along the integuments, one or a few cells in a cell layer of the inner or outer integument divided periclinally or sometimes obliquely (Fig. 1G). Similar aberrant cell divisions were observed at later stages as well. Furthermore, abnormally expanded cells could contribute to protrusion formation. However, not every aberrant cellular division seems to result in a protrusion. Generally, one outgrowth developed at a usually proximal integument position (18). Frequently, two protrusions, or occasionally three to four outgrowths, could be observed as well (Fig. 1 F and H). Protrusion formation on the inner integument did not depend on the presence of an outer integument (SI Appendix, Fig. S1). Expression of the class III HD-ZIP gene PHABULOSA (PHB) (19), an early marker for the adaxial cell layer of the inner integument (16), was not altered in ucn-1 ovules (Fig. 1 I–L; SI Appendix, Fig. S1). In addition, the abaxial layer of the outer integument of ucn-1 exhibited normal spatial expression of the abaxial cell fate marker gene INNER NO OUTER (INO) (refs. 14 and 20; Fig. 1 M–P; SI Appendix, Fig. S1). The result suggests that outer integument cells contributing to the protrusions exhibited at least partial outer integument identity and did not represent callus. In addition, the data indicate that UCN does not affect the establishment of adaxial–abaxial polarity in integuments.
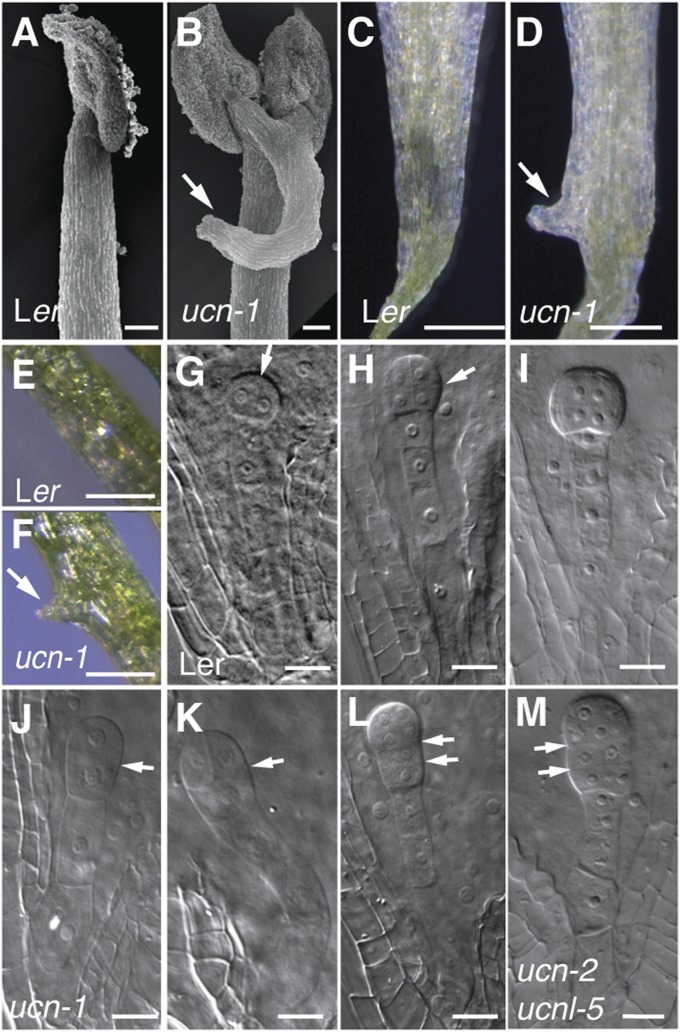
Further analysis revealed that stage 13 ucn-1 stamen filaments showed prominent outgrowths (Fig. 2 A and B), as did the petals (Fig. 2 C and D), which were also folded. In rare cases, we detected protrusions on petioles of cotyledons as well (Fig. 2 E and F), which superficially resembled leaf petiole outgrowths caused by allelic incompatibilities at the OAK locus (21). In addition, ucn-1 showed reduced fertility, and we observed aberrant cell divisions in ∼25% of embryos (Fig. 2 J–L; SI Appendix, Table S1). The asymmetric division of the zygote appeared normal in ucn-1. The first recognizable defect consisted of horizontal or oblique, rather than vertical, first and second symmetrical cell divisions in the proembryo. More advanced ucn-1 embryos showed abnormal cell proliferation, apparently concentrated to the basal part of the embryo. Thus, UCN affects the division plane of symmetrically dividing cells in integuments and early embryos. To test whether cell-cycle regulation was affected in ucn-1, we assayed the expression of several core cell-cycle genes in petals. Indeed, UCN influenced the cell cycle because we found complex changes in their expression levels (SI Appendix, Fig. S2). Together, the results indicate that UCN restrains abnormal cell proliferation and growth in ovules, filaments, petals, embryos, and cotyledons.
Fig. 2.
Aberrant growth in floral organs, cotyledons, and embryos of ucn-1 plants. (A–D) Floral defects (stage 13). (A and B) SEMs of stamens. (B) Protrusion on ucn-1 filament (arrow). (C and D) Petals. (D) ucn-1 petal with protrusion (arrow). (E and F) Cotyledons. (F) ucn-1. Note the protrusion (arrow). (G–M) Embryo defects in fertilized ovules (stage 12–14 flowers). (G–I) Wild type. (G) Proembryo with first vertical division (arrow). (H) Octant embryo after first horizontal division (arrow). (I) Globular embryo with protoderm formed. (J–L) ucn-1. Arrows indicate horizontal division planes. (J) Two-cell proembryo exhibiting aberrant horizontal division. (K) Proembryo (compare with H). (L) Proembryo with aberrant second horizontal division plane in the basal cell. (M) A mutant embryo in an ovule from a selfed ucn-2/ucn-2 ucnl-5/+ parent (compare with I). Note the aberrant cell proliferation in the basal proembryo. (Scale bars: A and B, 50 μm; C–F, 0.5 mm; G–M, 20 μm.)
UCN Encodes an AGC VIII Kinase.
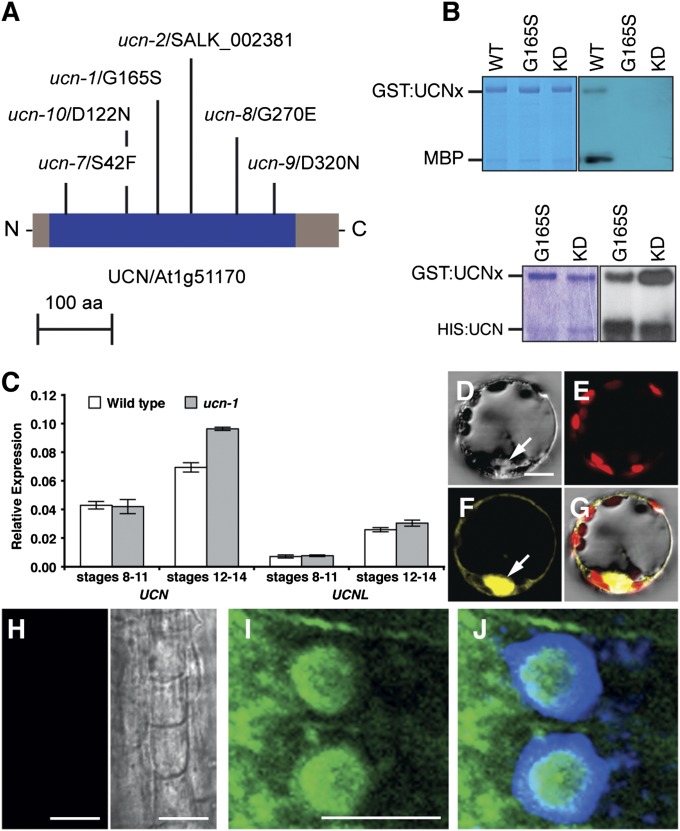
Map-based cloning, isolation of multiple mutant alleles, and genomic rescue experiments (SI Appendix, Fig. S3 A, B, and D, Table S2, and SI Materials and Methods) identified UCN as At1g51170 (Fig. 3A), encoding a member of the plant-specific AGC VIII family of protein kinases (22–25). UCN is a functional kinase that can autophosphorylate in trans as well as transphosphorylate the general kinase substrate myelin basic protein (Fig. 3B). The ucn-1 mutation resulted in the substitution of a conserved glycine at position 165 for a serine (G165S), which caused loss of kinase activity (Fig. 3B). UCN falls into the AGC2 subclass of the AGCVIII kinase family, which consists of two distantly related subclades with two members each (23). The closest relative of UCN (AGC2-3) is AGC2-4/At3g20830. UCN and AGC2-4 share 73% identity at the amino acid level, and thus AGC2-4 was renamed to UNICORN-LIKE (UCNL).
Fig. 3.
UCN is an active AGC kinase and localizes to the cytoplasm and the nucleus. (A) Scheme of predicted UCN protein (blue, kinase domain). Mutations are indicated. aa, amino acids. (B) In vitro kinase assay with purified GST:UCN or HIS:UCN fusion proteins. (Left) Coomassie blue gel. (Right) Corresponding autoradiographs. GST:UCNx denotes different GST:UCN variants indicated on top. (Upper) Autophosphorylation and phosphorylation of myelin basic protein (MBP). GST:UCNKD (K55E mutation, negative control). (Lower) Autophosphorylation and transphosphorylation of unfunctional GST:UCNG165S or GST:UCNKD by HIS:UCN. (C) A qRT-PCR-based comparison of floral UCN (Left) and UCNL (Right) mRNA levels in wild type and ucn-1. (D–G) BiFC-visualization of UCN dimerization. Different views of a protoplast cotransfected with pSPYNE:UCN and pSPYCE:UCN plasmids are shown. (D) Bright-field view. (E) Chloroplast autofluorescence (red signal). (F) YFP channel (yellow signal). Arrow indicates nucleus. (G) Merge. Cotransfecting empty pUC-SPYNE/SPYCE plasmids did not result in signal (SI Appendix, Fig. S4). (H–J) Immunolocalization of UCN/UCNL in whole-mount roots of 6-d-old seedlings. (H) Preimmune serum (Left) and bright-field view (Right). (I and J) Anti-UCN antibody (I) plus DAPI (J). (Scale bars: D–G, 30 μm; H–J, 10 μm.)
Detection of UCN expression in developing flowers by in situ hybridization or immunolocalization was unsuccessful, indicating that UCN is expressed at very low levels. However, quantitative real-time PCR (qRT-PCR) revealed UCN expression at floral stages 8–11 (Fig. 3C), which encompasses the period when ucn phenotype first becomes apparent. The results also showed that UCN is normally transcribed in stage 8–11 ucn-1 flowers but slightly higher in later stages (Fig. 3C).
To assess the subcellular localization of UCN, we performed bimolecular fluorescence complementation (BiFC) analysis (Fig. 3 D–G). The results indicated that full-length UCN can form homodimers in a plant cell, confirming results from the in vitro kinase assays. Furthermore, signals were broadly distributed throughout the cell, including the nucleus. Indeed, UCN carries a bipartite nuclear localization signal. Moreover, direct detection of endogenous UCN and UCNL protein (SI Appendix, Fig. S6) by using an anti-UCN antiserum revealed expression of UCN and/or UCNL in the epidermis and cortex of the transition zone of the root apex. Cells exhibited signal in the nucleus, the cytoplasm, and likely at the plasma membrane (Fig. 3 H–J). Computer prediction also suggested a chloroplast localization of UCN. However, the BiFC results (Fig. 3 D–G), as well as in vitro chloroplast import studies (SI Appendix, Fig. S3E) and chlorophyll fluorescence parameter measurements (SI Appendix, Table S3), failed to confirm this prediction. Together, the data indicate that UCN shows a broad subcellular distribution and may function in different cellular compartments.
UCN and UCNL Act Redundantly.
UCN is transcribed in ucn-1 (Fig. 3C), and UCNG165S protein may have residual biological activity. Furthermore, UCN and UCNL show a broad and overlapping expression profile (Fig. 3C; SI Appendix, Fig. S3F). In flowers, UCNL exhibited notably reduced transcript levels in comparison with UCN (Fig. 3C). Nevertheless, UCN and UCNL could act redundantly. Thus, we investigated the phenotype of ucn and ucnl null alleles as well as ucn ucnl double mutants. Two T-DNA insertion lines, ucn-2 and ucnl-5, carry null alleles (SI Appendix, Figs. S3 B and C, S5, and S6, Table S2, and SI Materials and Methods) but appeared phenotypically wild type. Upon selfing of a ucn-2/ucn-2 ucnl-5/+ plant, a total of 136 F1 plants were genotyped. Of those, 87 plants were heterozygous for ucnl-5, whereas 49 carried the wild-type UCNL allele, indicating that ucn-2 ucnl-5 double mutants were embryo lethal with full penetrance. Furthermore, embryo aberrations were essentially identical to the embryo defects observed in ucn-1 plants (Fig. 2 J–M; SI Appendix, Table S1). These results corroborate redundancy and reveal that complete loss of UCN and UCNL activity leads to fully penetrant embryo lethality. Interestingly, ucn-1 plants showed the null phenotype with a reduced penetrance of ∼25%, indicating that ucn-1 is a hypomorph, at least with respect to embryo development. A simple explanation for this genetic behavior is to assume that UCN and UCNL bind to the same protein complex and that UCN, as well as the partially functional UCNG165S protein, titrate out wild-type UCNL protein (SI Appendix, Fig. S7).
UCN Restrains Ectopic Growth in Integuments Through Negative Regulation of ATS.
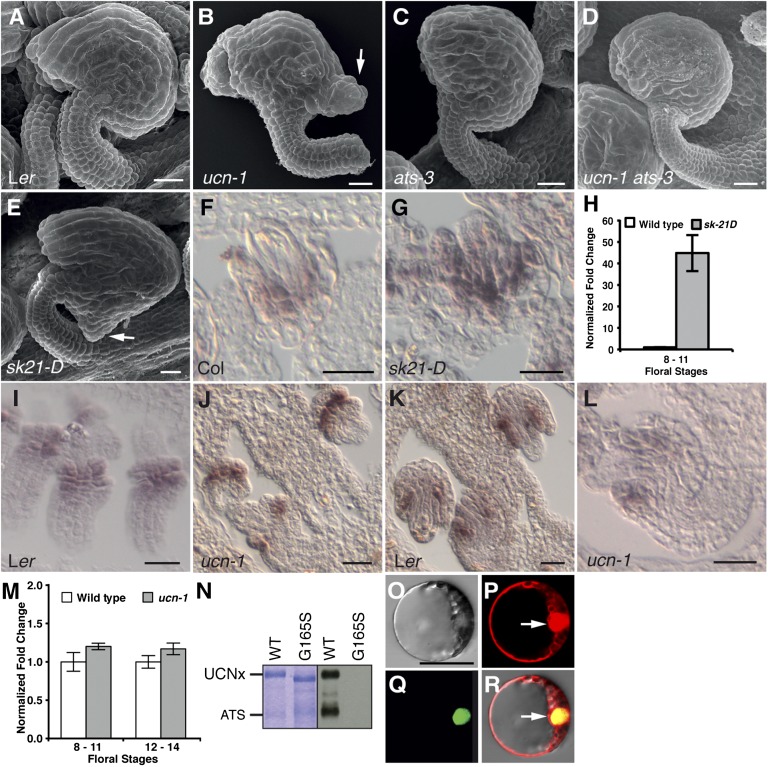
To address the mechanism of UCN-mediated ectopic growth suppression, we combined ucn-1 with several ovule mutants (SI Appendix, Figs. S1G and S9). Interestingly, the phenotype of ucn ats double mutants suggested that UCN suppresses protrusion formation in integuments by negatively regulating ATS (Fig. 4 A–D). ATS encodes a putative transcription factor (15) of the KANADI (KAN) family (26, 27). Defects in ats mutants are specific to ovules, which show congenitally fused integuments (15, 28, 29). In wild-type ovules, ATS expression is observed at the boundary between the two initiating integuments, at the abaxial side of the inner integument and the adaxial side of the outer integument, and eventually becomes restricted to the abaxial cells of the inner integument (15) (Fig. 4 I–J). Interestingly, ovules of ucn-1 ats-3 double mutants exhibited ats-like integuments that were essentially devoid of ucn-like protrusions (Fig. 4D; SI Appendix, Table S4). This result demonstrates that ats is epistatic to ucn. Moreover, it strongly suggests that UCN and ATS act in the same genetic pathway, that UCN is a negative regulator of ATS, and that ATS is the main direct or indirect downstream target of UCN in integument growth control.
Fig. 4.
UCN is a negative posttranscriptional regulator of ATS. (A–E) SEMs of early stage 4 ovules. (A) Wild type. (B) ucn-1. (C) ats-3. (D) ucn-1 ats-3. Note the absence of protrusions. (E) ATS activation tagging line sk21-D. Compare the protrusion with B (arrow). (F and G) Spatial ATS expression analyzed by in situ hybridization on sectioned ovules. Stage 2-IV wild-type (Left) and sk21-D (Right) ovules are shown. In wild type, detectable ATS expression ranges from the abaxial layer of the inner integument to the adaxial layer of the outer integument. ATS spatial expression appears normal in sk21-D. (H) Floral ATS mRNA levels in wild type and sk21-D measured by qRT-PCR. Note the high level of ATS expression at floral stages 8–11 in sk21-D. (I–L) Spatial ATS expression analyzed by in situ hybridization on sectioned ovules. (I and J) Stage 2-III wild-type (Left) and ucn-1 (Right) ovules are shown. No changes were observed. (K and L) Stage 2-V wild-type (Left) and ucn-1 (Right) ovules are shown. ATS expression is mainly observed in the inner integument. No changes were observed. (M) Floral ATS mRNA levels measured by qRT-PCR. No significant changes were observed. (N) In vitro kinase assay using purified GST:UCN and GST:ATS fusion proteins. (Left) Coomassie blue gel. (Right) Corresponding autoradiograph. UCNx denotes different GST:UCN variants indicated on top. (O–R) BiFC assay of UCN/ATS interaction. Different views of a protoplast cotransfected with pSPYNE:ATS, pSPYCE:UCN, and pGY-1:mCherry plasmids are shown. (O) Bright-field view. (P) Signal distribution of free mCherry. Arrow highlights the nucleus. (Q) YFP signal distribution indicating UCN/ATS interaction. (R) Merge. Cotransfecting of empty pUC-SPYNE/SPYCE plasmids did not result in signal (SI Appendix, Fig. S4). (Scale bars: A–G and I–L, 20 μm; O–R, 30 μm.)
We investigated how UCN regulates ATS. An ∼45-fold increase in ATS expression levels within the normal ATS expression domain in the activation-tagging line sk21-D (30) resulted in a ucn-like protrusion in ovules (Fig. 4 E–H; SI Appendix, Fig. S8 and Table S4), corroborating the notion that negative regulation of ATS is essential for regular planar growth of integuments. However, ATS expression was found to be unaltered in ucn-1 (Fig. 4 I–M), indicating that UCN regulates ATS posttranscriptionally. We thus explored whether the two proteins could interact physically. Although the low level of UCN expression in developing flowers precluded a direct test in planta, recombinant UCN phosphorylated ATS in in vitro kinase assays (Fig. 4N). Furthermore, we were able to show that the two proteins interacted in nuclei of plant cells in a BiFC assay (Fig. 4 O–R). These findings suggest that UCN may directly repress ATS protein activity in the nucleus, possibly through differential phosphorylation.
Discussion
Here we present evidence that UCN restrains aberrant growth in diverse plant organs. The ucn outgrowths are distinct from the wart-like epidermal cell swellings caused by several mutations in Arabidopsis and maize (31–33) or the multiplication of epidermal cell layers caused by the maize Xcl1 mutation (34). Furthermore, UCN is likely to function in a different pathway than ACR4 and ALE2 (SI Appendix, Fig. S9 M–P). Given its particular effects on cell division and cell differentiation, we propose UCN to be a unique plant growth regulator. Interestingly, our combined genetic and biochemical evidence suggests that UCN suppresses ectopic growth in integuments through the direct negative regulation of the transcription factor ATS. UCN acts in a context-dependent fashion, because outgrowth formation in other tissues, such as filaments and petals, still occurs in ucn ats double mutants. Thus, UCN constitutes a general growth suppressor that interacts with additional, yet to be identified factors.
Our results provide unique insight into the mechanism controlling planar orientation of symmetric cell divisions in plants. UCN seems to play an important role in this process, because a periclinal or oblique cell division is the first discernible cellular defect in integument outgrowth of ucn mutants. A role in division plane control of symmetrically dividing cells is corroborated by the defect observed in ucn proembryos. UCN is unlikely to directly mediate the asymmetric subcellular localization of cellular factors, as it is known for some AGC VIII kinases (35). Rather, our evidence indicates that UCN acts in a more indirect fashion by modulating transcriptional programs via ATS to control planar growth of integuments.
The ucn phenotype and the interaction between UCN and ATS also provide evidence for the notion that maintenance of planar growth needs to be coordinated with adaxial–abaxial polarity during integument development. Class III HD-ZIP, KAN, and YABBY transcription factors play central roles in the establishment of adaxial–abaxial polarity in leaves (36, 37) and ovules (38), with KAN genes promoting abaxial cell fate and lamina outgrowth in leaves (26, 27). ATS is a KAN gene that regulates integument boundary formation, inner integument outgrowth, and adaxial–abaxial polarity (15, 28, 29, 38). A role for UCN in establishing adaxial–abaxial polarity in integuments is unlikely given the unaltered spatial expression pattern of several marker genes in ucn. Rather, our results raise the possibility that abnormal up-regulation of ATS activity in ucn within its normal expression domain interferes with planar growth control during integument extension. The results further indicate that maintaining planar growth requires the negative regulation of a factor involved in the control of adaxial–abaxial polarity. In this scenario, UCN would contribute to the coordination of these two processes. A defect in the coordination would result in ectopic growth, perhaps not unlike the instances in which loss of control of asymmetric cell division results in aberrant cell divisions (2).
Apart from the genetic antagonism between KAN and class III HD-ZIP genes in the establishment of organ polarity, nothing is known about the mechanism restricting KAN function (36, 37). Interestingly, our data imply that UCN acts as a direct negative regulator of a KAN transcription factor. Thus, it is possible that UCN-dependent planar growth control in other organs may involve the regulation of different KAN family members. Furthermore, the molecular process with which hyperactive ATS interferes in ucn integuments remains unknown. Considering these aspects, additional exploration of UCN-mediated signaling and its role in maintaining planar growth is needed.
Materials and Methods
Plant Work, Genetics, and Transformation.
Arabidopsis thaliana (L.) Heynh. var. Columbia (Col-0) and var. Landsberg (erecta mutant) (Ler), were used as wild-type strains. Plants were grown essentially as described (18).
Detailed information about all ucn and ucnl mutants identified in this study is given in SI Appendix, Table S2 and SI Materials and Methods. The ucn-1 mutant was isolated previously in an ethane methyl sulfonate (EMS) mutagenesis (18) and outcrossed three times to Ler before further analysis. The EMS-induced mutations ucn-7 to -10 were identified in the UCN CDS in a Col er-105 background in conjunction with the Seattle Arabidopsis TILLING facility (39). The ucn-2 and -5 T-DNA insertion were predicted null alleles (SI Appendix, Fig. S3 B and C and Table S2). Moreover, in contrast to the other T-DNA lines, no wild-type UCN or UCNL transcripts, respectively, could be detected in these two mutants (SI Appendix, Fig. S5).
Plant transformation was performed by using the floral dip method (40).
Recombinant DNA Work.
For DNA and RNA work, standard molecular biology techniques were used (41). PCR fragments used for cloning were obtained by using Phusion high-fidelity DNA polymerase (New England Biolabs). Site-directed mutagenesis of plasmids was done by using the QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer’s recommendations. All PCR-based constructs were sequenced. All primers used in this study are listed in SI Appendix, Table S5.
PCR-Based Gene Expression Analysis.
To survey UCN and UCNL expression in plants, RNA from different organs was isolated by using the NucleoSpin RNA II kit (Macherey-Nagel). PCR was performed by using Taq DNA polymerase (New England Biolabs) and UCN and UCNL-specific primer pairs [UCN (sqRT)_F, UCN (sqRT)_R; UCNL (sqRT)_F, UCNL (sqRT)_R]. Between 19 and 32 thermal cycles were tested. The GAPC gene was used as positive control (42).
Tissue for qRT-PCR was harvested from 25-d-old plants grown in long-day conditions. With minor changes, tissue collection, RNA extraction, and quality control were performed as described (43). qRT-PCR was performed on a Roche LightCycler480 by using the iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s recommendations. By using the ∆∆-Ct method, all gene expression levels were normalized against At5g25760, At4g33380, and At2g28390 expression (44). Experiments were performed in biological and technical triplicates.
Generation, Expression, and Purification of Recombinant Proteins.
All recombinant proteins were fusions of either a 6×-His/X-press (HIS) or a GST tag to the amino terminus of full-length UCN and ATS. Detailed cloning information is available from the authors upon request. The various constructs were transformed into Escherichia coli BL21 (DE3) cells (Invitrogen) and grown to an OD600 of 0.6. Recombinant protein expression was then induced by 0.5 mM isopropyl-β-d-thiogalactoside (FLUKA) for 4 h at 37 °C. Cells were pelleted and subjected to solubilization and recombinant protein purification by using the Protino Ni-TED 2000 (Macherey-Nagel) and Protino Glutathione Agarose 4B kits (GE Healthcare), respectively, according to the manufacturer’s recommendations.
In Vitro Kinase Assays.
Kinase assays were done with 4 μg of purified fusion protein and in the absence or presence of 2 μg of myelin basic protein (Sigma-Aldrich). Proteins were incubated in 20 μL of 40 mM Hepes (pH 7.4), 40 mM MgCl2, 10 μM ATP, and 10 μCi [γ-32P]ATP (Hartmann Analytic). The reaction was stopped by adding 5 μL of 4× sample buffer [100 mM Tris, pH 6.8, 20% (wt/vol) glycerol, and 4% (wt/vol) SDS]. The samples were boiled and analyzed by SDS-PAGE. Coomassie blue-stained gels were washed, dried, and exposed to Kodak film using an intensifying screen (Sigma).
Antibody Generation and Immunohistochemistry.
Full-length recombinant HIS:UCN fusion protein purified from a SDS-PAGE gel was used to raise a polyclonal rabbit antiserum followed by IgG purification (Davids Biotechnologie). Goat anti-rabbit monoclonal antibody was coupled to Alexa-m488 (Invitrogen). Nuclei were stained with 1 mg/mL DAPI (Sigma). Antibody staining was performed as described (45) on the root tips of 6-d-old seedlings.
BiFC Assay.
UCN and ATS coding sequences were PCR amplified and cloned into pUC-SPYCE and pUC-SPYNE vectors (46). The plasmid pGY-1:mCherry (47) was used as transformation control. Four micrograms of each plasmid in desired combinations was used to transiently transfect mesophyll protoplasts that were generated from 2-wk-old Arabidopsis leaves (Col) (48).
Microscopy, In Situ Hybridization, and Artwork.
Preparation and analysis of samples for microscopy and histochemical localization of β-glucuronidase (GUS) activity in whole-mount tissue was done essentially as described (18, 49, 50). Confocal laser scanning microscopy was performed with an Olympus FV1000 setup and FluoView software (Olympus Europa) (SI Appendix, SI Materials and Methods). All images were obtained by sequential scanning to eliminate interference of lasers.
In situ hybridization with digoxigenin-labeled probes was done essentially as described (16). The INO and PHB probes were described (16). A 0.831-kb ATS antisense probe was obtained by PCR using a full-length cDNA clone (U8421; Arabidopsis Biological Resource Center) as template and the primer pair ATSas_831_F/ATSas_831_R. The sense control was obtained by using primer pair ATSsense_831_F/ATSsense_831_R. Slides were viewed with an Olympus BX61 upright microscope by using differential interference contrast optics. Images were adjusted for color and contrast by using Adobe Photoshop CS5 (Adobe) software.
Supplementary Material
Acknowledgments
We thank C. Gasser for the pINO::GUS reporter line; G. Ingram, H. Tanaka, and M. Gruber for providing the acr4-2, ale2-1, and sk21-D mutants, respectively; the Arabidopsis Biological Resource Center (ABRC) for providing materials; A. Schnittger for advice on the cell-cycle genes; and S. Balasubramanian, J. Lohmann, and M. Hülskamp for comments on the manuscript. B.E. was supported by the Technische Universität München Graduate School Graduiertenzentrum Weihenstephan. This work was supported by Deutsche Forschungsgemeinschaft Grants LE 1265/3 and 1265/9 (to D.L.) and SCHN 723/3-1 and SCHN 723/3-2 (to K.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205089109/-/DCSupplemental.
References
- 1.Abrash EB, Bergmann DC. Asymmetric cell divisions: A view from plant development. Dev Cell. 2009;16:783–796. doi: 10.1016/j.devcel.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 2.De Smet I, Beeckman T. Asymmetric cell division in land plants and algae: The driving force for differentiation. Nat Rev Mol Cell Biol. 2011;12:177–188. doi: 10.1038/nrm3064. [DOI] [PubMed] [Google Scholar]
- 3.Rasmussen CG, Humphries JA, Smith LG. Determination of symmetric and asymmetric division planes in plant cells. Annu Rev Plant Biol. 2011;62:387–409. doi: 10.1146/annurev-arplant-042110-103802. [DOI] [PubMed] [Google Scholar]
- 4.Tilney-Bassett RAE. Plant Chimeras. London: E. Arnold; 1986. [Google Scholar]
- 5.Besson S, Dumais J. Universal rule for the symmetric division of plant cells. Proc Natl Acad Sci USA. 2011;108:6294–6299. doi: 10.1073/pnas.1011866108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenik PD, Irish VF. Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development. 2000;127:1267–1276. doi: 10.1242/dev.127.6.1267. [DOI] [PubMed] [Google Scholar]
- 7.Schneitz K, Hülskamp M, Pruitt RE. Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 1995;7:731–749. [Google Scholar]
- 8.Gifford ML, Dean S, Ingram GC. The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development. 2003;130:4249–4258. doi: 10.1242/dev.00634. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka H, et al. Novel receptor-like kinase ALE2 controls shoot development by specifying epidermis in Arabidopsis. Development. 2007;134:1643–1652. doi: 10.1242/dev.003533. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe M, Tanaka H, Watanabe D, Machida C, Machida Y. The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 2004;39:298–308. doi: 10.1111/j.1365-313X.2004.02132.x. [DOI] [PubMed] [Google Scholar]
- 11.Colombo L, Battaglia R, Kater MM. Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci. 2008;13:444–450. doi: 10.1016/j.tplants.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Kelley DR, Gasser CS. Ovule development: genetic trends and evolutionary considerations. Sex Plant Reprod. 2009;22:229–234. doi: 10.1007/s00497-009-0107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson-Beers K, Pruitt RE, Gasser CS. Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell. 1992;4:1237–1249. doi: 10.1105/tpc.4.10.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balasubramanian S, Schneitz K. NOZZLE regulates proximal-distal pattern formation, cell proliferation and early sporogenesis in Arabidopsis thaliana. Development. 2000;127:4227–4238. doi: 10.1242/dev.127.19.4227. [DOI] [PubMed] [Google Scholar]
- 15.McAbee JM, et al. ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 2006;46:522–531. doi: 10.1111/j.1365-313X.2006.02717.x. [DOI] [PubMed] [Google Scholar]
- 16.Sieber P, et al. Pattern formation during early ovule development in Arabidopsis thaliana. Dev Biol. 2004;273:321–334. doi: 10.1016/j.ydbio.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 17.Skinner DJ, Gasser CS. Expression-based discovery of candidate ovule development regulators through transcriptional profiling of ovule mutants. BMC Plant Biol. 2009;9:29. doi: 10.1186/1471-2229-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneitz K, Hülskamp M, Kopczak SD, Pruitt RE. Dissection of sexual organ ontogenesis: A genetic analysis of ovule development in Arabidopsis thaliana. Development. 1997;124:1367–1376. doi: 10.1242/dev.124.7.1367. [DOI] [PubMed] [Google Scholar]
- 19.McConnell JR, et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 20.Villanueva JM, et al. INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 1999;13:3160–3169. doi: 10.1101/gad.13.23.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith LM, Bomblies K, Weigel D. Complex evolutionary events at a tandem cluster of Arabidopsis thaliana genes resulting in a single-locus genetic incompatibility. PLoS Genet. 2011;7:e1002164. doi: 10.1371/journal.pgen.1002164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bögre L, Okrész L, Henriques R, Anthony RG. Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 2003;8:424–431. doi: 10.1016/S1360-1385(03)00188-2. [DOI] [PubMed] [Google Scholar]
- 23.Galván-Ampudia CS, Offringa R. Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci. 2007;12:541–547. doi: 10.1016/j.tplants.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Hirt H, Garcia AV, Oelmüller R. AGC kinases in plant development and defense. Plant Signal Behav. 2011;6:1030–1033. doi: 10.4161/psb.6.7.15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, McCormick S. AGCVIII kinases: At the crossroads of cellular signaling. Trends Plant Sci. 2009;14:689–695. doi: 10.1016/j.tplants.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Eshed Y, Baum SF, Perea JV, Bowman JL. Establishment of polarity in lateral organs of plants. Curr Biol. 2001;11:1251–1260. doi: 10.1016/s0960-9822(01)00392-x. [DOI] [PubMed] [Google Scholar]
- 27.Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- 28.Balasubramanian S, Schneitz K. NOZZLE links proximal-distal and adaxial-abaxial pattern formation during ovule development in Arabidopsis thaliana. Development. 2002;129:4291–4300. doi: 10.1242/dev.129.18.4291. [DOI] [PubMed] [Google Scholar]
- 29.Léon-Kloosterziel KM, Keijzer CJ, Koornneef M. A seed shape mutant of Arabidopsis that is affected in integument development. Plant Cell. 1994;6:385–392. doi: 10.1105/tpc.6.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao P, et al. A new dominant Arabidopsis transparent testa mutant, sk21-D, and modulation of seed flavonoid biosynthesis by KAN4. Plant Biotechnol J. 2010;8:979–993. doi: 10.1111/j.1467-7652.2010.00525.x. [DOI] [PubMed] [Google Scholar]
- 31.Hunter CT, et al. Cellulose Synthase-Like D1 is integral to normal cell division, expansion, and leaf development in maize. Plant Physiol. 2012;158:708–724. doi: 10.1104/pp.111.188466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motose H, Tominaga R, Wada T, Sugiyama M, Watanabe Y. A NIMA-related protein kinase suppresses ectopic outgrowth of epidermal cells through its kinase activity and the association with microtubules. Plant J. 2008;54:829–844. doi: 10.1111/j.1365-313X.2008.03445.x. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds JO, Eisses JF, Sylvester AW. Balancing division and expansion during maize leaf morphogenesis: Analysis of the mutant, warty-1. Development. 1998;125:259–268. doi: 10.1242/dev.125.2.259. [DOI] [PubMed] [Google Scholar]
- 34.Kessler S, Seiki S, Sinha N. Xcl1 causes delayed oblique periclinal cell divisions in developing maize leaves, leading to cellular differentiation by lineage instead of position. Development. 2002;129:1859–1869. doi: 10.1242/dev.129.8.1859. [DOI] [PubMed] [Google Scholar]
- 35.Dhonukshe P, et al. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development. 2010;137:3245–3255. doi: 10.1242/dev.052456. [DOI] [PubMed] [Google Scholar]
- 36.Husbands AY, Chitwood DH, Plavskin Y, Timmermans MC. Signals and prepatterns: New insights into organ polarity in plants. Genes Dev. 2009;23:1986–1997. doi: 10.1101/gad.1819909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szakonyi D, Moschopoulos A, Byrne ME. Perspectives on leaf dorsoventral polarity. J Plant Res. 2010;123:281–290. doi: 10.1007/s10265-010-0336-3. [DOI] [PubMed] [Google Scholar]
- 38.Kelley DR, Skinner DJ, Gasser CS. Roles of polarity determinants in ovule development. Plant J. 2009;57:1054–1064. doi: 10.1111/j.1365-313X.2008.03752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Till BJ, et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. Plainview, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 42.Shih MC, Heinrich P, Goodman HM. Cloning and chromosomal mapping of nuclear genes encoding chloroplast and cytosolic glyceraldehyde-3-phosphate-dehydrogenase from Arabidopsis thaliana. Gene. 1991;104:133–138. doi: 10.1016/0378-1119(91)90242-4. [DOI] [PubMed] [Google Scholar]
- 43.Box MS, Coustham V, Dean C, Mylne JS. Protocol: A simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods. 2011;7:7. doi: 10.1186/1746-4811-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Völker A, Stierhof YD, Jürgens G. Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J Cell Sci. 2001;114:3001–3012. doi: 10.1242/jcs.114.16.3001. [DOI] [PubMed] [Google Scholar]
- 46.Walter M, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 47.Hoefle C, et al. A barley ROP GTPase ACTIVATING PROTEIN associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. Plant Cell. 2011;23:2422–2439. doi: 10.1105/tpc.110.082131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 49.Gross-Hardt R, Lenhard M, Laux T. WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 2002;16:1129–1138. doi: 10.1101/gad.225202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torres-Ruiz RA, Jürgens G. Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development. 1994;120:2967–2978. doi: 10.1242/dev.120.10.2967. [DOI] [PubMed] [Google Scholar]
- 51.Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.