Abstract
Holliday junctions can be formed during homology-dependent repair of DNA double-strand breaks and their resolution is essential for chromosome segregation and generation of crossover products. The Mus81–Mms4 and Yen1 nucleases are required for mitotic crossovers between chromosome homologs in Saccharomyces cerevisiae; however, crossovers between dispersed repeats are still detected in their absence. Here we show the Rad1–Rad10 nuclease promotes formation of crossover and noncrossover recombinants between ectopic sequences. Crossover products were not recovered from the mus81Δ rad1Δ yen1Δ triple mutant indicating that all three nucleases participate in processing recombination intermediates that form between dispersed repeats. We suggest a novel mechanism for crossovers that involves Rad1–Rad10 clipping and resolution of a single Holliday junction-containing intermediate by Mus81–Mms4 or Yen1 cleavage, or by replication. Consistent with the model, we show the accumulation of Rad1 dependent joint molecules in the mus81Δ yen1Δ mutant.
Homologous recombination is an important repair mechanism to eliminate double-strand breaks (DSBs) and to bypass lesions that result in formation of single-stranded DNA gaps during DNA replication1. Homologous recombination can result in the exchange of flanking markers to produce crossovers between chromosomes. Although crossovers are essential during meiosis they can have adverse consequences in somatic (mitotic) cells. A crossover between non-sister chromatids in G2 cells results in loss of heterozygosity (LOH) from the site of exchange to the telomere if the two recombinant chromatids segregate to opposite poles during mitosis2,3. It is estimated that 40% of the cases of hereditary retinoblastoma are caused by mitotic recombination leading to loss of the wild-type copy of the RB1 gene4. Furthermore, crossovers between dispersed repeats present on the same chromosome or non-homologous chromosomes generate deletions, duplications, inversions or translocations. Thus, crossovers can result in genome rearrangements and proteins that prevent mitotic crossovers function as cancer suppressors in humans5,6.
Homologous recombination initiates by the 5′ to 3′ nucleolytic degradation of the broken DNA ends to create 3′ single-stranded DNA (ssDNA) tails7. Rad51 binds to the resulting ssDNA tails to initiate pairing and strand invasion with homologous duplex DNA forming a displacement loop (D-loop). The invading 3′ end is used to prime leading strand DNA synthesis, templated by the donor duplex1. By the double-strand break repair (DSBR) model, the other end of the break anneals with the D-loop from the donor duplex to prime DNA synthesis and seal the break resulting in a double Holliday junction (dHJ) intermediate8. Intermediates containing a dHJ have been detected physically during mitotic and meiotic DSBR in S. cerevisiae9–12. By the synthesis-dependent strand annealing model, the invading strand that has been extended by DNA synthesis is displaced and anneals to complementary sequences exposed by 5′-3′ resection of the other side of the break yielding exclusively noncrossover products13,14.
Holliday junctions connecting homologous duplexes must be removed for accurate chromosome segregation. Recombination intermediates containing a dHJ can be dissolved by the yeast Sgs1–Top3–Rmi1 (STR) complex (BLM–TOPIIIα–RMI1–RMI2 in human) to yield noncrossover products, a mechanism favored in somatic cells to suppress crossover formation15–18. On the other hand, nucleolytic resolution of Holliday junctions can lead to crossover or noncrossover products. Several proteins, including Mus81–Mms4 (MUS81–EME1 in human), human SLX1–SLX4 and Yen1 (GEN1), able to cut HJs in vitro have been identified as candidate resolvases in yeast and mammals19. Mus81–Mms4 appears to be the primary resolvase activity in mitotically dividing S. cerevisiae, with Yen1 serving a back-up function, and it is essential for meiotic crossovers in Schizosaccharomyces pombe19–24. Most meiotic crossovers in S. cerevisiae result from biased resolution of dHJ intermediates by Mlh1-Mlh3-Exo1 and meiosis-specific ZMM factors25,26. However, Mus81-Mms4 is primarily responsible for the meiotic crossovers generated by the interference independent pathway, which might be considered analogous to mitotic crossovers and the S. pombe meiotic crossover pathway25,26. Mitotic crossover products are still detected between dispersed repeats in the mus81Δ yen1Δ mutant suggesting an additional activity or alternative mode of processing recombination intermediates operates in this context27. The Rad1–Rad10 (XPF–ERCC1) nuclease, which is essential for nucleotide excision repair28, is suggested to resolve meiotic recombination intermediates in some organisms29. The S. cerevisiae rad1Δ mutant exhibits normal meiotic recombination, but is defective for mitotic recombination between substrates that require removal of heterologous flaps30,31. In addition, rad1Δ and Ercc1−/− cells are defective for integration of linear DNA fragments to replace chromosomal sequences during gene targeting leading to the proposal that Rad1–Rad10 (XPF–ERCC1) cleaves at the boundary between homologous and heterologous sequences, represented by the selected marker, during this process32–34.
Here we sought to test the hypothesis that Rad1–Rad10 cleaves recombination intermediates formed at the heterology barrier resulting in the generation of crossover products between dispersed repeats. Using two different assays to measure recombination between homology-limited substrates we show Rad1 is required for normal levels of crossovers, and in the absence of Mus81, Rad1 and Yen1 crossovers are eliminated. In addition, we provide physical evidence that Rad1–Rad10 cleaves a recombination intermediate to generate a substrate that is dependent on Mus81–Mms4 or Yen1 for resolution.
RESULTS
Rad1 is required for plasmid integration
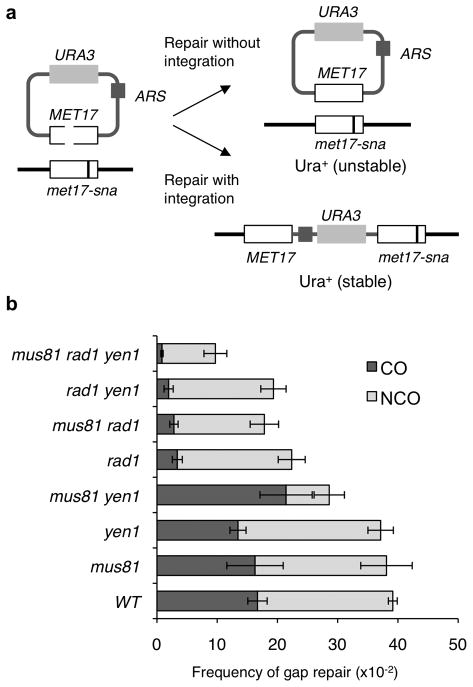
To test the role of Rad1 for recombination between homology-limited substrates we first analyzed integration of a linearized plasmid at the homologous chromosomal locus during transformation of yeast cells. An ARS-containing (autonomously replicating) plasmid containing URA3 and MET17 genes was digested with two restriction enzymes to create a 238-bp double strand DNA gap within the MET17 gene, which has 2.6 kb homology to the chromosomal locus (Figure 1a)35. Homology-dependent repair of the plasmid yields noncrossover (episomal) or crossover (integrated) products with an unstable or stable Ura+ phenotype, respectively. The overall frequency of gap repair was reduced in the rad1Δ mutant compared with wild type (P = 0.008), and decreased further in the mus81Δ rad1Δ (P = 0.04) and mus81Δ rad1Δ yen1Δ (P = 0.0008) mutants compared with the rad1Δ single mutant (Figure 1b). A previous study identified a role for Rad1–Rad10 in removal of heterologous sequences at the DSB ends during strand invasion36; however, the ends of the plasmid are homologous to the chromosomal donor in this assay indicating that the rad1Δ defect is at a later step of repair.
Figure 1. Rad1 is required for plasmid integration.
a, Noncrossover (NCO) repair of the dsDNA gap within the MET17 gene yields an episomal plasmid whereas repair associated with a crossover (CO) integrates the plasmid. b, Gap repair frequencies; WT refers to wild type, error bars show s.e.m. and significance was determined by the unpaired t test (N = 13 for WT, N = 5 for the mus81Δ rad1Δ yen1Δ mutant, N = 4 for mus81Δ, yen1Δ, mus81Δ rad1Δ, mus81Δ yen1Δ and rad1Δ yen1Δ mutants, and N = 3 for the rad1Δ mutant). The frequency of gap repair with or without integration was determined for 108 independent transformants from each trial.
Independent Ura+ transformants were scored for mitotic stability to determine the frequency of gap repair with or without plasmid integration (Figure 1b). In wild type, 44% of the Ura+ transformants were due to integration of the plasmid at the met17 locus. No crossover defect was found for the mus81Δ or yen1Δ single mutants, but crossovers were reduced in the rad1Δ mutant (P = 0.0018), consistent with previous studies33,37,38. Surprisingly, 75% of the Ura+ transformants recovered from the mus81Δ yen1Δ double mutant were due to plasmid integration, resulting in a significant decrease in the noncrossover class (P = 0.0001). This contrasts with our previous study using chromosome homologs demonstrating increased noncrossovers and decreased crossovers in the mus81Δ yen1Δ double mutant23. The mus81Δ rad1Δ yen1Δ mutant showed a lower frequency of integration than the rad1Δ single mutant (P = 0.008) indicating that Mus81–Mms4 and Yen1 contribute to plasmid integration in the absence of Rad1 and that the increased plasmid integration in the mus81Δ yen1Δ mutant is due to Rad1 activity (P = 0.0015). Southern blot analysis confirmed that the events scored genetically as crossovers from the mus81Δ yen1Δ and mus81Δ rad1Δ yen1Δ mutants were due to plasmid integration at the met17 locus (Supplementary Figure 1). In addition, the noncrossover class was reduced in the triple mutant compared with the rad1Δ single mutant (P = 0.016), but was not different to the mus81Δ yen1Δ double mutant. As discussed below, the decrease in formation of noncrossover products observed for the mus81Δ yen1Δ mutant suggests resolution of recombination intermediates contributes to the noncrossover class.
Crossovers between repeats require Mus81, Rad1 and Yen1
To assess the roles of Rad1, Mus81 and Yen1 in DSB-induced recombination between dispersed chromosomal repeats we used an assay that allows recovery of both noncrossover and crossover (translocation) products39. A previous study reported that DSB-induced translocations were significantly reduced in the rad1Δ mutant, but noncrossovers could not be detected by the system used40. The haploid strains have a 39 bp HO endonuclease cut site inserted within the native URA3 locus on Chromosome V (Chr. V) and a 5.6 kb ura3 fragment integrated at the LYS2 locus on Chr. II (Figure 2a). The donor ura3 allele includes a 39 bp insertion of the HOcs-inc (non-cleavable) site with a BamHI restriction site 6 bp from the non-cleavable HO recognition sequence to monitor noncrossover repair. Because the donor allele shares extensive homology to the cut locus (3 bp substitutions to create the BamHI site and 1 bp on the other side of the break due to the inc mutation) there should be no requirement for Rad1–Rad10 flap cleavage during strand invasion41. After induction of HO, regulated by the galactose-inducible GAL1 promoter, the DSB is repaired by gene conversion transferring the HOcs-inc allele and BamHI site to the recipient locus.
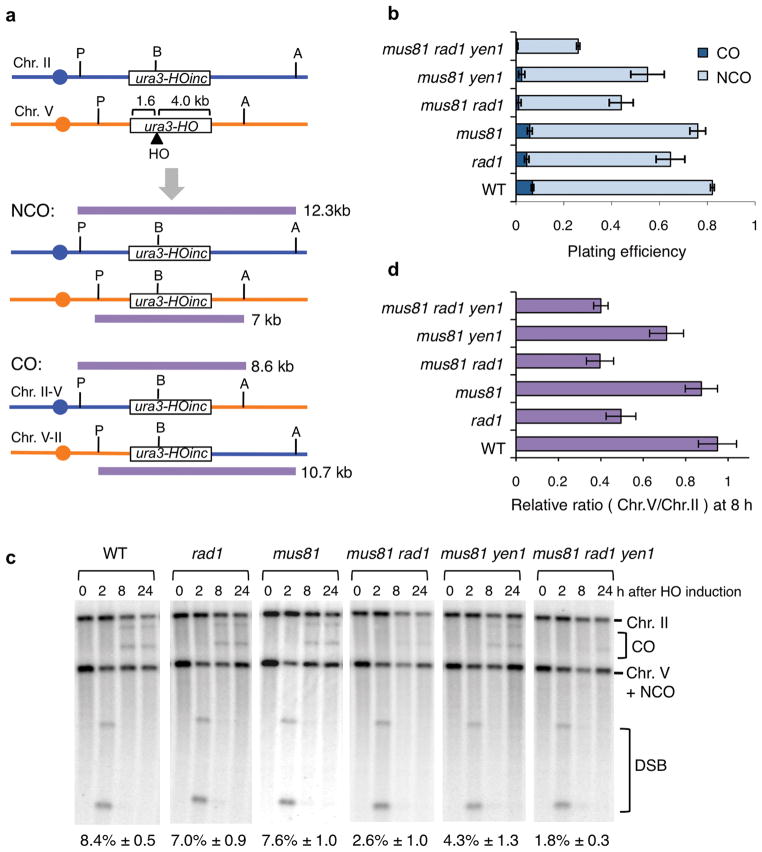
Figure 2. Rad1 and Mus81 cooperate to generate crossovers between dispersed repeats.
a, Schematic representation of the Chromosome II (Chr. II) – Chromosome V (Chr. V) ectopic recombination assay showing the distance between the HO endonuclease cut site (HO) and the heterology. Crossover repair is detected using ApaLI (A) and PvuII (P) sites outside of the region of shared homology, the sizes of parental/noncrossover and crossover fragments are indicated; the BamHI (B) site is used to monitor noncrossover repair. b, Repair efficiency was determined using at least three colonies for each strain. Errors bars show standard deviations (s.d.) and significance was determined by the t test. The distribution of noncrossover (NCO) and crossover (CO) recombinants was derived from Southern blot analysis 24 h after HO induction. c, Southern blot analysis of ApaLI-PvuII digested genomic DNA from the indicated strains. The crossover products of 10.7 and 8.6 kb, and DSB fragments of 4.5 and 2.5 kb are indicated, noncrossover repair restores the 7 kb Chr. V fragment. The mean values are from at least three independent trials for each strain with s.d. d, To quantify the noncrossover product the ratio of the Chr. V and Chr. II bands was determined at 8 h and normalized to the ratio at 0 h; the error bars show s.d.
As an overall evaluation of repair, the plating efficiency of each strain on galactose-containing medium (HO constitutively expressed) was compared with the plating efficiency on medium containing glucose (HO off) (Figure 2b). The rad1Δ mutant showed reduced plating efficiency on galactose-containing medium compared with wild type (P = 0.0075), and the plating efficiencies of the mus81Δ rad1Δ and mus81Δ rad1Δ yen1Δ mutants were lower than the rad1Δ single mutant (P = 0.015 and 0.0004, respectively). The yen1Δ and rad1Δ yen1Δ strains were not different to wild type and rad1Δ and were not analyzed further (Supplementary Figure 2b).
To determine the fraction of repaired products with an associated crossover HO was induced in liquid cultures and DNA isolated at different times after HO induction for ApaLI and PvuII restriction digestion followed by Southern blot hybridization to detect fragments diagnostic of crossover products (Figure 2a, c). In wild type, crossover products accumulated to 8.4% of the DNA products 24 h after HO induction. The mus81Δ single mutant displayed similar levels of crossover bands as the wild-type strain and crossovers were reduced by two-fold in the mus81Δ yen1Δ double mutant, as reported previously27. The rad1Δ single mutant showed a small decrease in the percent crossover bands compared with wild type (P = 0.011), and crossover products were reduced further in the mus81Δ rad1Δ and mus81Δ rad1Δ yen1Δ mutants compared with rad1Δ (P < 0.001 for both mutants). These data suggest Rad1–Rad10 and Mus81–Mms4 have partially redundant functions or cooperate to form crossovers between dispersed repeats.
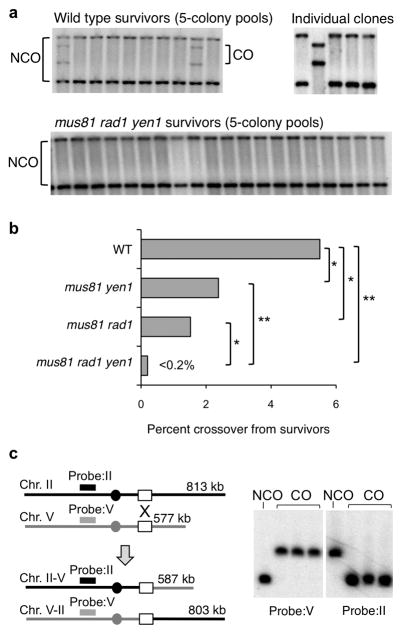
Faint bands corresponding to the sizes of crossover products were detected in the cell population of the mus81Δ rad1Δ yen1Δ mutant (Figure 2c). To determine whether these were bona fide reciprocal exchange products, cells of wild type, mus81Δ rad1Δ, mus81Δ yen1Δ and the triple mutant were plated on glucose-containing medium following an 8 h liquid induction of HO and DNA isolated from pools of five independent colonies was analyzed by restriction digestion and Southern blot hybridization to detect crossovers. For pools with crossover fragments, individual clones were analyzed to verify noncrossover or reciprocal crossover products. Eighteen of 324 colonies recovered from wild type, corrected for the 92% that were recombinants (transfer of the BamHI site to the cut locus), showed crossover bands (Figure 3a, b). To ensure that the crossover DNA fragments represent chromosome translocations, one noncrossover and three independently derived crossover recombinants from wild type were subjected to pulsed-field gel electrophoresis to separate intact chromosomes followed by Southern blot hybridization using probes specific to Chr. II or Chr. V. The 587 kb Chr. II–V and 803 kb Chr. V–II reciprocal translocation products were found in the three crossover clones (Figure 3c).
Figure 3. Absence of crossovers from survivors of the mus81Δ rad1Δ yen1Δ mutant.
a, Genomic DNA from pools of 5 independent survivors was digested with ApaLI and PvuII and analyzed by Southern blot hybridization. Individual clones from pools that exhibited crossover products were analyzed to verify formation of reciprocal crossovers. b, Percent crossovers from independent survivors of the indicated strains. Significance was determined by the Fisher’s exact test; comparisons with a P value of <0.05 are shown by *, and P values of <0.001 are shown by **. c, Schematic showing the sizes of parental (and noncrossover) and translocation products formed by reciprocal exchange between the ura3 repeats (white boxes). Probes specific for Chr. II or V were designed to hybridize to sequences on the opposite chromosome arm. PFGE to separate intact chromosomes from one noncrossover and three independently derived crossover products from wild type. The left panel shows hybridization of the blot with the Chr. V probe and the right panel shows the Chr. II probe.
No crossover products were found among 496 recombinant survivors of the mus81Δ rad1Δ yen1Δ triple mutant (P = 0.0001) (Figure 3a, b). The faint bands detected in populations could be due to break-induced replication (BIR) resulting in non-reciprocal LOH, a lethal event in haploid cells. Analysis of 263 colonies of the mus81Δ rad1Δ double mutant revealed three crossover clones, indicating a low level of Yen1-dependent cleavage to generate crossovers (P = 0.011 for mus81Δ rad1Δ vs. mus81Δ rad1Δ yen1Δ), even though the crossover bands were barely visible in the cell population (Figure 2c). Nine of 378 colonies analyzed from the mus81Δ yen1Δ double mutant exhibited crossovers, significantly higher than the triple mutant (P = 0.0005) and lower than wild type (P = 0.032). Thus, the same trends were found for the mutants when comparing crossover products detected in populations and crossovers among surviving colonies (Figures 2b and 3b).
Physical analysis of recombination products in the cell populations revealed a decrease in formation of the 7-kb noncrossover band in the rad1Δ derivatives at 8 h (Figure 2c, d). This does not appear to be due to a delay in HO cleavage or strand invasion because crossover products were present at 8 h and strand invasion intermediates detected by PCR were present at similar levels in the rad1Δ mutant and wild type (see below). The defect in formation of noncrossover products mirrors the reduced plating efficiency of the rad1Δ derivatives in response to the HO-induced DSB suggesting most of the lethality is due to loss of noncrossovers (Figure 2b, d). To confirm a defect in formation of noncrossover products in the absence of Rad1, we measured the plating efficiency of strains containing a 1.2 kb ura3-HO-inc donor on galactose-containing medium compared with glucose-containing medium (Supplementary Figure 2b). The rad1Δ strain with the 1.2 kb donor showed reduced plating efficiency on galactose compared with wild type (P = 0.0005) and the plating efficiency of the triple mutant was reduced to 0.11 (P = 0.0001 compared to rad1Δ). Because >99% of the products recovered from strains with the 1.2 kb substrate are noncrossover42, the reduced viability of the rad1Δ mutants with the short substrate is consistent with loss of noncrossover recombinants. Using primers (P2 and P3) to monitor strand invasion intermediates (as well as rare crossover products and unprocessed flap intermediates) the rad1Δ derivatives exhibited no defect, indicating that the requirement for Rad1–Rad10 is subsequent to strand invasion and extension of the invading 3′ end by DNA synthesis (Supplementary Figure 2c).
Rad1-dependent formation of ectopic joint molecules
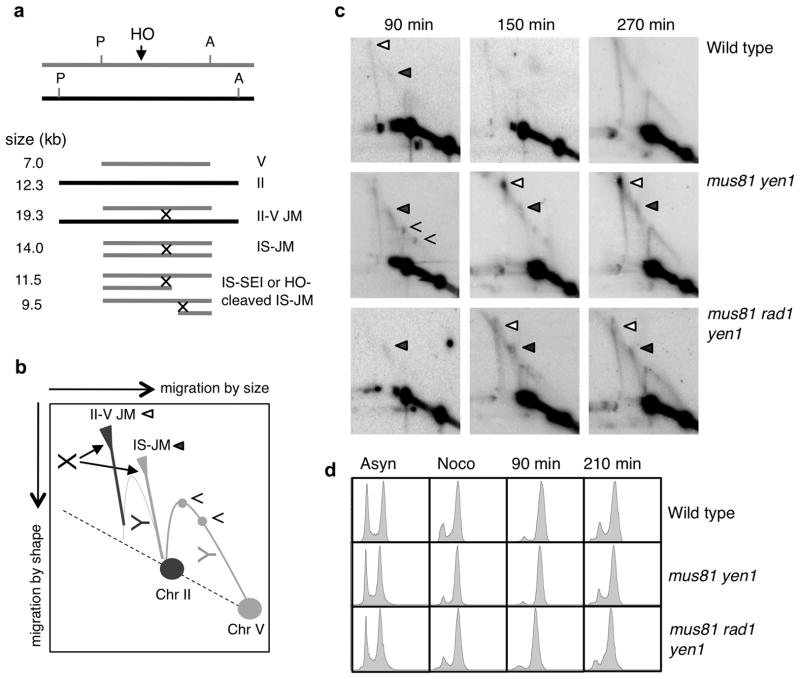
If Rad1–Rad10 functions redundantly with Mus81–Mms4 and Yen1 to resolve recombination intermediates we would predict a further increase in the accumulation of joint molecules in the triple mutant compared with the mus81Δ yen1Δ double mutant. Alternatively, if Rad1–Rad10 creates a joint molecule by cleaving the captured D-loop intermediate at the homology-heterology barrier, which is subsequently resolved by Mus81–Mms4 or Yen1, then the rad1Δ mutation should suppress the accumulation of ectopic joint molecules in the mus81Δ yen1Δ double mutant. To distinguish between these possibilities we used neutral two-dimensional (2D) gel electrophoresis to identify joint molecules during homology-dependent repair of the 5.6 kb repeat substrate (Figure 4). The analysis was performed with nocodazole-treated (G2-M arrested) cells to avoid replication intermediates, but at late times cells broke through the arrest and resumed cycling resulting in visible Y-shaped replication intermediates (Figure 4c and 4d).
Figure 4. Joint molecules accumulate in the mus81Δ yen1Δ double mutant.
a, Schematic showing the sizes of ApaLI-PvuII restriction fragments for the 5.6 kb ura3 repeats and expected inter-sister joint molecule (IS-JM), Chr. II–V joint molecule and branched molecules resulting from HO cleavage of the IS-JM or IS-SEI intermediates. Joint molecules could contain one or two Holliday junctions (one X shown for simplicity). b, Cartoon of 2D gel showing the migration of joint molecules and Y-shaped arcs; the open triangle marks the position of the II–V joint molecule, the filled triangle the position of the IS-JM and the open arrowheads the apparent IS-SEIs. c, Time course following HO induction for wild type, mus81Δ yen1Δ and mus81Δ rad1Δ yen1Δ strains. d, FACS profiles of the strains used for 2D gel analysis; asyn refers to the asynchronous population and noco to nocodazole-arrested cells.
Two distinct branched DNA species were observed that were identified as the Chr. II–V ectopic joint molecule (19.3 kb) and Chr. V-V inter-sister joint molecule (14 kb) (IS-JM) based on size and use of hybridization probes specific to each locus (Figure 4c and Supplementary Figure 3). Detection of the IS-JM indicates HO cleavage was asynchronous and one broken chromatid engaged with the uncut sister chromatid even though HO was continuously expressed during the time course. We presume both sisters are eventually cut forcing ectopic repair. Very faint spots corresponding to joint molecules were detected in the wild-type strain suggesting joint molecules are rapidly resolved. The IS-JM accumulated to similar levels in the mus81Δ yen1Δ and mus81Δ rad1Δ yen1Δ mutants indicating that its formation and resolution are unaffected by Rad1. In contrast, the Chr. II–V ectopic joint molecule accumulated to a much higher level (2.2% of total DNA) in the mus81Δ yen1Δ mutant compared with the mus81Δ rad1Δ yen1Δ triple mutant (0.1% of total DNA), supporting the hypothesis that Rad1–Rad10 cleaves the D-loop at the heterology barrier creating an intermediate that requires Mus81–Mms4 or Yen1 for resolution. The accumulation of joint molecules, coupled with the reduced DSB-induced viability of the mus81Δ yen1Δ mutant, suggests a substantial fraction of recombination intermediates remain unresolved causing mitotic catastrophe. A previous study reported the accumulation of IS-JMs and ensuing lethality in response to a replication-dependent (one-ended) DSB in the S. pombe mus81 mutant, consistent with an important role for Mus81–Mms4 in resolution of joint molecules in mitotic cells43.
We did not detect joint molecules corresponding to Y-shaped ectopic single-end invasion (SEI) intermediates. However, two discrete spots were observed on the Y-arc in mutants that accumulate the IS-JM that were of the sizes expected for inter-sister SEI intermediates (Figure 4c). Because we were unable to detect the ectopic SEI intermediate, even in strains that accumulated ectopic joint molecules, it is more likely that the 9.5 and 11.5 kb joint molecules are due to partial HO cleavage of the IS-JM followed by branch migration run-off of one of the junctions instead of SEI intermediates (Supplementary Figure 3d). This would suggest that after second end capture there is a delay in the DNA repair synthesis that is needed to form an intact dHJ intermediate, and it is possible that STR poorly dissolves such a structure. A previous study identified intact dHJ intermediates during repair of an I-SceI induced DSB and these persisted in the absence of Sgs1, suggesting STR is able to dissolve them9. Bzymek et al used cells synchronized at G1-S and they might be more proficient for repair synthesis than G2-M arrested cells9.
Rad1 is not required for LOH between chromosome homologs
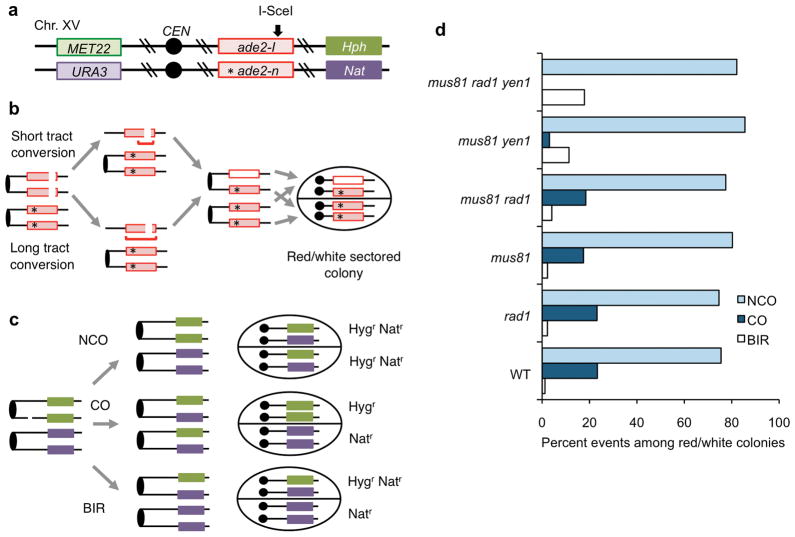
If Rad1 is only required for recombination events that involve processing of branched structures formed at heterology boundaries then the rad1Δ diploid should be proficient for crossovers between chromosome homologs. To test this, we used a previously described genetic assay to distinguish between noncrossover and crossover products induced by a site-specific DSB at the ade2 locus in diploid cells (Figure 5a)23. In this system an I-SceI induced DSB at the ade2-I allele is repaired from the homolog bearing a frame-shift mutation located 950 bp away from the ade2-I allele (ade2-n). The diploid has dominant drug resistance heterozygous markers 150 kb CEN distal to ade2 (Hph and Nat) and markers on the other chromosome arm (MET22 and met22::URA3) to distinguish LOH by mitotic recombination from chromosome loss. In these strains, I-SCEI is under the transcriptional control of the GAL1 promoter. Induction of I-SceI results in high frequency recombination to generate Ade+ (short tract gene conversion) or Ade−(long tract gene conversion) recombinants. Ade− events due to long tract gene conversion are distinguished from non-recombinants (cells in which I-SceI was not induced) by a re-induction assay23. Red/white sectored colonies result from G2 repair of one broken chromatid by short tract gene conversion and repair of the other broken chromatid by a long tract gene conversion event (Figure 5b). This class of colonies, representing 36% of the wild-type recombinants, is indicative of cells that were in G2 at the time the cells were plated and is the most informative to the mechanism of recombination as both products of the recombination event are recovered in the two halves of the colony. A crossover associated with repair of one of the broken chromatids is detected by reciprocal LOH of the Hph and Nat markers, whereas break-induced replication (BIR) results in non-reciprocal LOH (Figure 5c).
Figure 5. Rad1 is not required for crossovers between chromosome homologs.
a, Schematic of Chr. XV in the diploid strains used showing the I-SceI cut site and the location of the ade2-n mutation present on the other homolog (marked by *). b, Most recombination events result from I-SceI cutting both sister chromatids followed by repair from one or both non-sister chromatids. Short tract repair restores wild type ADE2, whereas long tract repair transfers the ade2-n allele to the cut locus. These events give rise to red/white-sectored colonies if the cells are plated prior to mitosis. c, Noncrossover (NCO) repair retains the markers in their original configuration; a crossover (CO) is detected by reciprocal LOH for the Nat and Hph markers. Break-induced replication (BIR) events are detected as sectored colonies in which one half retains heterozygosity for Nat and Hph, and the other half exhibits LOH for Hph or Nat. d, Distribution of NCO, CO and BIR products for red/white-sectored colonies.
I-SceI was induced in liquid culture for 1–3 h, glucose was added to the cells and appropriate dilutions were plated onto medium with glucose to repress expression of the nuclease. There was no increase in DSB-induced chromosome loss in the rad1Δ mutant indicating proficient homology-dependent repair, and the distribution of recombinants among the red/white-sectored colonies was the same as wild type (Figure 5d). In contrast to the ectopic system, the rad1Δ mutation did not decrease crossovers by itself (P = 0.96) or in the mus81Δ background (P = 0.66). Notably, the percent crossovers between homologs was reduced in the mus81Δ mutant compared with wild type (P = 0.003) and rad1Δ (P = 0.004), whereas crossovers between repeats were unaffected by the mus81Δ mutation (Figure 2). Crossovers between homologs were greatly reduced in the mus81Δ yen1Δ mutant and were lower than the mus81Δ rad1Δ mutant (P = 0.0001). The mus81Δ rad1Δ yen1Δ triple mutant exhibited poor vegetative growth and only a low yield of red/white-sectored colonies was recovered from several independent trials (Supplementary Figure 4). Although no crossovers were recovered from the triple mutant, this was not significantly different from the low number recovered from the mus81Δ yen1Δ double mutant (P = 0.09) (Supplementary Table 2). We cannot exclude the possibility that Rad1 plays a minor role in the formation of crossovers between chromosome homologs, but it is more important for recombination between dispersed repeats.
DISCUSSION
We propose the following model for Rad1–Rad10 function in DSB-induced recombination between ectopic sequences (Figure 6). After Rad51-dependent strand invasion to generate a D-loop, the 3′ end is extended by DNA synthesis. If the DNA synthesis tract were short then following displacement of the invading strand the nascent ssDNA would be able to pair with the other end of the break to complete repair by fill-in DNA synthesis and ligation. If the nascent strand were extended beyond the shared homology, then an unpaired flap would be generated after annealing of the displaced strand to the other side of the break. We suggest Rad1-Rad10 would cleave the unpaired flap to allow completion of repair to form noncrossover products, analogous to its role in the single-strand annealing mechanism to repair DSBs between repeats31. A role for Rad1–Rad10 in unpaired flap removal was previously suggested to explain the defect in gene conversion between non-tandem direct repeats in Ercc1−/− cells44. Unpaired flaps would be more likely to form between sequences with short homology and this could explain the reduced repair efficiency of the rad1Δ derivatives with the 1.2 kb substrate compared with the 5.6 kb substrate (Supplementary Figure 2).
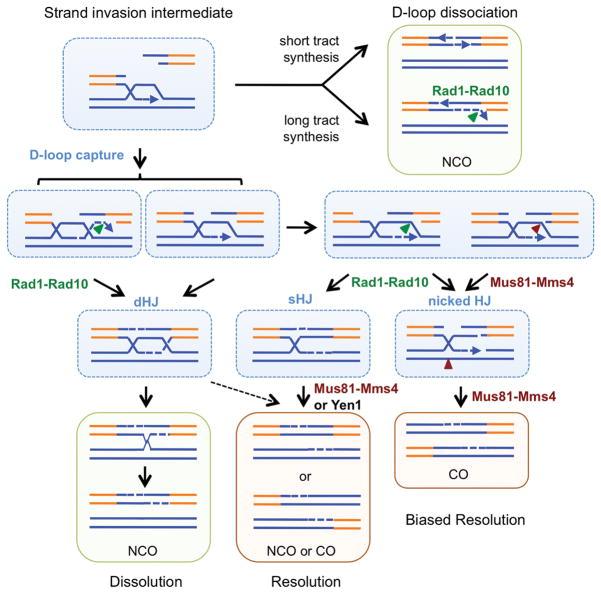
Figure 6. Model for the role of Rad1-Rad10 cleavage in formation of recombinants between dispersed repeats.
After Rad51-dependent strand invasion to generate a displacement loop (D-loop), the 3′ end is extended by DNA synthesis. Rad1–Rad10 is required to cleave an unpaired flap after strand displacement and annealing due to replication beyond the regions of homology. Capture of the D-loop followed by gap filling and ligation results in formation of a dHJ, which is preferentially removed by STR-mediated dissolution to form NCO products. Mus81–Mms4 could cleave the captured D-loop prior to ligation, or Rad1–Rad10 if resection had proceeded beyond the heterology boundary creating a single-stranded region adjacent to the branch point. Cleavage of the D-loop coupled with Mus81–Mms4 cutting of the nicked HJ would result in a CO product. Alternatively, clipping of the D-loop followed by gap filling and ligation would generate an sHJ intermediate that could not be dissolved by STR and would require Mus81–Mms4 or Yen1 for resolution. Intermediates are shown in blue boxes, NCO products in green boxes and CO products in oranges boxes.
Capture of the D-loop by the ssDNA formed at the other break end followed by gap filling and ligation would result in formation of a dHJ and subsequent dissolution by STR to generate noncrossover products (Figure 6). The dHJ could also be cleaved by endonucleases to form crossover or noncrossover products, but recent studies indicate this mechanism is inefficient in mitotic cells17. Mus81–Mms4 could directly cleave the captured D-loop prior to ligation or Rad1–Rad10 could cut if resection had proceeded beyond the heterology boundary creating a structure with a single-stranded region adjacent to the branch point19,45. In vitro studies have shown that Mus81–Mms4 preferentially cleaves DNA structures with an ssDNA gap of <4 nt adjacent to the branch point, whereas Rad1-Rad10 cut structures with more extensive ssDNA, consistent with the model proposed (Supplementary Figure 5)46,47. Furthermore, the reduced frequency of ectopic crossovers in the mus81Δ rad1Δ mutant is consistent with the nucleases acting on similar substrates. Cleavage of the D-loop coupled with Mus81–Mms4 cutting of the nicked Holliday junction would result in a crossover product. Alternatively, clipping of the D-loop followed by gap filling and ligation would generate a single Holliday junction (sHJ) intermediate that could not be dissolved by STR and would require Mus81-Mms4 or Yen1 for resolution. As suggested previously, resolution of a sHJ intermediate would explain the pattern of hDNA observed in crossover products from plasmid-chromosome recombination48. Rad1-dependent ectopic joint molecules accumulated in the absence of Mus81 and Yen1 supporting the hypothesis that Rad1–Rad10 cleaves the D-loop intermediate converting it to a sHJ intermediate (Figure 4). Thus, in the absence of Mus81, Rad1 and Yen1, second-end capture intermediates can only be channeled through the dHJ pathway and dissolved by STR yielding exclusively noncrossover products.
The triple mutant showed low viability after DSB induction, and delayed colony formation on galactose-containing medium compared with the other mutants, suggesting persistent poorly repaired structures. These could be unresolved second-end capture intermediates, or unpaired flaps due to replication beyond the shared homology that persist after D-loop dissociation, second end capture or STR mediated dissolution. If not removed by nucleases the ssDNA flaps could be bound by Rad51 and engage in secondary recombination events, such as BIR, a lethal event in haploids. Unpaired flaps might also be generated between homologous sequences if the tract of DNA repair synthesis exceeded the length of the resection tract. In this case, Mus81-Mms4 might remove the 3′ flap, or Yen1 if the structure converted to a 5′ flap by branch migration.
To explain the high frequency plasmid integration observed in the mus81Δ yen1Δ double mutant we suggest a sHJ intermediate covalently linking the plasmid to Chr. XII persists and is not recognized as aberrant during mitosis because the plasmid lacks a centromere, and is resolved by replication through the Holliday junction in the next cell cycle, resulting in episomal and integrated plasmid products (Supplementary Figure 6). Replication of a dHJ intermediate would not result in plasmid integration. This mechanism might explain the higher percent of crossovers observed during plasmid gap repair compared with ectopic recombination between chromosomal repeats35,38,42,49,50. Resolution by replication could also explain why only a minor defect was observed for the mus81Δ yen1Δ mutant when cells were transformed with a plasmid containing a sHJ51. The high frequency of crossovers associated with plasmid gap repair, and dependence on Rad1, might also be explained if there was less of a constraint on second end capture.
Surprisingly, ectopic crossover products were recovered from the mus81Δ yen1Δ double mutant even though unresolved joint molecules persist in this strain (Figures 3 and 4). The covalently linked non-sister chromatids would be expected to co-segregate to the same daughter cell in 50% of mitoses and the sHJ connecting them might be replicated in the next cell cycle resulting in products indistinguishable from crossovers (as suggested for plasmid integration in the absence of Mus81 and Yen1). Replication has also been proposed to explain the resolution of Holliday junction-containing chromosomal DNA in E. coli ruv mutants52. Breakage of the joint molecule during mitosis and repair of fragments by break-induced replication in the next cell cycle would lead to inviable products and might contribute to the reduced viability of the mus81Δ yen1Δ mutant following DSB induction23. Alternatively, it is conceivable that after cleavage of the D-loop by Rad1–Rad10 the branch point migrates back to the other heterology boundary and Rad1–Rad10 cuts again yielding a crossover (Supplementary Figure 6).
In summary, we have identified a novel function for the Rad1–Rad10 nuclease in promoting DSB-induced crossovers between dispersed repeats by converting a strand invasion intermediate to an intermediate that can only be resolved by Mus81–Mms4 or Yen1. The function of XPF–ERCC1 in heterologous flap trimming and gene targeting is conserved in mammals suggesting the role in crossover formation is also likely to be conserved34,44,53. Mammals have a much larger repertoire of repeated sequences than budding yeast raising the possibility that XPF–ERCC1 plays a significant role in the formation of gross chromosomal rearrangements leading to genomic disorders in higher eukaryotes54. Given the potential of structure-selective nucleases to generate detrimental crossovers in mitotic cells it is important for their activities to be highly regulated during the cell cycle and to be used as a last resort when less mutagenic options are available55.
METHODS
Yeast strains
Saccharomyces cerevisiae strains used in this study are listed in Supplementary Table 1. All strains are in W303 background and were generated by crossing strains with different recombination reporters to strains with mus81::KanMX6, rad1::LEU2 or yen1::HIS3 alleles23,35,56.
Plasmid DNA gap repair assay
The gap repair plasmid, pSB110, and transformation protocol were described previously35. The frequency of gap repair is the number of Ura+ transformants per microgram of linearized DNA transformed divided by the number of Ura+ transformants per microgram of circular plasmid DNA transformed. To test for mitotic stability, all of the Ura+ transformants were picked from one region of a plate and transferred to water-filled 96-microtiter plate wells, and spotted onto synthetic complete medium lacking uracil (SC-Ura). The cells were grown for 3 days then replica plated to rich medium (1% yeast extract, 2% peptone, 2% glucose; YPD) and grown for 2 more days to allow loss of the plasmid. The cells were then replica plated to medium containing 5-fluoroorotic acid (5-FOA). Confluent growth on 5-FOA indicated that the Ura+ phenotype was mitotically unstable due to plasmid repair without integration.
DSB-induced ectopic recombination assay
Plating efficiency, Southern blot analysis, and PCR methods to analyze recombination intermediates were as described previously, except cells were pre-grown with lactate as a carbon source prior to galactose induction27,56. Cells were plated at the 8 h time point following HO induction for analysis of survivors. 1 ml cultures of 5 independent survivors were pooled for DNA extraction and Southern blot analysis. If crossover products were detected in a pool, individual clones form the pool were analyzed to distinguish between crossover and noncrossover recombinants. Independent noncrossover and crossover recombinants clones were analyzed by pulsed-field gel electrophoresis (Bio-Rad CHEF II system) followed by Southern blot hybridization using probes specific for Chr. II or Chr. V. Between twenty and forty independent colonies obtained after 8 h HO induction were also analyzed by PCR and BamHI digestion to calculate the number of recombinants for each of the strains used in Figure 3. PCR using primers P2 and P3 was used to detect strand invasion intermediates, CO products and NCO products with an unrepaired heterologous flap. 50 ng template of the indicated strains isolated from cells at different times after HO induction was amplified for 27 cycles.
Neutral 2D gel electrophoresis
Samples for neutral 2D-gel electrophoresis were obtained from 30ml aliquots of cultures arrested with nocodazole (15 ug/ml) and induced with 2% galactose. Cells were embedded in agarose and lysed as described previously57. Agarose plugs were melted and treated with β-agarase after digestion with ApaLI-PvuII and the genomic DNA was precipitated, three plugs were combined to load each lane of the first dimension gel. Conditions for 2D gels were as described57. Filters were hybridized with URA3, Chr. II specific or Chr. V specific probes.
DSB-induced ade2 recombination assay
I-SceI induction and analysis of recombinant colonies were as described previously23. If both broken chromatids are repaired by short tract gene conversion a solid white colony is produced, two long tract gene conversions produce a solid red colony and red/white sectored colonies are due to one short and one long tract conversion. Because the sectored colonies are diagnostic of two independent repair events a colony that retained heterozygosity for Nat and Hph was scored as two noncrossover events, similarly a sectored colony with reciprocal LOH was scored as one noncrossover and one crossover event. In a previous study each sectored colony was scored as only one event and this resulted in a larger number of crossovers than scored with the current method23. Crossover events are only detected if the two recombinant chromatids segregate to opposite poles at mitosis3, thus the numbers presented derive from doubling the number of crossover events and subtracting an equivalent number from the noncrossover class (Figure 5d). Statistical significance for the distribution of recombination events between given strains was calculated by Fisher’s exact test (Supplementary Table 2). Independent inductions were performed at least three times for each strain.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R01GM041784 and R01GM094386 to LSS, and from the Israeli Ministry of Science and Technology and the Israel Science Foundation to MK.
Footnotes
AUTHOR CONTRIBUTIONS
GM, CKH, MK and LSS designed experiments and LSS wrote the paper; experiments in Figure 1 and Supplementary Figure 1 were carried out by AFL and CKH, in Figures 2, 3, 5 and Supplementary Figure 2 by GM and AFL, and in Figure 4 and Supplementary Figure 3 by GM.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 2.Barbera MA, Petes TD. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2006;103:12819–12824. doi: 10.1073/pnas.0605778103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chua P, Jinks-Robertson S. Segregation of recombinant chromatids following mitotic crossing over in yeast. Genetics. 1991;129:359–369. doi: 10.1093/genetics/129.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagstrom SA, Dryja TP. Mitotic recombination map of 13cen-13q14 derived from an investigation of loss of heterozygosity in retinoblastomas. Proc Natl Acad Sci U S A. 1999;96:2952–2957. doi: 10.1073/pnas.96.6.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair (Amst) 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009;34:264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 9.Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins I, Newlon CS. Meiosis-specific formation of joint DNA molecules containing sequences from homologous chromosomes. Cell. 1994;76:65–75. doi: 10.1016/0092-8674(94)90173-2. [DOI] [PubMed] [Google Scholar]
- 11.Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 12.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson DO, Holloman WK. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci U S A. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P- element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010:44. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 17.Dayani Y, Simchen G, Lichten M. Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS Genet. 2011;7:e1002083. doi: 10.1371/journal.pgen.1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanco MG, Matos J, Rass U, Ip SC, West SC. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst) 2010;9:394–402. doi: 10.1016/j.dnarep.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 21.Munoz-Galvan S, et al. Distinct roles of Mus81, Yen1, Slx1-Slx4, and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol Cell Biol. 2012;32:1592–1603. doi: 10.1128/MCB.00111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boddy MN, et al. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 23.Ho CK, Mazon G, Lam AF, Symington LS. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell. 2010;40:988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashton TM, Mankouri HW, Heidenblut A, McHugh PJ, Hickson ID. Pathways for Holliday junction processing during homologous recombination in Saccharomyces cerevisiae. Mol Cell Biol. 2011;31:1921–1933. doi: 10.1128/MCB.01130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Muyt A, et al. BLM Helicase Ortholog Sgs1 Is a Central Regulator of Meiotic Recombination Intermediate Metabolism. Mol Cell. 2012;46:43–53. doi: 10.1016/j.molcel.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agmon N, Yovel M, Harari Y, Liefshitz B, Kupiec M. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 2011;39:7009–7019. doi: 10.1093/nar/gkr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fagbemi AF, Orelli B, Scharer OD. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst) 2011;10:722–729. doi: 10.1016/j.dnarep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekelsky JJ, McKim KS, Chin GM, Hawley RS. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics. 1995;141:619–627. doi: 10.1093/genetics/141.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dowling EL, Maloney DH, Fogel S. Meiotic recombination and sporulation in repair-deficient strains of yeast. Genetics. 1985;109:283–302. doi: 10.1093/genetics/109.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyndaker AM, Alani E. A tale of tails: insights into the coordination of 3′ end processing during homologous recombination. Bioessays. 2009;31:315–321. doi: 10.1002/bies.200800195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langston LD, Symington LS. Opposing roles for DNA structure-specific proteins Rad1, Msh2, Msh3, and Sgs1 in yeast gene targeting. EMBO J. 2005;24:2214–2223. doi: 10.1038/sj.emboj.7600698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiestl RH, Prakash S. RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol Cell Biol. 1988;8:3619–3626. doi: 10.1128/mcb.8.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niedernhofer LJ, et al. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 2001;20:6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartsch S, Kang LE, Symington LS. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol Cell Biol. 2000;20:1194–1205. doi: 10.1128/mcb.20.4.1194-1205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fishman-Lobell J, Haber JE. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 37.Symington LS, Kang LE, Moreau S. Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in nuclease-deficient strains of Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:4649–4656. doi: 10.1093/nar/28.23.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welz-Voegele C, Jinks-Robertson S. Sequence divergence impedes crossover more than noncrossover events during mitotic gap repair in yeast. Genetics. 2008;179:1251–1262. doi: 10.1534/genetics.108.090233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aylon Y, Kupiec M. The checkpoint protein Rad24 of Saccharomyces cerevisiae is involved in processing double-strand break ends and in recombination partner choice. Mol Cell Biol. 2003;23:6585–6596. doi: 10.1128/MCB.23.18.6585-6596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholson A, Fabbri RM, Reeves JW, Crouse GF. The effects of mismatch repair and RAD1 genes on interchromosomal crossover recombination in Saccharomyces cerevisiae. Genetics. 2006;173:647–659. doi: 10.1534/genetics.105.055244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inbar O, Liefshitz B, Bitan G, Kupiec M. The relationship between homology length and crossing over during the repair of a broken chromosome. J Biol Chem. 2000;275:30833–30838. doi: 10.1074/jbc.C000133200. [DOI] [PubMed] [Google Scholar]
- 43.Roseaulin L, et al. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 46.Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol Cell Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ehmsen KT, Heyer WD. A junction branch point adjacent to a DNA backbone nick directs substrate cleavage by Saccharomyces cerevisiae Mus81-Mms4. Nucleic Acids Res. 2009;37:2026–2036. doi: 10.1093/nar/gkp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchel K, Zhang H, Welz-Voegele C, Jinks-Robertson S. Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol Cell. 2010;38:211–222. doi: 10.1016/j.molcel.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orr-Weaver TL, Szostak JW. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci U S A. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tay YD, Wu L. Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J Biol Chem. 2010;285:11427–11432. doi: 10.1074/jbc.M110.108399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wardrope L, Okely E, Leach D. Resolution of joint molecules by RuvABC and RecG following cleavage of the Escherichia coli chromosome by EcoKI. PLoS One. 2009;4:e6542. doi: 10.1371/journal.pone.0006542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sargent RG, et al. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc Natl Acad Sci U S A. 1997;94:13122–13127. doi: 10.1073/pnas.94.24.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matos J, Blanco MG, Maslen S, Skehel JM, West SC. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aylon Y, Liefshitz B, Bitan-Banin G, Kupiec M. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:1403–1417. doi: 10.1128/MCB.23.4.1403-1417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hyppa RW, Smith GR. Using Schizosaccharomyces pombe meiosis to analyze DNA recombination intermediates. Methods Mol Biol. 2009;557:235–252. doi: 10.1007/978-1-59745-527-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.