Abstract
The transcription factor p53 regulates genes associated with a wide range of functions, including the Toll-like receptor (TLR) set of innate immunity genes, suggesting that p53 also modulates the human immune response. The TLR family comprises membrane glycoproteins that recognize pathogen-associated molecular patterns (PAMPs) and mediate innate immune responses, and TLR agonists are being used as adjuvants in cancer treatments. Here, we demonstrate that doxorubicin, 5-fluorouracil, and UV and ionizing radiation elicit changes in TLR expression that are cell line- and damage-specific. Specifically, treatment-induced expression changes led to increased downstream cytokine expression in response to ligand stimulation. The effect of DNA stressors on TLR expression was mainly mediated by p53, and several p53 cancer-associated mutants dramatically altered the pattern of TLR gene expression. In all cell lines tested, TLR3 induction was p53-dependent, while induction of TLR9, the most stress-responsive family member, was less dependent on status of p53. In addition, each of the 10 members of the innate immune TLR gene family tested was differentially inducible. Our findings therefore demonstrate that the matrix of p53 status, chromosome stress, and responsiveness of individual TLRs should be considered in TLR-based cancer therapies.
INTRODUCTION
The p53 tumor suppressor is a master regulatory transcriptional factor activated in response to several stress signals including DNA damage, resulting in transactivation of target genes. It functions in a variety of biological processes including cell cycle arrest, apoptosis, senescence, embryo implantation and nutritional stress ((1)). p53 binds in vitro as a tetramer to a DNA motif composed of two RRRCWWGYYY decamer half-sites (R=A, G; W=A,T; Y=C, T) separated by a spacer of up to 14 bases (2). Using a combination of yeast and human cell systems (3) we established “rules” whereby p53 was also found to function from noncanonical sequences comprising only a decamer 1/2-site or a 3/4-site (4–6). Employing these rules for p53 binding and transactivation we recently identified p53 binding sites in the promoter region of most human Toll-like receptors (TLRs) and demonstrated a direct role for p53 in regulation of this entire family of genes in primary human immune cells (7).
Toll-like receptors (TLRs) are a group of highly conserved integral membrane glycoproteins that recognize a variety of distinct pathogen-associated molecular patterns (PAMPs) (8). The recognition of PAMPs by TLR expressing cells leads to quick and acute responses required to eliminate pathogens in the host. Upon ligand stimulation, TLRs recruit adaptor molecules MyD88, TIRAP, TRIF and TRAM leading to activation of NF-kappaB, interferon responsive factors and MAP kinases that, in turn, results in ligand-dependent patterns of gene expression including inflammatory cytokines, chemokines and interferons (reviewed in (8)). In addition, endogenously produced ligands, referred to as damage-associated molecular patterns (DAMPs) or alarmins that include self-DNA and intracellular proteins, signal tissue injury through TLRs when DAMPs are released from cells (9). In both cases TLR pathway activation mediates immune/inflammatory responses. Ten TLRs (TLR 1–10) have been identified in humans. TLRs are expressed in several types of immune cells including spleen, T and B lymphocytes, dendritic cells and macrophages (10). In addition, TLRs function in non-immune tissues such as airway and gut epithelial cells which have direct contact with pathogens (11, 12).
Altered TLR expression has been associated with autoimmune and chronic inflammatory diseases including atherosclerosis, type I diabetes, inflammatory bowel disease, liver diseases, rheumatoid arthritis, and systemic lupus erythematosus (13, 14). In recent years cancer-related functions of the human TLR gene family have become apparent. Impaired expression and signaling by TLR7 and TLR9 may contribute to reduced innate immune responses during chronic viral infections and oncogenesis (15). In addition, the loss-of-function allele Asp299Gly of TLR4 is associated with breast cancer (16).
For cancer cells expressing TLRs, activation of the pathway can negatively affect growth and viability (17) including direct killing of tumor cells via induction of TNFalpha (18, 19) and, therefore, can be employed in treatment. For instance, TLR7 agonist Imiquimod is used in treating basal cell and cutaneous malignancies; also, synthetic oligodeoxynucleotides containing unmethylated CpG motifs that activate TLR9 are in clinical trials (20, 21). Stimulation of the TLR pathway in antigen-presenting cells (APCs) can induce and potentiate an effective immune response against tumor cells (22–25). In addition, TLR ligands conjugated to tumor-derived proteins or immunogenic peptides are used for therapeutic vaccination to treat cancer (reviewed in (26)).
Surprisingly, despite the importance of TLRs in human health, disease and therapy, relatively little is known about their transcriptional regulation. Only a few studies suggest involvement of transcriptional factors such as PU.1, AP-1, interferon response factors, and Forkhead Box P3 in regulation of specific TLRs in defined cellular systems (27, 28).
Since we have shown in primary cells that activated p53 can regulate expression of the various TLRs differentially, depending in part on the agent, the p53 status could influence expression and function of TLRs in cancer cells (7). The TP53 gene is mutated in over half of human tumors (29, 30). Most mutations are missense and are found within the central core DNA binding domain of the protein resulting in loss of transcriptional function. However, approximately one-third of cancer-associated p53 mutants retain transactivation capability and exhibit change-in-spectrum of transactivated genes (4, 31). Therefore, differential effects on TLR gene expression might be expected from the various p53 mutations.
So far, only TLR3 was shown to be a p53 target (32). Based on our results with primary human cells where nearly all TLR genes are in the p53 network, we have investigated p53-dependent regulation of the entire TLR gene family in a panel of cancer cell lines and the consequences of p53 mutations on TLR expression. We focused on human cells because there are significant differences in regulation, expression and specificity of TLRs between humans and rodents (reviewed by (33)), especially as relates to p53 responsiveness (7). Here, we establish that most TLR genes are responsive to p53 and genotoxic stress in human cancer cells. The considerable variability in responses of specific TLR genes that is dependent on cell line, specific p53 mutations and stressors underscores the importance of considering these factors in TLR-based cancer treatments.
MATERIALS AND METHODS
Reagents and antibodies
All reagents were from Sigma (St. Louis, MO) unless stated otherwise. The primary antibodies used were against p53 (DO-1), TLR5 (H1-27), actin (C-11), β-tubulin (H-300) and lamin B1 (H-90) (Santa Cruz Biotechnology, Santa Cruz, CA), p21 (SXM30, BD Biosciences Pharmigen, San Diego, CA), TLR2 (#2229, Cell Signaling, Boston, MA).
Cell lines and treatments
The source and maintenance conditions of cell lines are described in Supplementary Materials and Methods. Where indicated, cells were treated for 24–48 h with the following: Nutlin-3 (10 µM), doxorubicin (0.3–0.6 µg/mL), 5-fluoruracil (300 µM). For ionizing radiation treatment and UV cells were irradiated at 1.56 Gy/min or UV at 1 J/m2/s, respectively, Pretreatment with the p53 inhibitor pifithrin-α (40 µM) was for 4 h. Doses of ionizing radiation, UV, doxorubicin and Nutlin-3 were chosen based on preliminary optimization as shown in Supplementary Fig. 1. None of the treatments resulted in noticeable cell death during the course of the experiments, based on visual observation.
Plasmid constructs, transfections, luciferase assays and chromatin immunoprecipitation assays, immunoblot and gene expression analysis by Real Time PCR were performed as previously described (7). For details see Supplementary Materials and Methods.
RESULTS
p53 modulates expression of TLR genes in breast adenocarcinoma and osteosarcoma cells
The impact of changes in p53 protein levels on TLR gene expression was examined in several cancer cell lines. Increases were accomplished using the p53 inducer Nutlin-3 or p53 overexpression. Nutlin-3 inhibits p53 interaction with its ubiqutin ligase MDM2 leading to p53 stabilization and accumulation (34). The p53 inhibitor pifithrin-α or RNA interference were employed to reduce p53-mediated activity. Although pifithrin-α can suppress p53-mediated transactivation, the underlying mechanism of p53 inhibition remains to be established (35).
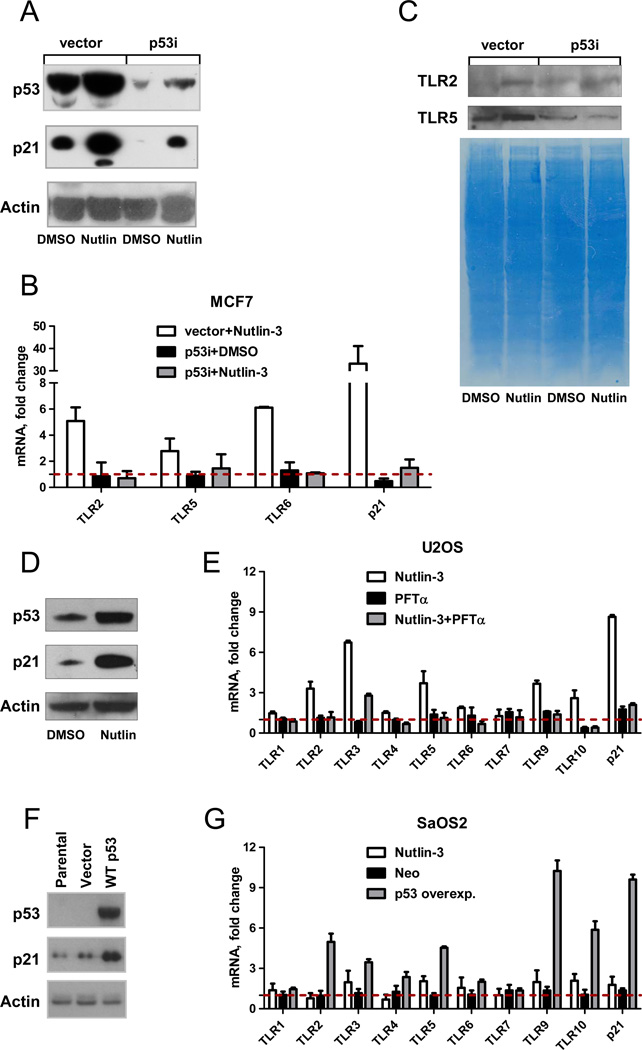
Shown in Fig. 1A are representative Western blots of p53 and p21 proteins in MCF7 breast adenocarcinoma cells (wild type p53) and an MCF7 cell line stably expressing shRNAi to p53 (designated as “vector” and “p53i”, respectively) treated with Nutlin-3 or vehicle control DMSO for 48 h. As expected, Nutlin-3 greatly increased expression of p53 and its target gene p21in MCF7-vector cells and only to a small extent in MCF7-p53i cells (less than the basal level of p53 in the MCF7-vector cells). Nutlin-induced changes in expression of TLR genes in MCF7-vector and MCF7-p53i cells are presented in Fig. 1B. Expression of only three of the ten TLR genes was detected in MCF7 cells: TLR2, 5 and 6 under our experimental conditions. The TLR genes TLR2, 5 and 6 were induced 3.5 to 6-fold by Nutlin-3 compared to DMSO-treated control in a p53-dependent manner in MCF7 cells. There was a corresponding increase in TLR2 and TLR5 proteins in the membrane fraction of the MCF7 vector cell lysate following Nutlin-3 treatment (a reliable human TLR6 antibody was not available) as described in Fig1C.
Figure 1. The p53 activator Nutlin-3 induces expression of TLR genes.
MCF7-vector or MCF7-p53i cells were incubated with 10 µM of Nutlin-3 or 0.1% DMSO for 48 h. Following treatment, cells were harvested and total RNA and protein were isolated. A. p53 and p21 activation were assessed by Western blot analysis. The exposure was extended long enough to allow visualization of and direct comparison between all samples. Actin was used as a loading control. B. Expression of TLRs 2, 5 6 and p21 following Nutlin-3 treatment was assessed using real time PCR and normalized to expression of the housekeeping gene GUSB. Expression of each gene in DMSO treated control cells was set as one. C. Cell lysates were subjected to subcellular fractionation and 100 µg of protein from each membrane fraction was resolved by SDS PAGE. Protein levels of TLR2 and TLR5 were detected by Western blot; gel staining was used for the loading control. D. p53 and p21 induction in U2OS cells following 24 h of incubation with Nutlin-3 was assessed by Western blot analysis. E. TLR gene expression in U2OS cells incubated with either Nutlin-3, pifithrin-α, a combination of both for 24 h relative to DMSO treated control was assessed by real time PCR. F. SaOS2 cells were transfected either with empty vector or with wild p53 or left untreated. Expression levels of p53 and p21 were examined by Western blot 24 h post-transfection. G. TLRs mRNA expression levels following this transfection or Nutlin-3 (10 µM, 24 h) treatment were examined by qPCR; p21 was used as a positive control. Gene expression in DMSO treated sample was set as one. Shown are averages of 3 independent experiments.
In contrast to MCF7 cells, all TLRs except TLR8 are expressed in the osteosarcoma cell line U2OS (p53+) and most TLRs were induced by Nutlin-increases in p53 (see p53 and p21 proteins in Fig. 1D). There was a 2 to 6.5-fold increase in expression of TLR2, 3, 5, 6, 9 and 10 genes by 24 h Nutlin-3 treatment compared to the DMSO treated control in U2OS cells (Fig. 1E). This induction was prevented by pretreatment with pifithrin-α, confirming a specific role for p53 in TLR gene expression.
To further investigate a direct connection between p53 and TLR gene expression, wild type p53 was overexpressed in p53-null osteosarcoma SaOS2 cells (Fig. 1F). The pattern of induction of TLR genes differed from both U2OS and MCF7 cells. At 24 h post-transfection, the levels of TLR2–6, 9 and 10 mRNAs were increased 2 to 10-fold (Fig. 1G) relative to untransfected cells. The TLR1 and 7 gene expression was not changed and TLR8 mRNA was not detected in SaOS2 cells.
Thus, we establish that many TLR genes in cancer cells are responsive to p53. However, there are dramatic differences in the spectra of TLRs transactivated suggesting that a profile of individual cancers needs to be considered if TLR-related therapies are to be used.
Functional evaluation of proposed p53 response elements (REs) in TLR promoters
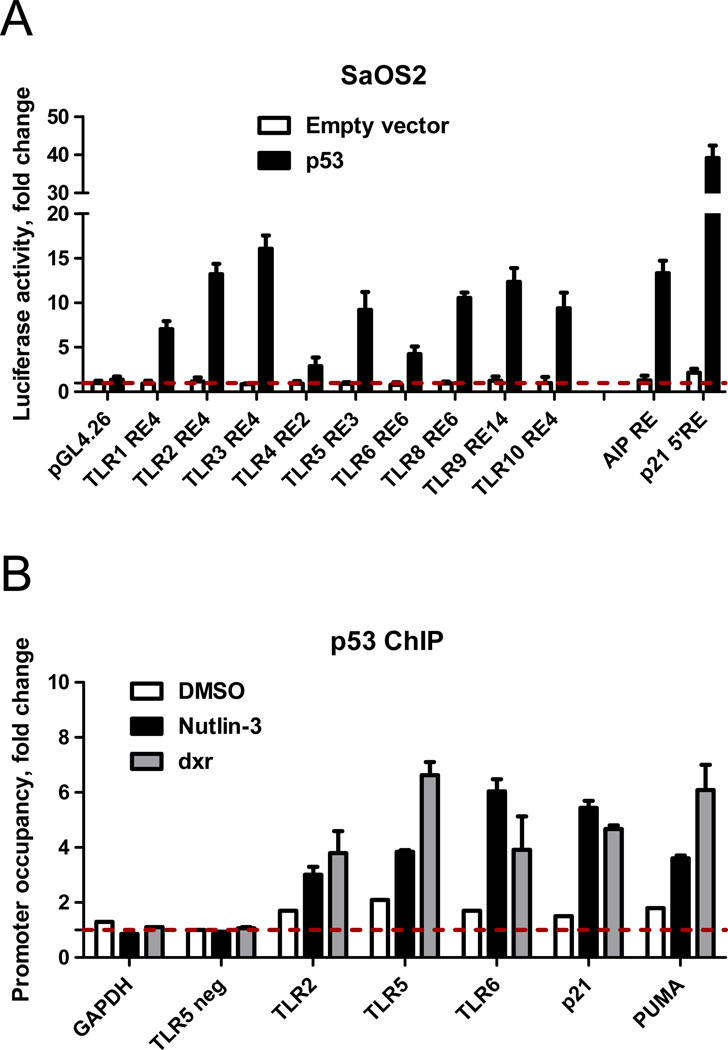
Based on rules for functional p53 binding we identified several canonical and noncanonical p53-REs located in the promoter vicinity of almost all human TLR genes (summarized in (7) and Supplementary Table 1). Luciferase reporter constructs containing selected REs from TLR promoter areas were transfected into SaOS2 cells in the presence (solid bars) or absence (open bars) of a vector expressing wild type p53 and reporter induction was assessed 48 h post-transfection. Consistent with our previous report, all proposed p53RE sequences from the TLRs could support p53-driven transcription of a luciferase reporter. For most the levels induced were similar to the moderately responsive p53 target response element of AIP which was used as an internal control (Fig. 2A).
Figure 2. Functional evaluation of proposed p53 response elements (REs) in TLR promoters.
A. SaOS2 cells were transfected with luciferase reporter constructs containing p53 RE sequences from different TLRs in the presence (solid bars) or absence (open bars) of the vector expressing wild type p53. At 48 h post-transfection, induction of the luciferase reporter was assessed. Relative luciferase activity was compared to the pGL4 plasmid lacking the p53 RE (empty vector) or constructs that contained REs of established p53 targets. Shown is an average of three independent experiments each performed in triplicate. B. MCF7 cells were treated with 0.3 µg/ml of doxorubicin (dxr) or 10 µM of Nutlin-3 for 24 h. Occupancy of p53 at promoters of TLRs 24 h later was assessed by ChIP assay and quantified by qPCR.
The ability of several of these p53-REs to function as p53 target sequences was investigated further using chromatin immunoprecipitation (ChIP) assays of MCF7 cells treated for 24 h with Nutlin-3 or doxorubicin (dxr), another well-established activator of p53. The p53 occupancy at the TLR promoters was assessed by ChIP analysis and quantified by SYBR Green qPCR, as described in Fig. 2B and Supplementary File 1. The PCR reaction specificity was confirmed using product dissociation curve analysis and by visualization of PCR product on ethidium bromide stained gels (Supplementary Fig.2). Binding to the GAPDH promoter or a region in the TLR5 promoter distant from the p53 RE provided a negative control. The binding of p53 to RE sequences in the promoter regions of the TLR2, TLR5 and TLR6 genes was enriched 3 to 6-fold at 24 h after Nutlin-3 or doxorubicin treatments compared to the control (untreated or DMSO-treated cells). This is similar to the binding enrichment at the well-established p53 REs in p21 or PUMA promoters. Thus, as found in primary cells, TLRs can be direct transcriptional targets of p53 in cancer cells.
Role of p53 in doxorubicin and 5-fluoruracil-induced up-regulation of TLRs in MCF7 and U2OS cells
Many clinical anticancer treatments rely on induction of DNA damage as well as p53activation. Since several TLR ligands have been employed as adjuvant treatments in combination with anti-cancer agents (36), profiling TLR gene expression in response to p53 in different cancer cell lines is expected to be relevant to therapeutic treatments. We, therefore, determined whether DNA damage induced by the commonly used anti-cancer agents doxorubicin, 5-fluoruracil (5-FU) and ionizing radiation (IR) can alter expression of TLR genes in cancer cell lines and what role is played by p53.
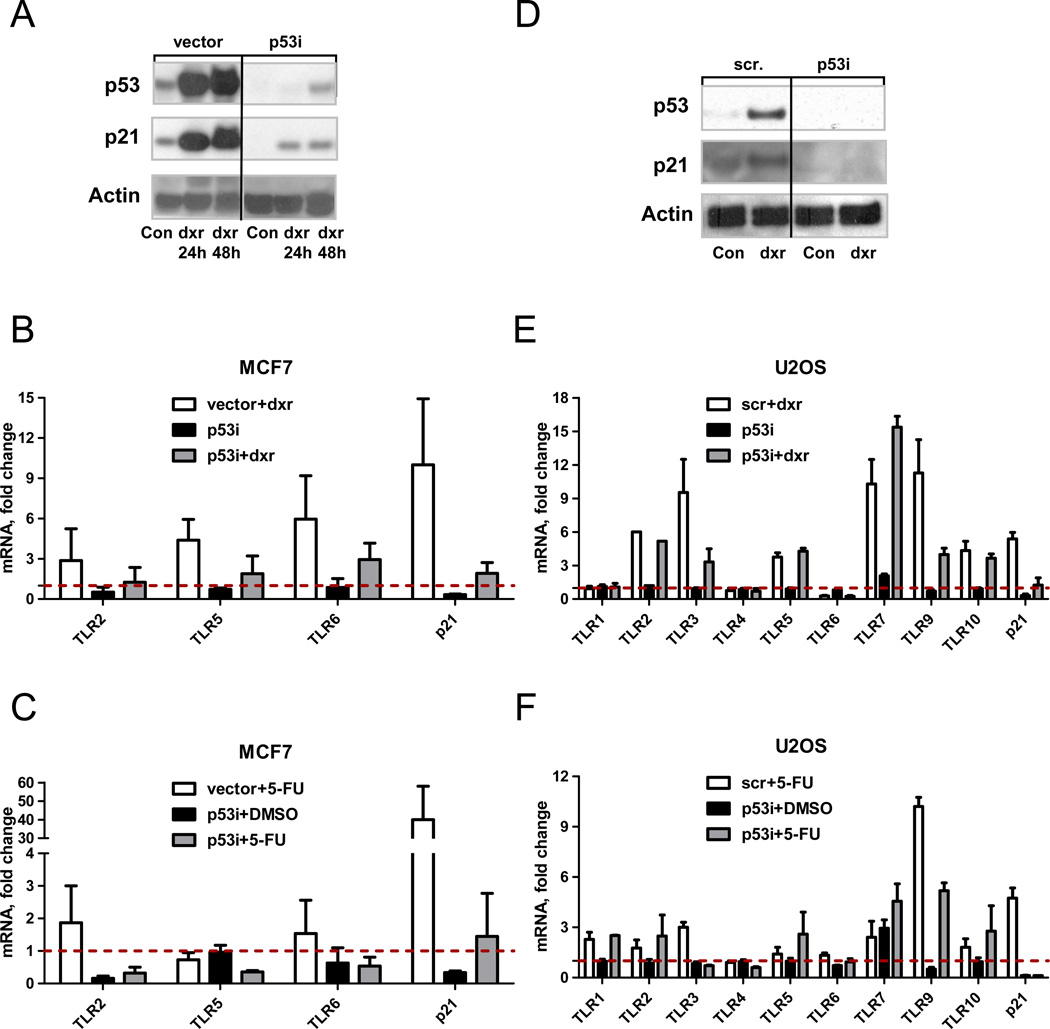
MCF7-vector and MCF7-p53i cells were treated with 0.3 µg/ml doxorubicin for 48 h and protein (Fig. 3A) or total RNA (Fig. 3B) were isolated. Similar to Nutlin-3, the TLR2, 5, and 6 genes were induced by doxorubicin in a p53-dependent fashion. Treatment with 5-fluoruracil for 24 h (Fig. 3C) or IR (Supplementary Fig. 3) also caused p53-dependent induction of TLR2 and TLR6. The lack of TLR5 induction by 5-fluoruracil demonstrates that there are differences between agents in ability to induce TLRs in this cancer cell line.
Figure 3. Role of p53 in doxorubicin and 5-fluoruracil induced up-regulation of TLRs in MCF7 and U2OS cells.
A. MCF7-vector and MCF7-p53i cells were treated with 0.3 µg/ml of doxorubicin for 48 h or left untreated. Following incubation, cells were harvested and subjected to SDS PAGE analysis to detect p53 and p21 induction. Actin was used as a loading control. B. Gene expression of TLRs 2, 5 and 6 and p21 following treatment with doxorubicin (dxr, 0.3 µg/ml, 48 h) or C. with 5-fluoruracil (5-FU, 300 µM, 24 h) was assessed using real time PCR. Shown are averages of 2 to 6 independent experiments.
D. U2OS cells transiently transfected with 25 nM of Dharmacon p53 smart pool (p53i) or scrambled oligos (scr) were treated at 24 h post-transfection with 0.6 µg/ml of doxorubicin for 24 h. The p53 and p21 protein levels were assessed by Western blot; actin was used as a loading control. E. Presented is the gene expression of TLRs 1–10 and p21 following treatment with doxorubicin (0.6 µg/ml, 24 h) or (F) with 5-fluoruracil (300 µM, 24 h).
Gene expression was assessed by real time PCR and normalized to GUSB housekeeping gene expression. Expression of each gene in the DMSO treated control cells was set to one.
To confirm the role of p53 in regulating of TLRs gene expression after DNA damage, U2OS cells were transiently transfected with 25 nM of p53 smart pool (p53i) or scrambled oligos (scr) and subsequently treated for 24 h with doxorubicin. There was substantial induction of p53 and p21 protein levels upon doxorubicin treatment which was prevented by transfection with p53 siRNA (Fig. 3D). As shown in Fig. 3E, there was a strong (9 to11-fold) p53 dependent induction of TLRs 3 and 9. However, there also may be p53-independent induction of TLRs 2, 5, 7, and 10. The pattern of induction by 5-fluoruracil was similar to doxorubicin except that there was p53-independent induction of TLR1 (Fig. 3F). Altogether, the results show that even though an increase in p53 is sufficient to drive expression of TLRs in U2OS cells (as shown for Nutlin-3, Fig. 1) there are cell-specific as well as additional stress–specific factors that can contribute to induced expression of the TLR genes.
Doxorubicin-mediated increase in TLR5 enhances flagellin-induced cytokine responses
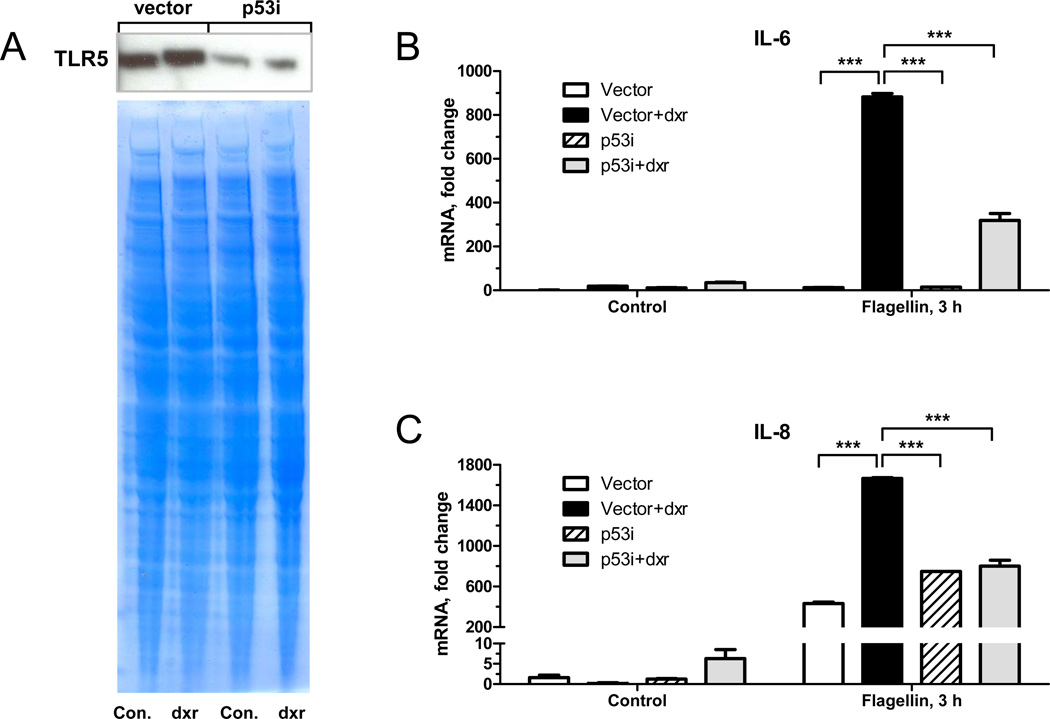
Next, we determined whether a p53-dependent increase in TLR expression affects functional response to TLR ligands downstream of receptor activation. MCF7 cells were treated with doxorubicin to induce TLR5 protein or left untreated for 48 h (Fig. 4A) and then exposed to the TLR5 ligand flagellin for 3 h. There was a significant 2- to 3-fold increase in response to flagellin in p53-positive MCF7-vector cells pretreated with doxorubicin as shown by increased production of cytokines IL-6 and IL-8 mRNA (Fig. 4B– C). Importantly, this effect was p53-dependent based on reduction in the MCF7-p53i cells.
Figure 4. Doxorubicin-mediated increase in TLR5 enhances flagellin-induced cytokine responses.
MCF7-vector or p53i cells were incubated with 0.3 µg/ml doxorubicin or left untreated for 48 h. Cell lysates were subjected to subcellular fractionation and equal amounts of protein from membrane fractions (50 µg per lane) were resolved by SDS PAGE. TLR5 protein was detected by Western blot; gel staining was used for the loading control (A). Flagellin (10 ng/ml) was present in the medium for the last 3 h. mRNA was purified and IL-6 (B) and IL-8 (C) cytokine gene expression was assessed by real time PCR and normalized to expression of the housekeeping gene GUSB. Expression of each gene in control cells was set as one. Statistical analysis was performed by Graph PadPrizm using 2-way ANOVA and Bonferroni posttests. *** corresponds to P-value<0.001.
Differential expression profile of TLR genes across cancer cell lines
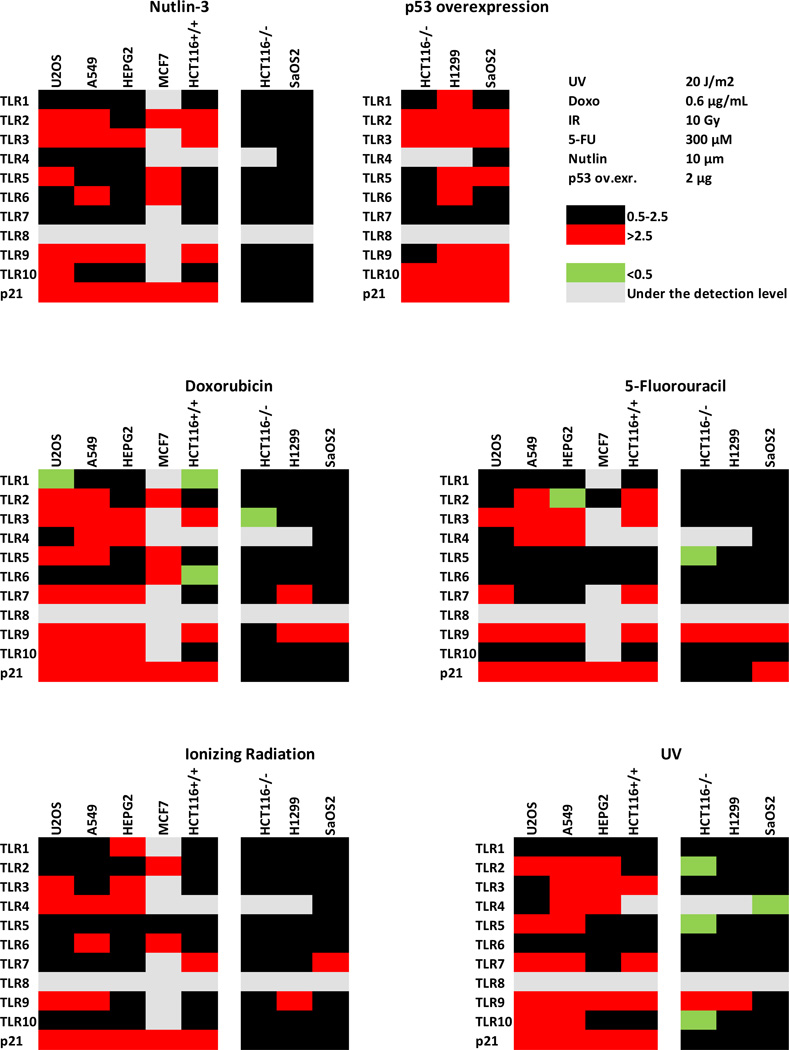
As shown above, several TLR genes were subject to p53 regulation in the three cell lines examined; however, the extent of induction and response to specific agents was variable between cell lines. Since induction of innate immune genes may be important in cancer treatments and differences between cancers could be meaningful, we expanded the panel of cell lines examined for TLR responses to DNA damaging agents and p53 activation. Changes in TLR gene expression following 24 h treatments with Nutlin-3, doxorubicin, 5-fluorouracil, IR and UV (all known to activate p53) are presented in a heat map format in Fig. 5 and in Supplementary Table 2 (conditions for exposure of MCF7 cells to Nutlin-3 and doxorubicin were modified as described in the table). The red, green, and black squares correspond, respectively, to >2.5-fold induction; less than 0.5 expression (i.e., repression) and modest or no change in expression (0.5 to 2.5-fold).
Figure 5. Genotoxic stress and p53 activation differentially affect expression of TLR1–10 genes across a variety of cancer cell lines.
Presented is a heat map of changes in TLR gene expression after the following treatments: 24 h incubation with Nutlin-3, 10 µM; doxorubicin, 0.6 µg/ml; 5-fluoruracil, 300 µM. Cells were also exposed to ionizing radiation (IR), 10 Gy; UV, 20J/ m2 and harvested 24 h after the treatment. MCF7 cells were exposed to Nutlin-3, 10 µM and to 0.3 µg/ml doxorubicin in MCF7 cells for 48 h. Gene expression is relative to the untreated control cells, set as one. The red, green, and black squares correspond, respectively, to >2.5-fold induction; less than 0.5 expression (i.e., repression) and modest or no change in expression (0.5 to 2.5-fold). The values are averages of 3–6 independent experiments.
The TLR 2, 3 and 9 genes were subject to damage/p53-mediated induction in almost all p53 expressing cell lines. Re-introduction of functional wild type p53 in p53 null cancer cells (colon carcinoma HCT116 p53−, osteosarcoma SaOS2 and lung cancer H1299) also led to induction of most TLRs in at least 2 of the 3 cell lines evaluated. Clearly, there is considerable variation in responsiveness of the TLR genes between agents across the many cell lines tested. Importantly, induction is largely dependent on p53.
Effect of p53 mutations on TLR expression
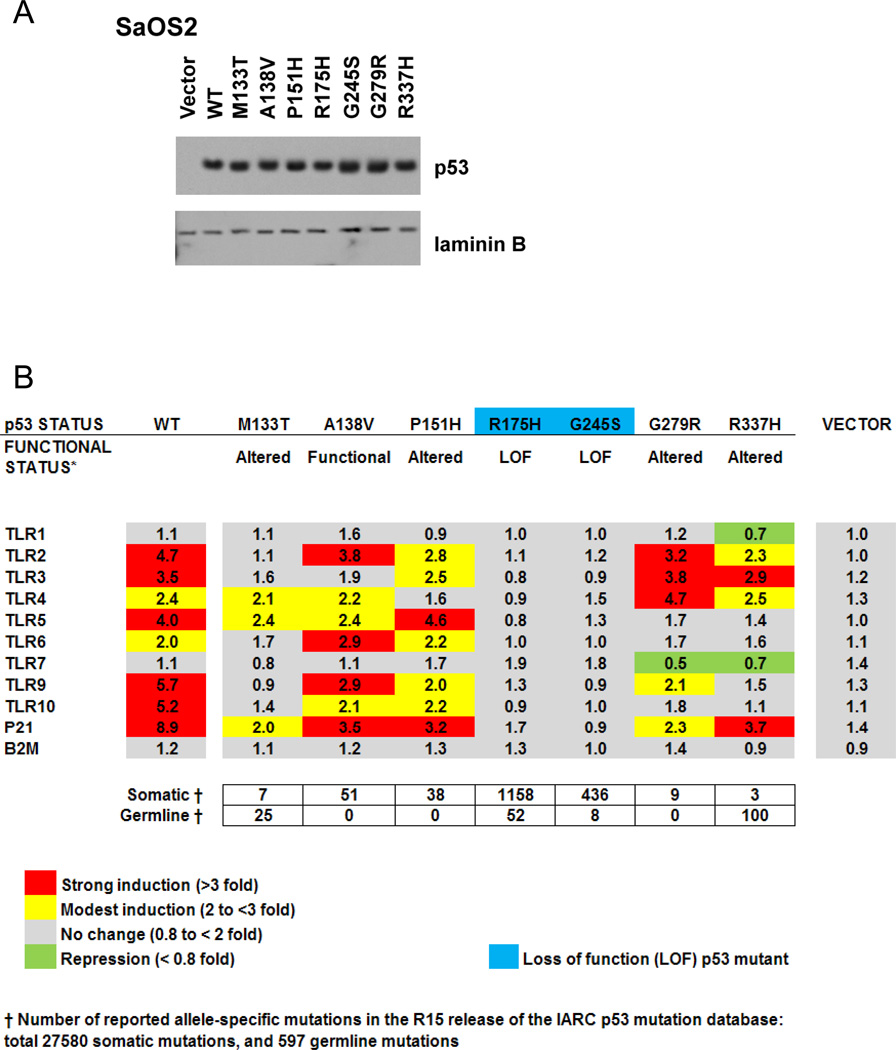
Having established that wild type p53 can regulate expression of TLR genes, we assessed the potential for mutant p53 to activate endogenous TLR genes since mutants that retain function are frequent in cancers. The osteosarcoma SaOS2 p53 null cells were transiently transfected with vectors that express p53 mutant proteins at comparable levels (Fig. 6A).
Figure 6. Mutations in p53 differentially modify p53 effects on expression of the TLR gene family.
A. p53 null SaOS2 cells were transfected with a panel of cancer-related p53 mutants. At 24 h post-transfection, cells were harvested and p53 nuclear levels were evaluated by Western blot. B. Presented is a heat map of fold-change in mRNA expression for TLRs, p21 and B2M genes following transfection with WT or mutant p53 as compared to the mock transfected control (set as one). Gene expression was assessed by real time PCR and normalized to GAPDH expression.
Seven cancer-associated p53 mutants were analyzed, and the TLR expression results are presented as a heatmap in the Fig. 6B. Included is a brief description of the functional transactivation status for each p53 mutant based on yeast reporter assays (37, 38). Also presented is the frequency of these mutations in human somatic and germline tumors. The levels of mRNA from all the TLR genes were evaluated 24 h post-transfection. Consistent with our previous findings, TLR8 expression was not detected. B2M and p21 gene expression were used as p53 negative and positive controls, respectively.
As shown in Fig. 6, three patterns of gene expression were observed. Mutants A138V, P151H, and G279R retain the ability to drive expression of most TLRs at levels similar to wild type p53. However, both A138V and P151H have decreased ability to drive expression of TLR3 gene, a strong target of the wild type protein. The P151H was unable to induce TLR4. A fourth mutant in this group, G279R, had a profile similar to wild type for TLRs 1, 2, 3, and 4 but reduced capacity to transactivate TLR5, TLR9 and TLR10 along with p21.
A much greater change-of-spectrum for TLR expression was observed for M133T and R337H. The former exhibited limited transactivation for TLR4 and TLR5 only. The R337H mutant showed a clear change-in-spectrum of TLR expression when compared to wild type p53. In the third group, the cancer-associated loss-of-function hotspot p53 mutants R175H and G245S were transcriptionally inactive toward all TLR genes as well as p21. Differences in transactivation profiles for the p53 mutants were also evident for the p53 target gene p21. Thus, cancer-associated p53 mutants can have dramatically different effects on endogenous expression of the TLR family of genes and potentially affect their transcriptional response to stress and downstream signaling.
DISCUSSION
Here, we demonstrate that expression of most members of the TLR gene family in human cancer cells can be responsive to p53 activation and genotoxic stress. This is expected to increase opportunities in TLR-based cancer treatments, especially since therapies may include both DNA damaging agents and increased levels of p53. In support of this, we showed that treatment of p53-positive MCF7 cells with doxorubicin prior to activation of TLR5 by its ligand flagellin leads to increased downstream expression of cytokines IL-6 and IL-8. Interestingly, treatment of breast cancer cells with flagellin was recently shown to reduce cell proliferation in culture and xenograft growth in mice. This effect was in part mediated by flagellin-induced soluble factors (39). Similarly, a combined treatment with flagellin along with a class I phosphoinositide 3-kinase inhibitor resulted in delayed tumor growth and increased animal survival in several mouse models (40). Therefore, an increased expression of cytokines in response to Nutlin-3/flagellin combination might predict better treatment outcome.
We have shown that there is considerable variation in transcriptional responses of specific TLR genes between cell lines and stressors. For example, TLR3 expression was induced by all genotoxic agents tested in almost all the p53-positive cell lines examined as well as by overexpression of p53 protein in p53-null cells. TLR3 was previously shown to induce apoptosis in cancer cells (18, 41), suggesting that a combination of TLR3 ligands with genotoxic drugs could be beneficial for treatment of tumors containing wild type p53. Alternatively, since DNA damage can induce TLR9 expression in p53 dependent and independent manners, it may be possible to amplify TLR9-based treatment by genotoxic agents regardless of tumor p53 status. Similarly, enhanced expression of TLR1 and TLR7 may be associated with cell type as well as with DNA damage response mechanism. We did not see a dependence on p53 status for treatments leading to enhanced TLR7. The TLRs 2, 4, 5 and 6 appeared to be less subject to induction across the cell lines examined; however, the induction generally required the presence of functional p53. Previously, we reported that a wide diversity in responses was also seen for p53-induced expression of TLR genes in stimulated lymphocytes from healthy individuals (7). Similar differences between individuals in terms of transcriptional response to ionizing radiation have been observed in derived lymphoblastoid cell lines (42). Therefore, variability between cell lines may originate from either inter-individual or tissue type-dependent difference.
The altered-function p53 mutations can result in cellular responses that impact genome stability, repair, replication, and programmed cell death (37, 43). Single amino acid changes in p53 that differentially impact transactivation may alter the efficacy of chemotherapeutic agents and diversify cell responses to stress (44). Here, we evaluated the effect of several p53 mutants associated with both somatic and germline tumors, particularly breast and colon cancers (Fig. 5 and the IARC database R15), on expression of the TLR genes. The changes in TLR transactivation patterns, including change-of-spectrum, are consistent with our previous report in which each of mutant was tested for transactivation at 11 validated human p53 REs using a yeast-based system (4).
While the classic loss-of-function mutants R175H and G245S were unable to induce any TLR gene, the profile of TLR-expressed genes for the remaining five p53 mutants was altered. The R337H mutation resulted in the most dramatic changes including loss or gain of transactivation for different TLRs (Fig. 5). Located on the surface of the tetramerization domain of the p53 protein, the R337H mutation is considered to have a subtle functional transactivation change causing a pH dependent effect on folding (45). The differential response of TLR genes to this p53 mutant, which is associated with pediatric adrenal cortical carcinoma (ACC), could inform treatments that are TLR ligand-based. In our study we observed that the R337H p53 mutant might have the ability to induce expression of TLR2. Other factors also may play a role in the regulation of the adrenal response as shown for TLR9 and its relation with sepsis response in the presence of bacterial DNA (46). These observations support the view that the specific status of p53 should be considered in designing adjuvant cancer therapies that utilize TLR pathways.
Overall, we suggest that a profile of TLR gene expression patterns in specific tumors in response to p53 and DNA damaging agents combined with knowledge of p53 expression and mutation status in these tumors can be an important tool in cancer diagnosis and in strategies that target TLR pathway for cancer therapy. For example, the presence of a p53 mutation that can change a damage-induced TLR expression pattern could determine which TLR agonists or antagonists to employ for specific tumors as well as the choices of p53 adjuvant inducers of TLRs. Also, we suggest that there are unique opportunities to capitalize on chromosome stress responses in human cancer therapies that might not be apparent from studies in other organisms since the responsiveness of the human set of TLR genes to p53 is unique to primates (7).
Supplementary Material
ACKNOWLEDGEMENTS
We appreciate discussions with David Schwartz at early stages of work and critical comments of Carl Anderson, Michael Fessler, and Stavros Garantziotis during manuscript development. Support was provided NIEHS intramural research funds, Project Z01-ES065079.
Footnotes
Authors declare that no conflict of interests exists
REFERENCES
- 1.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 2.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 3.Jegga AG, Inga A, Menendez D, Aronow BJ, Resnick MA. Functional evolution of the p53 regulatory network through its target response elements. P Natl Acad Sci USA. 2008;105:944–949. doi: 10.1073/pnas.0704694105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jordan JJ, Menendez D, Inga A, Nourredine M, Bell D, Resnick MA. Noncanonical DNA Motifs as Transactivation Targets by Wild Type and Mutant p53. Plos Genetics. 2008;4 doi: 10.1371/journal.pgen.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menendez D, Inga A, Resnick MA. Estrogen receptor acting in cis enhances WT and mutant p53 transactivation at canonical and noncanonical p53 target sequences. P Natl Acad Sci USA. 2010;107:1500–1505. doi: 10.1073/pnas.0909129107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menendez D, Shatz M, Azzam K, Garantziotis S, Fessler MB, Resnick MA. The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks. PLoS Genet. 2011;7:e1001360. doi: 10.1371/journal.pgen.1001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Bauer S, Muller T, Hamm S. Pattern recognition by Toll-like receptors. Adv Exp Med Biol. 2009;653:15–34. doi: 10.1007/978-1-4419-0901-5_2. [DOI] [PubMed] [Google Scholar]
- 10.Kabelitz D. Expression and function of Toll-like receptors in T lymphocytes. Curr Opin Immunol. 2007;19:39–45. doi: 10.1016/j.coi.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Uematsu S, Jang MH, Chevrier N, Guo Z, Kumagai Y, Yamamoto M, et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nature immunology. 2006;7:868–874. doi: 10.1038/ni1362. [DOI] [PubMed] [Google Scholar]
- 12.Muir A, Soong G, Sokol S, Reddy B, Gomez MI, Van Heeckeren A, et al. Toll-like receptors in normal and cystic fibrosis airway epithelial cells. Am J Respir Cell Mol Biol. 2004;30:777–783. doi: 10.1165/rcmb.2003-0329OC. [DOI] [PubMed] [Google Scholar]
- 13.Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 14.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nature immunology. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch I, Caux C, Hasan U, Bendriss-Vermare N, Olive D. Impaired Toll-like receptor 7 and 9 signaling: from chronic viral infections to cancer. Trends Immunol. 2010;31:391–397. doi: 10.1016/j.it.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 17.Taura M, Fukuda R, Suico MA, Eguma A, Koga T, Shuto T, et al. TLR3 induction by anticancer drugs potentiates poly I:C-induced tumor cell apoptosis. Cancer science. 2010 doi: 10.1111/j.1349-7006.2010.01567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salaun B, Coste I, Rissoan MC, Lebecque SJ, Renno T. TLR3 can directly trigger apoptosis in human cancer cells. J Immunol. 2006;176:4894–4901. doi: 10.4049/jimmunol.176.8.4894. [DOI] [PubMed] [Google Scholar]
- 19.Garay RP, Viens P, Bauer J, Normier G, Bardou M, Jeannin JF, et al. Cancer relapse under chemotherapy: why TLR2/4 receptor agonists can help. Eur J Pharmacol. 2007;563:1–17. doi: 10.1016/j.ejphar.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Bong AB, Bonnekoh B, Franke I, Schon MP, Ulrich J, Gollnick H. Imiquimod, a topical immune response modifier, in the treatment of cutaneous metastases of malignant melanoma. Dermatology. 2002;205:135–138. doi: 10.1159/000063904. [DOI] [PubMed] [Google Scholar]
- 21.Krieg AM. Development of TLR9 agonists for cancer therapy. J Clin Invest. 2007;117:1184–1194. doi: 10.1172/JCI31414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nat Rev Cancer. 2009;9:57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 23.Akazawa T, Ebihara T, Okuno M, Okuda Y, Shingai M, Tsujimura K, et al. Antitumor NK activation induced by the Toll-like receptor 3-TICAM-1 (TRIF) pathway in myeloid dendritic cells. Proc Natl Acad Sci U S A. 2007;104:252–257. doi: 10.1073/pnas.0605978104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He H, Genovese KJ, Nisbet DJ, Kogut MH. Synergy of CpG oligodeoxynucleotide and double-stranded RNA (poly I:C) on nitric oxide induction in chicken peripheral blood monocytes. Mol Immunol. 2007;44:3234–3242. doi: 10.1016/j.molimm.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 25.Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat Med. 2007;13:552–559. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- 26.Zom GG, Khan S, Filippov DV, Ossendorp F. TLR Ligand-Peptide Conjugate Vaccines: Toward Clinical Application. Adv Immunol. 2012;114:177–201. doi: 10.1016/B978-0-12-396548-6.00007-X. [DOI] [PubMed] [Google Scholar]
- 27.Bell MP, Svingen PA, Rahman MK, Xiong Y, Faubion WA., Jr FOXP3 regulates TLR10 expression in human T regulatory cells. J Immunol. 2007;179:1893–1900. doi: 10.4049/jimmunol.179.3.1893. [DOI] [PubMed] [Google Scholar]
- 28.Tsatsanis C, Androulidaki A, Alissafi T, Charalampopoulos I, Dermitzaki E, Roger T, et al. Corticotropin-releasing factor and the urocortins induce the expression of TLR4 in macrophages via activation of the transcription factors PU. 1 and AP-1. J Immunol. 2006;176:1869–1877. doi: 10.4049/jimmunol.176.3.1869. [DOI] [PubMed] [Google Scholar]
- 29.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 30.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 31.Menendez D, Inga A, Resnick MA. Estrogen receptor acting in cis enhances WT and mutant p53 transactivation at canonical and noncanonical p53 target sequences. Proc Natl Acad Sci U S A. 2010;107:1500–1505. doi: 10.1073/pnas.0909129107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taura M, Eguma A, Suico MA, Shuto T, Koga T, Komatsu K, et al. p53 regulates Toll-like receptor 3 expression and function in human epithelial cell lines. Mol Cell Biol. 2008;28:6557–6567. doi: 10.1128/MCB.01202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz DA, Cook DN. Polymorphisms of the Toll-like receptors and human disease. Clin Infect Dis. 2005;41(Suppl 7):S403–S407. doi: 10.1086/431985. [DOI] [PubMed] [Google Scholar]
- 34.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 35.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 36.Adams S. Toll-like receptor agonists in cancer therapy. Immunotherapy. 2009;1:949–964. doi: 10.2217/imt.09.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Resnick MA, Inga A. Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity. Proc Natl Acad Sci U S A. 2003;100:9934–9939. doi: 10.1073/pnas.1633803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jordan JJ, Inga A, Conway K, Edmiston S, Carey LA, Wu L, et al. Altered-function p53 missense mutations identified in breast cancers can have subtle effects on transactivation. Mol Cancer Res. 2010;8:701–716. doi: 10.1158/1541-7786.MCR-09-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai Z, Sanchez A, Shi Z, Zhang T, Liu M, Zhang D. Activation of Toll-like receptor 5 on breast cancer cells by flagellin suppresses cell proliferation and tumor growth. Cancer Res. 2011;71:2466–2475. doi: 10.1158/0008-5472.CAN-10-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall NA, Galvin KC, Corcoran AM, Boon L, Higgs R, Mills KH. Immunotherapy with PI3K inhibitor and Toll-like receptor agonist induces IFN-gamma+IL-17+ polyfunctional T cells that mediate rejection of murine tumors. Cancer Res. 2012;72:581–591. doi: 10.1158/0008-5472.CAN-11-0307. [DOI] [PubMed] [Google Scholar]
- 41.Paone A, Starace D, Galli R, Padula F, De Cesaris P, Filippini A, et al. Toll-like receptor 3 triggers apoptosis of human prostate cancer cells through a PKC-alpha-dependent mechanism. Carcinogenesis. 2008;29:1334–1342. doi: 10.1093/carcin/bgn149. [DOI] [PubMed] [Google Scholar]
- 42.Correa CR, Cheung VG. Genetic variation in radiation-induced expression phenotypes. Am J Hum Genet. 2004;75:885–890. doi: 10.1086/425221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menendez D, Inga A, Resnick MA. The biological impact of the human master regulator p53 can be altered by mutations that change the spectrum and expression of its target genes. Molecular and Cellular Biology. 2006;26:2297–2308. doi: 10.1128/MCB.26.6.2297-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Temam S, Flahault A, Perie S, Monceaux G, Coulet F, Callard P, et al. p53 gene status as a predictor of tumor response to induction chemotherapy of patients with locoregionally advanced squamous cell carcinomas of the head and neck. J Clin Oncol. 2000;18:385–394. doi: 10.1200/JCO.2000.18.2.385. [DOI] [PubMed] [Google Scholar]
- 45.Ribeiro RC, Sandrini F, Figueiredo B, Zambetti GP, Michalkiewicz E, Lafferty AR, et al. An inherited p53 mutation that contributes in a tissue-specific manner to pediatric adrenal cortical carcinoma. Proc Natl Acad Sci U S A. 2001;98:9330–9335. doi: 10.1073/pnas.161479898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran N, Koch A, Berkels R, Boehm O, Zacharowski PA, Baumgarten G, et al. Toll-like receptor 9 expression in murine and human adrenal glands and possible implications during inflammation. J Clin Endocrinol Metab. 2007;92:2773–2783. doi: 10.1210/jc.2006-2697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.