Abstract
Affinity purification is a useful approach for purification of recombinant proteins. Eukaryotic expression systems have become more frequently used at the expense of prokaryotic systems since they afford recombinant eukaryotic proteins with post-translational modifications similar or identical to the native ones.
Here, we present a one-step affinity purification set-up suitable for the purification of secreted proteins. The set-up is based on the interaction between biotin and mutated streptavidin. Drosophila Schneider 2 cells are chosen as the expression host, and a biotin acceptor peptide is used as an affinity tag. This tag is biotinylated by E. coli biotin-protein ligase in vivo. We determined that localization of the ligase within the ER led to the most effective in vivo biotinylation of the secreted proteins. We optimized a protocol for large-scale expression and purification of AviTEV-tagged recombinant human glutamate carboxypeptidase II (Avi-GCPII) with milligram yields per litre of culture. We also determined the 3D structure of Avi-GCPII by X-ray crystallography and compared the enzymatic characteristics of the protein to those of its non-tagged variant. These experiments confirmed that AviTEV tag does not affect the biophysical properties of its fused partner.
Purification approach, developed here, provides not only a sufficient amount of highly homogenous protein but also specifically and effectively biotinylates a target protein and thus enables its subsequent visualization or immobilization.
Keywords: affinity purification, biotin acceptor peptide, recombinant protein expression, biotin-protein ligase (BirA), co-localization, PSMA
Introduction
Purification of recombinant proteins via diverse affinity tags has recently largely replaced approaches utilizing the biophysical features of the proteins. Affinity tags based on the well-characterized biotin–avidin interaction (or its analogues) were among the first candidates for this application since this interaction is the strongest (KD ~ 10−15 M) non-covalent bond known in biology [1].
Ironically, the strength of the binding is a major drawback for its use as a purification technique, since the elution conditions would have to be so harsh that they would lead to destruction of the purified protein. One possible way to overcome this problem is to use site-directed mutagenesis to modify the streptavidin molecule to have a KD for biotin in the micromolar range, which is more suitable for purification purposes [2–3].
Enzymatic biotinylation of target proteins by biotin-protein ligase from E. coli (BirA; EC 6.3.4.15) is often used instead of the less specific in vitro chemical biotinylation of free amino groups. In E. coli, this ligase recognizes and biotinylates the ε-amino group of a specific lysine residue within the biotin carboxyl carrier protein subunit of acetyl-CoA carboxylase [4–5]. Using combinatorial peptide libraries, a 15 amino acid peptide with one lysine residue specifically biotinylated by BirA was identified. The sequence was referred to as biotin acceptor peptide (BAP) or, more frequently, AviTag and it bears little similarity to the natural BirA substrate [6–7].
The biotinylation via BirA can proceed either in vitro or in vivo. The in vivo biotinylation, being a part of intracellular, post-translational modification of target proteins represents an elegant, high-yield approach [8–10]. AviTag is specifically recognized only by the BirA, therefore if other expression systems than E. coli are used for production of biotinylated proteins, the expressing cells have to be co-transfected with plasmids coding for both targeted protein and BirA. Proteins biotinylated in vivo were successfully expressed in both mammalian [8, 10–13] and insect [9, 14–15] expression systems. In some of these studies, different cellular localizations of BirA (in cytoplasm, within the ER, or in the secretory pathway) were investigated showing a strong dependency of BirA localization on its biotinylation efficiency. In all these experiments, the biotinylated proteins were expressed as secreted proteins [10–11, 15].
Besides the AviTag/BirA/mutated streptavidin system, there are other affinity purification approaches utilizing avidin or its analogues. Above all, the Strep-tag II/Strep-Tactin system is commonly used [16–17]. This system does not require additional transfection with BirA since the Strep-tag II binds directly to the Strep-Tactin molecule. On the other hand, the Strep-tag II affinity to Strep-Tactin is in the micromolar range, which is suitable for purification but might represent a drawback during visualization, immobilization, or specific uptake of a target protein. Generally, purification approaches based on the avidin-biotin interaction are very specific, ensuring a high homogeneity of the purified proteins. Achieving such a homogeneity may be an occasional problem with the use of other affinity tags, e.g. His-tag [18].
In this paper, we present an optimized one-step protocol for affinity purification of recombinant proteins expressed via the secretory pathway in insect cells with BirA localized within the ER. The purified protein, the extracellular portion of glutamate carboxypeptidase II (rhGCPII, amino acids 44–750), is a 90 kD N-glycosylated metalloprotease [19–20]. GCPII (EC 3.4.17.21) belongs to the family of type II transmembrane proteins and is an interesting pharmaceutical target for prostate cancer imaging and treatment [21–23]. It is also implicated in neuropathological disorders [24] and has an unknown function in angiogenesis [25–26].
Materials and Methods
Preparation of expression plasmid for N-terminally AviTEV-tagged proteins
DNA encoding the AviTEV tag was prepared de novo from six individual oligonucleotides with complementary overlaps. Short 5’ overhangs were filled in with Phusion polymerase (Finnzymes). The following primers were used: Avi0-F (5’-aaaaatgatcaggcctgaacgacatc-3’), Avi1-R (5’-atcttctgggcctcgaagatgtcgttcaggcc-3’), Avi2-F (5’-ttcgaggcccagaagatcgagtggcacg-3’), Avi3-R (5’-ggttctcgctgccgctgccgctgccctcgtgccactcg-3’), Uni3-F (5’-gcagcggcagcgagaacctgtacttccagggcagatctgaattcaaaaa-3’), and Uni4-R (5’-tttttgaattcagatctgc-3’). Sequences encoding AviTag™ (Avidity) are underlined, and sequences encoding the TEV cleavage site are in bold. Two restrictions sites introduced into the DNA are shown in italics, BclI at the 5’ end and BglII at the 3’ end of the sequence.
A standard PCR reaction was performed with a mixture of all primers. The resulting DNA construct was cleaved by BclI and BglII and inserted into the pMNAEXST, pre-cleaved with BglII, to create a plasmid for expression of N-terminally AviTEV-tagged rhGCPII. Both correct orientation and sequence of the prepared plasmid were verified by sequencing. The resulting plasmid was denoted pMT/BiP/AviTEV/rhGCPII and contained AviTEV tag immediately downstream of the BiP peptide signal sequence for secretion. Additionally, the DNA encoding rhGCPII can be easily excised with BglII and XhoI/XbaI restriction enzymes, and DNA encoding a different secreted protein can be substituted.
Preparation of plasmids encoding differently localized BirA
The commercially available plasmid pACYLBirA (Avidity) was used as a template for PCR reactions. To obtain a plasmid encoding cytoplasmic BirA, primers BirAKpnI-F (5’-atcggggtaccatgaaggataacaccgtgcc-3’) and BirAnoKDELXhoI-R (5’-tctagactcgagttattctgcactacgcaggg-3’) were used (restriction sites are italicized). The DNA amplicon was cleaved with KpnI and XhoI and ligated into the vector pMT/V5-HisA (Invitrogen). The resulting plasmid was denoted pMT/BirA. To obtain DNA encoding BirA retained within the endoplasmic reticulum (ER), primers BirABglII-F (5’-ctcgggagatctatgaaggataacaccgtgcc-3’) and BirAXhoI-R (5’-tctagactcgagttacagctcatctttttctgcactacgcaggg-3’) were used to amplify DNA from pACYLBirA. The DNA was cleaved with BglII and XhoI and ligated into the vector pMT/BiP/V5-HisA (Invitrogen). The resulting plasmid was named pMT/BiP/BirA/KDEL. A plasmid encoding secreted BirA was obtained similarly, but primer BirAXhoI-R was substituted with BirAnoKDELXhoI-R. The resulting plasmid encoding secreted BirA was denoted pMT/BiP/BirA.
The correct sequences of all three plasmids were subsequently verified by sequencing.
Preparation of stable Drosophila S2 cell lines expressing Avi-GCPII and BirA
Drosophila S2 cells were transfected consecutively, first with 9 µg of pMT/BiP/AviTEV/rhGCPII together with 0.5 µg of pCoBlast (Invitrogen) and selected with the appropriate antibiotic. Afterwards, they were transfected with 9 µg of one of the three different plasmids encoding differently localized BirA together with 0.5 µg of pCoHygro (Invitrogen), as previously published [27]. The transfected cells were selected by cultivation with both Blasticidin (5 µg/mL, Invitrogen) and Hygromycin B (300 µg/mL, Invitrogen) until they regained growth. Two transfections were performed in case of BirA localized within ER to prepared two individual polyclonal stable cell lines of S2 cells.
2.106 cells of each stable transfectant were transferred to 35 mm Petri dish supplemented with 2 ml SF900II medium (Invitrogen). The protein expression was induced next day by 1 mM CuSO4 (Sigma). Three days post-induction, the cells were harvested by centrifugation, and the medium was analyzed by Western blot.
Large-scale expression of Avi-GCPII and rhGCPII in Drosophila S2 cells
The protocol for large-scale expression of Avi-GCPII was almost identical to that previously described [19]. The only modification was the final volume of the cell suspension, which changed from 0.5 L to 1 L. Different volumes of cell culture supplements were used accordingly. The protocol for large-scale expression and purification of rhGCPII was identical to that described previously [19].
Purification of Avi-GCPII on Streptavidin Mutein Matrix
The commercial protocol for Streptavidin Mutein Matrix™ (Roche) was used as a starting point for optimization of the purification protocol. Phosphate buffer was replaced with Tris-HCl buffer, since phosphate acts as a weak inhibitor of GCPII [28]. Additional optimization experiments were performed on a small-scale with 9 mL of conditioned medium and 200 µl of mutein resin (i.e. 400 µl of resin mixture with 20% ethanol). Following optimization, large-scale purification of Avi-GCPII was performed.
Conditioned medium from S2 cells (0.8 L) was centrifuged at 3400 g for 30 minutes and then concentrated to 70 ml at 4°C using a LabScale™ TFF system (Millipore) with a Pellicon XL Biomax50 cassette (Millipore). All subsequent purification steps were performed at 4°C. The concentrated medium was mixed with equilibration buffer (450 mM NaCl, 300 mM Tris-HCl, pH 7.2) in a 2:1 ratio (fraction L). Equilibrated medium was mixed with 1.6 mL of mutein resin, which had been washed several times with washing buffer (150 mM NaCl, 100 mM Tris-HCl, pH 7.2), and incubated overnight on a rocker. The next day, a disposable gravity column (Thermo Scientific) was used to separate the resin from the equilibrated medium (fraction FT). The mutein resin was washed with 30 ml of washing buffer and then 1.6 ml of elution buffer (150 mM NaCl, 2 mM D-biotin, 100 mM Tris-HCl, pH 7.2) was applied to the column to replace the remaining wash buffer (fraction W). The column was sealed and the resin was incubated with elution buffer for 1 hour. Then, the bound Avi-GCPII protein was eluted with 7 ml of elution buffer to obtain seven 1 ml elution fractions (fractions E1-7). Following the purification procedure, the Streptavidin Mutein Matrix™ was regenerated and stored according to the manufacturer’s protocol.
After the first purification and regeneration, the flow-through fraction and regenerated resin were mixed again and the whole purification procedure was repeated. All fractions were analyzed by reducing SDS-PAGE. The E1 and E2 fractions were dialyzed against appropriate buffer overnight, aliquoted and stored at −80°C until further use.
SDS-PAGE and Western blotting
Protein samples were resolved by reducing, 0.1% sodium dodecyl sulphate (SDS), polyacrylamide gel electrophoresis (PAGE). Following SDS-PAGE, gels were either stained using Coomassie Brilliant Blue R-250 (Thermo Scientific), silver-stained, or electroblotted. For N-terminal sequencing, the proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane, which was stained with Coomassie Brilliant Blue G-250 (Thermo Scientific).
Proteins were transferred onto a nitrocellulose membrane for detection by a specific antibody against rhGCPII (GCP-04) or Neu-HRP (Thermo Scientific). The membrane was blocked with Blocker™ Casein in TBS (Thermo Scientific) for 1 hour. For detection of rhGCPII, GCP-04 (1 mg/mL) was diluted 1:5000 in casein blocker, incubated overnight, washed several times with 0.05% Tween 20 in PBS, and incubated for 1 hour with HRP-conjugated goat anti-mouse antibody (1 mg/mL; Thermo Scientific) diluted in casein blocker in a 1:25000 ratio. For detection of biotinylated proteins, Neu-HRP (1 mg/mL) was diluted 1:2500 in casein blocker and incubated with the membrane for 1 hour.
Afterwards, the membranes were washed with 0.05% Tween 20 in PBS several times to remove either Neu-HRP or HRP-conjugated goat anti-mouse antibody. The blots were developed using SuperSignal West Dura Chemiluminiscence Substrate (Thermo Scientific) according to the manufacturer’s protocol. Reactive bands were visualized on a LAS-3000 CCD camera (FujiFilm). If necessary, densitometry analysis was performed using ImageJ software v1.43 [29].
Cleavage of AviTEV tag from Avi-GCPII by TEV protease
Purified Avi-GCPII was diluted to a final concentration of 1 µg/mL and mixed with His-TEV(S219V)-Arg protease in an equimolar ratio or with a 100-fold molar excess of TEV protease. Equivalent reference mixture, without TEV protease, was also prepared. 5 µl of concentrated TBS buffer (1.5 M NaCl, 0.5 M Tris-HCl, pH 7.6) was added to each reaction, and the final volume was adjusted to 50 µl with distilled water. Mixtures were incubated overnight at 4°C and analyzed on Western blots. Similar experiments were performed under identical conditions substituting concentrated TBS buffer with concentrated TEV buffer (10 mM β-mercaptoethanol, 5 mM EDTA, 0.5 M Tris-HCl, pH 8.0) or concentrated MOPS buffer (0.2 M NaCl, 0.2 M MOPS, pH 7.4).
Inhibition of Avi-GCPII degradation
20 µl of purified Avi-GCPII (40 µg/mL) was mixed with 5 µl of inhibitor or water (negative control) to achieve a final concentration of 200 µM for EDTA (Sigma-Aldrich), 2-PMPA, Pefabloc SC (Roche) and E-64 (Sigma-Aldrich) or 2 µM for Pepstatin A (Sigma-Aldrich). Reactions were incubated at room temperature and 6 µl samples were taken sequentially every other day and analyzed by SDS-PAGE.
Crystallization and data collection
The Avi-GCPII stock solution (10 mg/mL) was mixed with 1/10 volume of 1 mM 2-PMPA inhibitor, and the crystallization droplets were prepared by combining 2 µl of the complex solution with 2 µl of the reservoir solution containing 33% pentaerythritol propoxylate (Hampton Research), 1% polyethylene glycol 3350 (Fluka), and 100 mM Tris-HCl, pH 8.0. Crystals were grown by the hanging drop vapour diffusion method at 293 K.
Crystals were flash-frozen in liquid nitrogen directly from the reservoir solution, and the diffraction data were collected at 100 K using synchrotron radiation at the Southeast Regional Collaborative Access Team sector 22 beamlines BM of the Advanced Photon Source (Argonne, IL, USA) at an X-ray wavelength of 1.0 Å. The complete dataset was collected from a single crystal using the MarMosaic 225 mm CCD detector. Data processing was performed with the HKL2000 software package [30].
Structure determination and refinement
Since the crystal of the Avi-GCPII/2-PMPA complex was isomorphous with the crystals of rhGCPII containing the same inhibitor (pdb entry 2PVW) [31], the latter (without the inhibitor and water molecules) was used as the starting model for the structural refinement based on the X-ray data collected from a crystals of the Avi-GCPII/2-PMPA complex. Calculations were performed with the program Refmac 5.1 [32], and the refinement protocol was interspersed with manual corrections to the model using Coot [33]. The stereochemical quality of the final model was evaluated using MolProbity [34] and the final model, together with experimental amplitudes, deposited in the RCSB Protein Data Bank (entry code 3RBU). During the refinement process, 1363 of the randomly selected reflections were kept aside for cross-validation (Rfree).
Radioenzymatic assay of carboxypeptidase activity
To determine the kinetic constants of Ac-Asp-Glu cleavage, radioenzymatic assays using [3H]-Ac-Asp-Glu (radiolabelled on the terminal glutamate; 50 Ci/mmol in Tris buffer, Perkin–Elmer) were performed as previously described [27–28] with several modifications.
Briefly, appropriate amount of enzyme solution in reaction buffer (20 mM NaCl, 20 mM MOPS, pH 7.4) was pre-incubated for 10 min at 37°C in a final volume of 90 µl. The reaction was started by addition of a 10 µl mixture of unlabelled and radioactive Ac-Asp-Glu with different overall concentration ranging from 200 nM to 50 µM (containing 50 nM 3H-Ac-Asp-Glu together with corresponding amount of unlabelled Ac-Asp-Glu) and 100 µM to 400 µM (containing 100 nM 3H-Ac-Asp-Glu together with corresponding amount of unlabelled Ac-Asp-Glu). Reactions were then incubated at 37°C and after 20 min were stopped with 100 µl of ice-cold stopping solution (2 mM β-mercaptoethanol, 200 mM sodium phosphate, pH 7.4). Free glutamate was separated from the uncleaved substrate by ion exchange chromatography, and quantified by liquid scintillation using the Rotiszint ECO Plus scintillation cocktail (Roth) and Tri-Carb 2900TR liquid scintillation counter (PerkinElmer).
Reactions were performed in duplicate for each measurement, and substrate turnovers ranged between 14–20%. For Avi-GCPII, two separate measurements were performed. The Km and kcat values were determined from reaction rate versus substrate concentration plots using the GraFit program [35].
HPLC assay of carboxypeptidase activity
Kinetic constants of N-acetyl-L-aspartyl-L-methionine (Ac-Asp-Met) cleavage by Avi-GCPII and rhGCPII were determined using HPLC for quantification of the hydrolysis products. An appropriate amount of enzyme was added to reaction buffer (20 mM NaCl, 20 mM MOPS, pH 7.4) to final volume of 45 µl. The reaction mixtures were pre-incubated for 10 min at 37°C. Then, 5 µl of substrate was added into each reaction (final concentration of substrate ranged from 2.8 µM to 360 µM). The reaction mixtures were incubated for 15 min at 37°C and then stopped by adding 125 µl of ice-cold 0.2 M sodium borate, pH 10.0, and 4 µM L-glutamate (internal standard). Subsequently, the amount of free L-methionine was determined following a previously described method based on o-phthaldialdehyde (OPA) derivatization [36] and analyzed on an Agilent 1200 Series system using an AccQ-Tag Ultra column (2.1 × 100 mm; Waters).
Reactions were performed with substrate turnovers ranging from 10–15%. Methionine quantification was performed using a calibration curve constructed from known concentrations of methionine standard. The measurements were performed in duplicate for both proteins. The Km and kcat values were determined from reaction rate versus substrate concentration plots using the GraFit program [35].
Active site titration of Avi-GCPII and rhGCPII
Mixtures of an appropriate amount of enzyme (to achieve 10–15% substrate turnover for the non-inhibited reaction) and the tight-binding inhibitor 2-PMPA (final concentration ranging from 0–500 nM) in reaction buffer (20 mM NaCl, 20 mM MOPS, pH 7.4) were pre-incubated for 10 min at 37°C. Reactions were started by addition of Ac-Asp-Met (final concentration 100 µM) and stopped after 15 minutes by addition of ice-cold 0.2 M sodium borate, pH 10.0, and 4 µM L-glutamate (internal standard). Reaction volumes and determination of free L-methionine were the same as for the carboxypeptidase activity assay.
The active site concentrations and Ki' values were determined from proportional velocity versus 2-PMPA concentration plots using the GraFit program [35].
Determination of the inhibition constant of 2-PMPA for Avi-GCPII and rhGCPII
The Ki for 2-PMPA was calculated from the Ki' value obtained during active site titration measurements following the formula: Ki = Ki'/(1+[S]/Km) where [S] stands for Ac-Asp-Met concentration and Km for the Michaelis constant of the enzyme towards Ac-Asp-Met.
Amino acid analysis
The total protein concentration of Avi-GCPII and rhGCPII stock solution was determined by analysis performed on a Biochrom30 amino acid analyzer (Biochrom) following the manufacturer’s protocol.
Results
Preparation of the stable insect cell line expressing biotinylated Avi-GCPII
The DNA encoding AviTEV tag was prepared from oligonucleotides by the gene fusion approach and inserted into the plasmid pMNAEXST, which enables inducible expression in Drosophila Schneider 2 (S2) cells [19]. Cells stably transfected with this plasmid expressed rhGCPII N-terminally fused with AviTEV tag (Avi-GCPII) into the medium (the composition of the tag is schematically shown in Figure 1, Panel A). The cells were subsequently transfected with a plasmid encoding E. coli biotin-protein ligase (BirA). In order to analyze the efficiency of Avi-GCPII in vivo biotinylation, three different plasmids were prepared encoding BirA localized in cytoplasm (pMT/BirA), within the ER (pMT/BiP/BirA/KDEL), and secreted into the media (pMT/BiP/BirA). Two peptide signal sequences for BirA targeting were used: the BiP sequence for the secretory pathway and the KDEL sequence for retaining BirA within the ER.
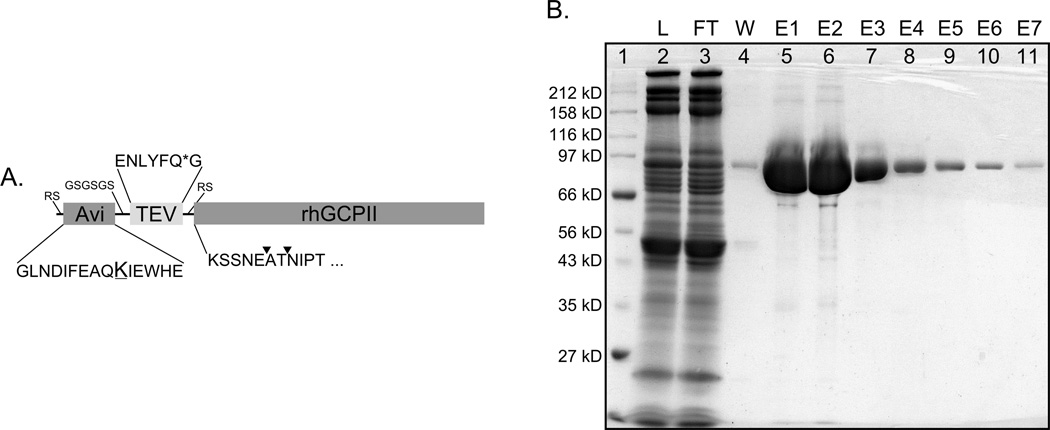
Figure 1. Schematic representation of Avi-GCPII and analysis of its purification.
Panel A: Schematic representation of Avi-GCPII with amino acid sequences detailed for all AviTEV tag parts; Avi – sequence of 15 amino acids known as AviTag, which is specifically recognized by biotin-protein ligase and biotinylated on the ε-amino group of the lysine residue (underlined); TEV – protein sequence specifically recognized by TEV protease (cleavage site is marked with *); rhGCPII - extracellular portion of GCPII consisting of amino acids 44–750. ▼ - identified cleavage site within Avi-GCPII recognized by an unknown host protease which is co-purified with Avi-GCPII. Spacer sequence and amino acids introduced during molecular cloning are depicted in a smaller font size. Panel B: Analysis of Avi-GCPII purification on Streptavidin Mutein Matrix. Equilibrated concentrated conditioned medium from S2 cells was mixed with resin and incubated overnight at 4°C. The resin was separated from the medium on a gravity-flow column, and purified protein was eluted with an excess of D-biotin. Individual fractions obtained during purification were subsequently analyzed by SDS-PAGE and stained by Coomassie blue. 1. Molecular weight marker; 2. Load; 3. Flow-through; 4. Wash; 5–11. Elutions 1–7. 4 µl of the sample was loaded to each lane. Detailed descriptions of individual fractions can be found in Materials and Methods.
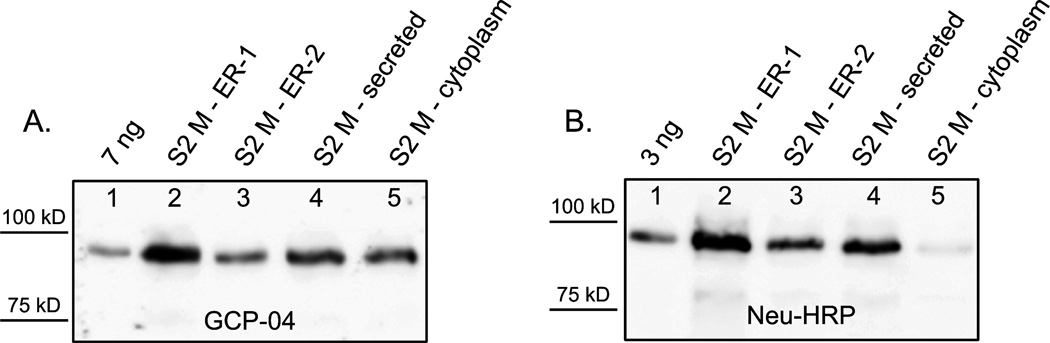
The expression of Avi-GCPII in the cell lines stably transfected with BirA and Avi-GCPII was monitored by the Western blot (see Figure 2). To determine the biotinylation efficiency, two visualizing agents were used: the monoclonal antibody GCP-04, which specifically recognizes the extracellular portion of GCPII (rhGCPII) [37], and NeutrAvidin conjugated with horseradish peroxidase (Neu-HRP), which visualizes only the biotinylated fraction of Avi-GCPII (and potential other endogenously biotinylated proteins). The cytoplasmic localization of BirA led to a very low biotinylation yield. On the other hand, the localization of BirA either within the ER or the secretory pathway caused the effective biotinylation of Avi-GCPII. Densitometry analysis of the Western blot revealed that BirA localization within the ER resulted in a slightly higher biotinylation yield compared to BirA localized in the secretory pathway (the ratio of the signals of biotinylated and total Avi-GCPII for BirA localized within the ER was approximately 15% higher than for BirA localized in secretory pathway). Based on these results, the cell line containing BirA within the ER (stable cell line 1) was used for large-scale expression of Avi-GCPII.
Figure 2. Comparison of the biotinylation efficiency of BirA in different cellular compartments of S2 cells.
Conditioned media from the cells, expressing Avi-GCPII and BirA (targeted to three different cellular compartments - cytoplasmic, secreted, or within the ER), were harvested 3 days post-induction and analyzed on Western blot. Western blots were visualized with the monoclonal antibody GCP-04, which recognizes total Avi-GCPII (Panel A), and Neu-HRP, which recognizes only the biotinylated Avi-GCPII (Panel B). 1. 7 ng (Panel A) or 3 ng (Panel B) of Avi-GCPII standard; 2. medium from cells with BirA localized within the ER (stable cell line 1); 3. medium from cells with BirA localized within the ER (stable cell line 2); 4. medium from cells with secreted BirA; 5. medium from cells with BirA localized in cytoplasm. Lanes 2–5 in both panels contained identical volumes of the analyzed samples.
Previous work suggested that the addition of exogenous D-biotin into the cell medium might influence the BirA biotinylation efficiency [15]. However, this approach did not lead to any significant improvement in biotinylation efficiency in our hands (data not shown).
Avi-GCPII is successfully purified on Streptavidin Mutein Matrix
The purification protocol given by the manufacturer of Streptavidin Mutein Matrix (Roche) was changed for the purpose of Avi-GCPII purification. The appropriate amount of resin, buffer composition and its ionic strength, and incubation times were all optimized to provide the highest possible yield while sustaining high homogeneity of the purified protein.
After optimization of the purification protocol, large-scale purification of concentrated conditioned media was performed (see Materials and Methods for details). Fractions collected during purification were analyzed and their purity assessed by reducing SDS-PAGE (see Figure 1, Panel B for analysis of fractions from first purification). To increase the yield of purified Avi-GCPII, the affinity purification was performed in two rounds, with the flow-through from the first round used as an input for the second. The yields from the first and second round of purification were 4.3 mg and 0.7 mg of Avi-GCPII protein, respectively. The homogeneity of the purified protein was more than 95% and more than 90% of the protein was present in the first two elution fractions (E1, E2). The amount of Avi-GCPII in the cell conditioned medium before concentration and purification was estimated by densitometry analysis to be approximately 20 mg of overall and 12 mg of biotinylated Avi-GCPII in 0.8 L of the cell culture, suggesting that the purification yield was approximately 40%.
The purified Avi-GCPII was characterized and compared to its non-tagged version, the rhGCPII. We analyzed the influence of AviTEV tag on Avi-GCPII activity and crystallizability. We also investigated the possibility of removing the AviTEV tag with TEV protease.
The Avi-GCPII structure is not influenced by the attachment of the AviTEV tag
Purified Avi-GCPII was concentrated to 10 mg/mL and co-crystallized with a potent GCPII inhibitor, 2-(phosphonomethyl)-pentanedioic acid (2-PMPA), using conditions identical to those used for rhGCPII crystallization [38]. The Avi-GCPII structure was obtained by the structural refinement of isomorphous structure of rhGCPII (pdb entry 2PVW) at 1.60 Å resolution, using the X-ray data collected for Avi-GCPII crystals. The new structure was deposited in the Protein Data Bank (pdb entry 3RBU). Detailed information about data collection and refinement statistics are summarized in Table I.
Table I.
Data collection and refinement statistics for the Avi-GCPII structure (3RBU)
| Data Collection Statistics | |
| wavelength (Å) | 1.000 |
| temperature (K) | 100 |
| space group | I222 |
| unit cell parameters a, b, c (Å) | 101.3, 130.6, 158.9 |
| resolution limits (Å) | 30.0-1.60 (1.66-1.60)* |
| number of unique reflections | 136479 (12595) |
| redundancy | 7.0 (4.4) |
| completeness (%) | 99.0 (91.9) |
| I/meanI | 19.8 (2.4) |
| Rmerge | 0.081 (0.517) |
| Refinement Statistics | |
| resolution limits (Å) | 28.46-1.60 (1.64-1.60) |
| total no. of reflections | 134416 |
| no. of reflections in working | 133053 (8872) |
| no. of reflections in test set | 1363 (68) |
| R | 0.158 (0.251) |
| Rfree | 0.182 (0.255) |
| total no. of non-H atoms | 6770 |
| no. of protein atoms | 6050 |
| no. of inhibitor atoms | 14 |
| no. of ions | 4 |
| no. of water molecules | 702 |
| average B-factor (Å2) | 30.31 |
| protein atoms | 28.32 |
| waters | 42.11 |
| inhibitor | 28.00 |
| RMS deviations | |
| bond lengths (Å) | 0.018 |
| bond angles (deg) | 1.689 |
| Ramachandran plot (%) | |
| favoured region | 97.8% |
| allowed region | 2.05% |
| disallowed region | 0.15% (Gly335) |
| missing residues | T1-T32, 44–54, 541–544, 655 |
values in parentheses correspond to the highest resolution shell
The amino acids which belong to the AviTEV tag are denoted as Txy.
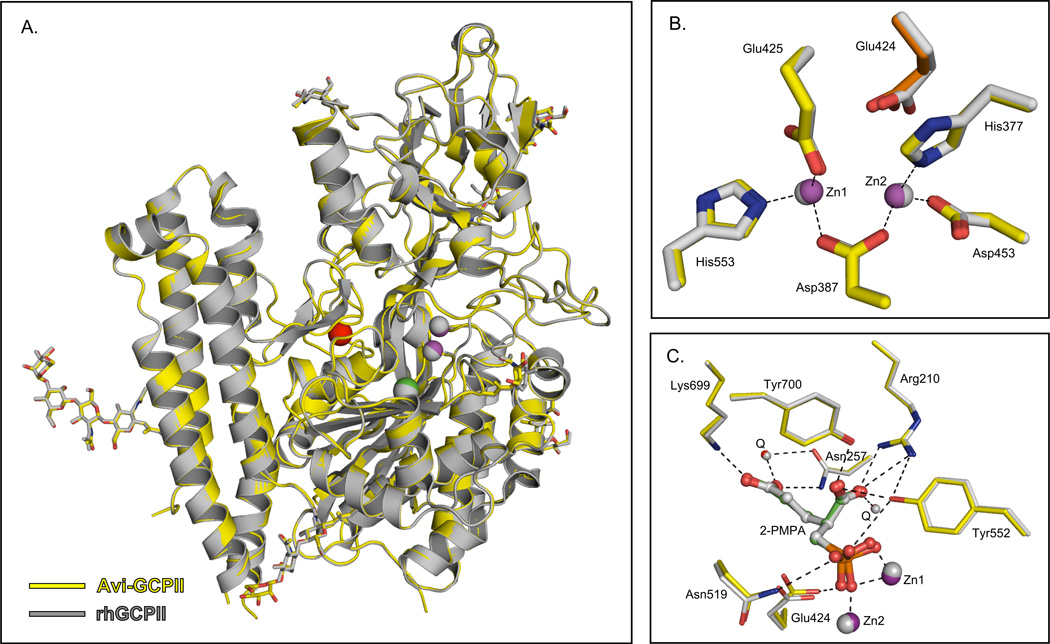
The structure is virtually identical to that of rhGCPII (pdb entry 2PVW) with r.m.s.d. of 0.14 Å for 649 equivalent Cα atoms (see Figure 3, Panel A). In particular, residues participating in Zn2+ chelation and inhibitor binding retain their spatial arrangement (see Figure 3, Panels B and C). The AviTEV tag and the first 11 residues of rhGCPII are not visible in the electron density map, suggesting a high flexibility of the N-terminal part of the protein.
Figure 3. Superposition of Avi-GCPII (3RBU) and rhGCPII (2PVW) structure.
Panel A: The overall fold of Avi-GCPII and rhGCPII is identical. The protein backbone is shown in cartoon representation, sugar units in stick representation, and ions as spheres (Zn2+– magenta, Cl− – green, Ca2+ – red for Avi-GCPII). Avi-GCPII is shown in yellow, rhGCPII in gray (including ions and sugars). Sugar oxygens and nitrogens are shown in red and blue, respectively. Panel B: Superposition of the active site residues/atoms. Avi-GCPII is shown in yellow (Zn2+ ions as magenta spheres) and rhGCPII in gray (including ions). Oxygen atoms are depicted in red and nitrogen atoms in blue. Coordination of the active site zinc ions in the Avi-GCPII structure is shown as dashed lines. Panel C: 2-PMPA interactions in the Avi-GCPII/rhGCPII active sites. Avi-GCPII is shown in yellow [Zn2+ ions as magenta spheres, 2-PMPA in green ball-and-stick representation, waters Q as red spheres]; rhGCPII in gray (including ions, 2-PMPA, and waters). Oxygen atoms are depicted in red, nitrogen atoms in blue, and the phosphorus atom in orange. Direct H-bonding and coordination to Zn2+ ions in the Avi-GCPII structure are shown as dashed lines. The figures were created using the PyMOL Molecular Graphics System [43].
Avi-GCPII activity is not affected by the presence of the AviTEV tag
As a prerequisite for a careful quantitative comparison of Avi-GCPII and rhGCPII enzymatic activity, the total protein concentrations and the active site concentrations of both enzymes were determined by quantitative amino acid analysis and active site titration by a specific inhibitor. Relative portion of the enzymatically active molecules in the stock solution was assessed to be 102 ± 2% for Avi-GCPII and 72 ± 2% for the rhGCPII, respectively. The rhGCPII was purified according to the protocol established in our laboratory [19].
Kinetic constants characterizing cleavage of the prototypical GCPII substrate Ac-Asp-Glu [28], the less-efficient substrate Ac-Asp-Met [19], and the potent inhibitor 2-PMPA [39] were determined in order to characterize the effect of AviTEV tag on Avi-GCPII enzymatic activity. As shown in Table II, all kinetic constants are comparable for both Avi-GCPII and rhGCPII, indicating that AviTEV tag does not affect the enzymatic characteristics of Avi-GCPII.
Table II.
Direct comparison of Avi-GCPII and rhGCPII enzymatic activity.
| Avi-GCPII | rhGCPII | |||||
|---|---|---|---|---|---|---|
| Substrate |
Km [µmol/L] |
kcat [per second] |
kcat/Km [105/s.(mol/L)] |
Km [µmol/L] |
kcat [per second] |
kcat/Km [105/s.(mol/L)] |
| Ac-Asp-Glu | 0.53 ± 0.08* | 0.46 ± 0.01* | 8.66 ± 1.4 | 1.06 ± 0.18* | 0.41 ± 0.02* | 3.88 ± 1.06 |
| Ac-Asp-Met | 20.8 ± 3.3§ | 0.29 ± 0.01§ | 0.14 ± 0.02 | 25.2 ± 4.0§ | 0.34 ± 0.02§ | 0.13 ± 0.02 |
| Inhibitor | Ki [pmol/L] | Ki [pmol/L] | ||||
| 2-PMPA | 370 ± 80§ | 540 ± 120§ | ||||
measurements were performed by radioactive assay
measurements were performed by HPLC assay
Avi-GCPII is ineffectively processed by TEV protease
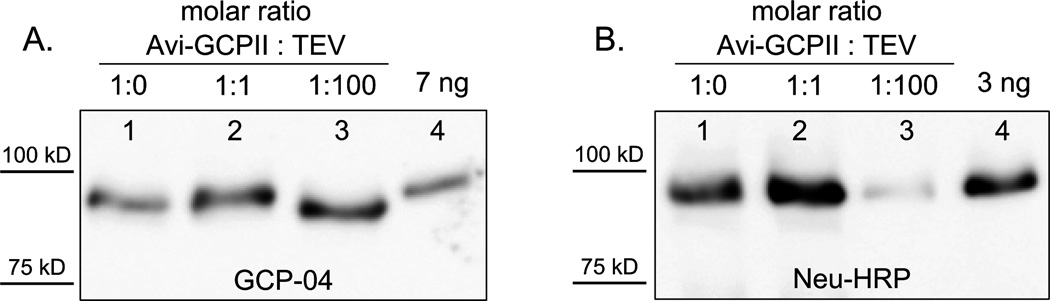
The AviTEV tag was designed with the specific cleavage site for the tobacco etch virus (TEV) protease to enable the removal of the tag from Avi-GCPII (see Figure 1, Panel A). TEV protease is known as one of the most specific proteolytic enzymes and is widely used for the processing of recombinant proteins [40]. The purified, tagged TEV protease [His-TEV(S219V)-Arg] was incubated with purified Avi-GCPII in two different ratios (equimolar and with a 100-fold molar excess of TEV protease to Avi-GCPII). Cleavage was performed in three different buffers (see Materials and Methods). The results were identical for all buffers used, and for simplicity, only the experiment using TBS is shown in Figure 4. It is visible from the Western blot analysis that a 100-fold molar excess of TEV protease is needed to efficiently remove the AviTEV tag, suggesting that the cleavage by TEV protease proceeds inefficiently, and its use in large-scale experiments would not be feasible.
Figure 4. Avi-GCPII cleavage by TEV protease.
Purified Avi-GCPII was mixed with TEV protease in different ratios. The reactions were incubated overnight at 4°C and then analyzed using Western blots. Blots were visualized with the monoclonal antibody GCP-04, which recognizes total Avi-GCPII (Panel A), and Neu-HRP, which recognizes only biotinylated Avi-GCPII (Panel B). 1. purified Avi-GCPII without TEV protease; 2. equimolar mixture of Avi-GCPII and TEV protease; 3. Avi-GCPII with a 100-fold molar excess of TEV protease; 4. 7 ng (panel A) or 3 ng (Panel B) of Avi-GCPII standard. Lanes 1–3 in both panels contained identical volumes of the analyzed samples.
Avi-GCPII is processed by host protease
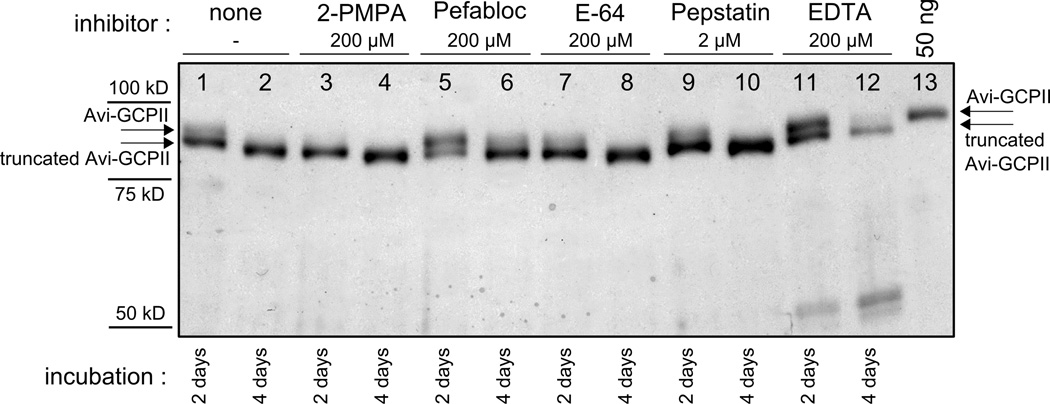
Interestingly, a cleavage within the N-terminal part of purified Avi-GCPII itself was observed. N-terminal sequencing analysis identified two cleavage sites within the Avi-GCPII molecule (see Figure 1, Panel A). A series of experiments was performed in order to determine whether the processing is due to auto-proteolysis or hydrolysis by some host contaminating protease. The data shown in Figure 5 suggested that Avi-GCPII is probably cleaved by a contaminant serine protease, since addition of a specific serine protease inhibitor, Pefabloc, slowed down the processing. The intensive degradation of Avi-GCPII observed in the experiment with EDTA was probably caused by chelation of zinc and calcium ions within Avi-GCPII and the protein’s subsequent structural destabilization by which more cleavage sites became accessible for the contaminant host protease. This contaminant protease is probably not co-purified during the standard purification of rhGCPII since no N-terminal cleavage was observed for rhGCPII (data not shown).
Figure 5. Inhibition of Avi-GCPII cleavage by protease class-specific inhibitors.
Purified Avi-GCPII was incubated at room temperature, and samples were taken after 2 days (lanes 1,3,5,7,9,11) and 4 days (lanes 2,4,6,8,10,12) and analyzed on silver-stained reducing SDS-PAGE. Inhibitors tested: 2-PMPA (specific GCPII inhibitor; final concentration 200 µM; lanes 3,4), Pefabloc SC (serine protease inhibitor; final concentration 200 µM; lanes 5,6), E-64 (cysteine protease inhibitor; final concentration 200 µM; lanes 7,8), Pepstatin A (aspartate protease inhibitor; final concentration 2 µM; lanes 9,10), EDTA (metalloprotease inhibitor; final concentration 200 µM; lanes 11,12). The experiment without inhibitor was also performed (lanes 1, 2). 3 µl of the sample was loaded to each lane. 50 ng of purified Avi-GCPII was loaded to line 13 as a standard.
Additional experiments showed that the cleavage is also effectively slowed at 4°C and no hydrolysis of purified Avi-GCPII was detected after 2 days incubation at this temperature. To ensure the integrity of our protein preparation, the potential degradation of Avi-GCPII was examined under the conditions of enzymatic assays and protein crystal growth. In both these conditions no Avi-GCPII processing whatsoever was observed (data not shown).
Discussion
Simple, efficient, and robust recombinant protein expression and purification is indispensable for many areas of molecular biology, from structural genomics to protein-protein interaction studies. Even though expression and purification in prokaryotic expression systems is reasonably characterized, the eukaryotic systems still remain a challenge. We aimed to develop a well-functioning one-step purification procedure utilizing a biotin acceptor peptide (BAP) as a purification tag, for its well-characterized properties of binding to streptavidin and its analogues, and Drosophila S2 cells as an expression system, for their relatively easy handling, possibility of large-scale growth in suspension, and high expression yields. As a target protein, we chose glutamatecarboxypeptidase II. Even though a purification protocol for this protein has already been developed[19], it is quite laborious and not much versatile. Even a single amino acid mutation influences the purification yield and requires some optimization [41] and for the GCPII homologs, however close, the method needs to be re-developed. [27]. We decided to locate the AviTEV-tag on the N-terminus of rhGCPII since its 3D structure indicates that it is more flexible than C-terminus of the molecule.
This purification set-up requires preparation of stable cell line expressing both E. coli biotin-protein ligase (BirA) and the target recombinant protein. Previously published results describing the influence of BirA cellular localization (cytoplasmic, within the ER, or in the secretory pathway) on the biotinylation efficiency of secreted recombinant proteins fused with BAP were not complete. For instance, only two subcellular localizations were compared in parallel, and the results varied significantly between mammalian [10–11] and insect systems [15]. Therefore, we decided to prepare three stable insect cell lines with BirA localized in cytoplasm (BirA), within the ER (BiP-BirA-KDEL), or in secretory pathway (BiP-BirA), and compared them in terms of the biotinylation efficiencies of secreted Avi-GCPII.
The cytosolic localization of BirA was shown to be the least effective (see Figure 2). This finding contradicts the results of Yang et al., who determined that cytosolic BirA had a higher biotinylation yield than the secreted form of BirA in insect cells [15]. This discrepancy might be due to the use of different peptide leading sequences for the secretion of BirA and target protein (BiP versus HLA-DR β leader sequence), by different localization of AviTag in the target molecule (N-terminal versus C-terminal), or by less obvious properties of a target protein or expression system. On the other hand, localizations of BirA within the ER and in the secretory pathway led to efficient Avi-GCPII biotinylation, indicating that the biotinylation in both cases probably proceeds within the ER. Only a slightly better relative biotinylation efficiency was determined for BirA localized within the ER compared to BirA localized in the secretory pathway. These data were consistent with the observations of Barat et al. in mammalian cells (even though we determined a much smaller difference between cells with secreted BirA and BirA retained within the ER) [11]. Additionally, it should be noted that using presented protocol for creation of stable S2 cell transfectants we obtained a polyclonal cell lines. When the transfection was done in duplicates, it showed higher variability in overall expression yields of biotinylated Avi-GCPII (Figure 2, lanes 2–3) than was observed between cell lines with differently localized BirA (Figure 2, lanes 2, 4). These findings suggest that the careful optimization of the expression system is at least as important as proper localization of biotin ligase within the cell.
The purification procedure itself was optimized to provide the highest possible yield while preserving high homogeneity of the purified protein. Surprisingly, the use of non-dialyzed conditioned medium as a starting material for purification was shown to be the best option. Dialysis of the medium followed by optimization of ionic strength and/or addition of exogenous D-biotin resulted in lower yields and/or purity (data not shown). This effect might have been caused by some unknown additive present in the SF900II medium. We have also observed that by concentrating the conditioned medium prior to affinity purification, we improved greatly the purification yield. The explanation of this observation is not straightforard: it might be caused by increasing the effective concentration of both Avi-GCPII and mutein resin beads within the incubation mixture. Similar behaviour was also observed during purifications of several Avi-GCPII mutants and homologues (data not shown).
Tags covalently attached to purified protein are useful tools for subsequent visualization or immobilization and are often retained with the target protein. In the case of AviTEV tag, the covalently attached biotin enables easy visualization, immobilization, or specific uptake of the target protein. Since the AviTEV tag is not effectively cleaved from its fusion partner by TEV protease, we focused on a detailed analysis of its influence on the biochemical features of rhGCPII.
We were able to crystallize Avi-GCPII under conditions identical to those used for the non-tagged enzyme, suggesting that the AviTEV tag does not influence the ability of Avi-GCPII to crystallize. In fact, we showed that the structure of Avi-GCPII is virtually identical to that of rhGCPII. A lack of interpretable electron density for the AviTEV tag indicates that this motif is likely disordered. Furthermore, we solved a structure of the AviTEV-tagged extracellular part of Naaladase L (amino acids 28–740), a GCPII homolog, in which the AviTEV tag was also not visible (manuscript in preparation). Those findings, together with a previously reported study of the structure of maltodextrin-binding protein fused with biotin acceptor peptide [42], supports the conclusion that the presence of AviTEV tag does not significantly affect the 3D structure of its fusion partner.
To determine the effect of AviTEV tag on Avi-GCPII enzymatic activity, kinetic constants for cleavage of GCPII substrates Ac-Asp-Glu and Ac-Asp-Met and the inhibition constant for the potent GCPII inhibitor 2-PMPA were determined for both Avi-GCPII and rhGCPII (see Table II). The slightly improved catalytic efficiency of Avi-GCPII compared to rhGCPII might be explained by the milder conditions of the purification procedure used for Avi-GCPII. This notion was also supported by active site titration measurements. Purification via AviTEV tag led to Avi-GCPII molecules that were nearly 100% enzymatically active, while the standard four-step purification method yielded only 72% enzymatically active rhGCPII. The Lower relative representation of enzymatically active enzyme in rhGCPII preparation compared to Avi-GCPII may be caused by complexity of purification (4 steps versus 1 step) and overall duration of purification procedure (approximately 5 days versus 2 days) in the former case. The inactive fraction of rhGCPII might partially bind the substrate/inhibitor and thus lower the observed catalytic efficiency of rhGCPII. The kcat values for rhGCPII presented in Table II are not quite in agreement with previously published data [19, 27]. This discrepancy might be caused by different methods of protein concentration determination (active site titration versus Bradford assay). Taken together, we can conclude that in this purification set-up AviTEV tag does not compromise the enzymatic activity of its fusion partner.
Even though the N-terminus part of Avi-GCPII molecule is disordered and accessible for host contaminant protease, the TEV protease showed to be very inefficient in cleaving off the tag. The poor processing may be caused by the arginine residue located at the P2’ position of the TEV cleavage site in Avi-GCPII molecule (see Figure 1, Panel A). In support of this hypothesis, we checked the enzymatic activity of the same preparation of TEV protease and determined it successfully cleaved different fusion protein containing TEV cleavage site (data not shown).
A potential versatility of this purification set-up was confirmed by successful purification of Avi-GCPII site-directed mutants and also several of its human paralogs and animal orthologs (data not shown). Furthermore, a secreted version of a different enzyme, carbonic anhydrase IX, with the AviTEV tag on its C-terminus was also successfully expressed and purified (P. Mader et al.; manuscript in preparation). It might be relevant to mention that carbonic anhydrase XI is a type I transmembrane protein, and, consequently, the topology of the AviTEV-tag location was the same as for Avi-GCPII)
In conclusion, we prepared a cell line of stably transfected Drosophila S2 cells with BirA localized within the ER and compared it with stable cell lines with the cytosolic and secreted forms of BirA. We showed that BirA localizations within both the ER and the secretory pathway lead to efficient biotinylation of target protein while the localization within ER showed slightly better biotinylation yield. Using this stable cell line we expressed, purified and characterized recombinant human GCPII. We achieved milligrams yields of pure protein per litre of culture and showed that AviTEV tag does not compromise the features of its fusion partner. Since the interaction is independent of target protein properties, we believe this purification set-up may represent a versatile and facile method for efficient expression and purification of secreted recombinant proteins in Drosophila S2 cells.
A method of expression of biotinylated proteins in insect cells is described.
Biotin protein ligase has to colocalize with the target for proper biotinylation.
Tagged and biotinylated protein crystallizes after single step purification.
Tag does not compromise enzymatic activity of the protein.
Acknowledgements
The authors thank Karolína Šrámková for excellent technical assistance, Klára Hlouchová for preparation of rhGCPII protein, Zdeněk Voburka for N-terminal sequencing analysis, Martin Hradílek for synthesis of tested N-acetylated dipeptides, Radko Souček for expert HPLC analysis, and Hillary Hoffman for preparation of TEV protease and insightful proofreading of the manuscript. The plasmid encoding TEV protease was a kind gift from David Waugh (National Cancer Institute at Frederick, NIH, MD, USA), and the inhibitor 2-PMPA was a kind gift from Barbara Slusher (School of Medicine, John Hopkins University, MD, USA).
This work (performed under the research project Z4 055 905) was supported by grants 1M0508 and LC 512 from the Ministry of Education of Czech Republic, EMBO Installation grant #1978 (C.B), the IBT institutional support (AV0Z50520701), by research support from Guilford Pharmaceuticals, and in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (J.L.). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng38.
Abbreviations
- GCPII
glutamate carboxypeptidase II
- rhGCPII
extracellular portion of GCPII consisting of amino acids 44–750
- Avi-GCPII
molecule of rhGCPII fused at its N-terminus with AviTEV tag
- TEV protease
Tobacco Etch Virus protease
- Ac-Asp-Glu
N-acetyl-L-aspartyl-L-glutamate
- Ac-Asp-Met
N-acetyl-L-aspartyl-L-methionine
- 2-PMPA
2-(phosphonomethyl)-pentanedioic acid
- BAP
biotin acceptor peptide
- BirA
biotin-protein ligase from E. coli
- ER
endoplasmic reticulum
- SDS-PAGE
sodium dodecyl sulphate polyacrylamide gel electrophoresis
- FBS
fetal bovine serum
- PBS
phosphate buffered saline
- TBS
Tris buffered saline
- Tris
tris(hydroxymethyl)aminomethane
- HPLC
high-performance liquid chromatography
- HRP
horseradish peroxidase
- Neu
NeutrAvidin
- S2 cells
Schneider 2 cells
- r.m.s.d.
root mean square deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Green NM. Avidin. 1. The Use of (14-C)Biotin for Kinetic Studies and for Assay. Biochem J. 1963;89:585–591. doi: 10.1042/bj0890585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qureshi MH, Wong SL. Design, production, and characterization of a monomeric streptavidin and its application for affinity purification of biotinylated proteins. Protein Expr Purif. 2002;25:409–415. doi: 10.1016/s1046-5928(02)00021-9. [DOI] [PubMed] [Google Scholar]
- 3.Wu SC, Wong SL. Engineering soluble monomeric streptavidin with reversible biotin binding capability. J Biol Chem. 2005;280:23225–23231. doi: 10.1074/jbc.M501733200. [DOI] [PubMed] [Google Scholar]
- 4.Lane MD, Rominger KL, Young DL, Lynen F. The Enzymatic Synthesis of Holotranscarboxylase from Apotranscarboxylase and (+)-Biotin. Ii. Investigation of the Reaction Mechanism. J Biol Chem. 1964;239:2865–2871. [PubMed] [Google Scholar]
- 5.Lane MD, Young DL, Lynen F. The Enzymatic Synthesis of Holotranscarboxylase from Apotranscarboxylase and (+)-Biotin. I. Purification of the Apoenzyme and Synthetase; Characteristics of the Reaction. J Biol Chem. 1964;239:2858–2864. [PubMed] [Google Scholar]
- 6.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N Y) 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 8.de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy S, Tsao KL, Waugh DS. Site-specific, enzymatic biotinylation of recombinant proteins in Spodoptera frugiperda cells using biotin acceptor peptides. Anal Biochem. 1998;262:122–128. doi: 10.1006/abio.1998.2770. [DOI] [PubMed] [Google Scholar]
- 10.Predonzani A, Arnoldi F, Lopez-Requena A, Burrone OR. In vivo site-specific biotinylation of proteins within the secretory pathway using a single vector system. BMC Biotechnol. 2008;8:41. doi: 10.1186/1472-6750-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barat B, Wu AM. Metabolic biotinylation of recombinant antibody by biotin ligase retained in the endoplasmic reticulum. Biomol Eng. 2007;24:283–291. doi: 10.1016/j.bioeng.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driegen S, Ferreira R, van Zon A, Strouboulis J, Jaegle M, Grosveld F, Philipsen S, Meijer D. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res. 2005;14:477–482. doi: 10.1007/s11248-005-7220-2. [DOI] [PubMed] [Google Scholar]
- 13.Sung K, Maloney MT, Yang J, Wu C. A novel method for producing monobiotinylated, biologically active neurotrophic factors: An essential reagent for single molecule study of axonal transport. J Neurosci Methods. 2011 doi: 10.1016/j.jneumeth.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juzumiene D, Chang CY, Fan D, Hartney T, Norris JD, McDonnell DP. Single-step purification of full-length human androgen receptor. Nucl Recept Signal. 2005;3:e001. doi: 10.1621/nrs.03001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, Jaramillo A, Shi R, Kwok WW, Mohanakumar T. In vivo biotinylation of the major histocompatibility complex (MHC) class II/peptide complex by coexpression of BirA enzyme for the generation of MHC class II/tetramers. Hum Immunol. 2004;65:692–699. doi: 10.1016/j.humimm.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt TG, Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc. 2007;2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 17.Skerra A, Schmidt TG. Applications of a peptide ligand for streptavidin: the Strep-tag. Biomol Eng. 1999;16:79–86. doi: 10.1016/s1050-3862(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 18.Lichty JJ, Malecki JL, Agnew HD, Michelson-Horowitz DJ, Tan S. Comparison of affinity tags for protein purification. Protein Expr Purif. 2005;41:98–105. doi: 10.1016/j.pep.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Barinka C, Rinnova M, Sacha P, Rojas C, Majer P, Slusher BS, Konvalinka J. Substrate specificity, inhibition and enzymological analysis of recombinant human glutamate carboxypeptidase II. J Neurochem. 2002;80:477–487. doi: 10.1046/j.0022-3042.2001.00715.x. [DOI] [PubMed] [Google Scholar]
- 20.Barinka C, Sacha P, Sklenar J, Man P, Bezouska K, Slusher BS, Konvalinka J. Identification of the N-glycosylation sites on glutamate carboxypeptidase II necessary for proteolytic activity. Protein Sci. 2004;13:1627–1635. doi: 10.1110/ps.04622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buhler P, Wolf P, Elsasser-Beile U. Targeting the prostate-specific membrane antigen for prostate cancer therapy. Immunotherapy. 2009;1:471–481. doi: 10.2217/imt.09.17. [DOI] [PubMed] [Google Scholar]
- 22.Elgamal AA, Holmes EH, Su SL, Tino WT, Simmons SJ, Peterson M, Greene TG, Boynton AL, Murphy GP. Prostate-specific membrane antigen (PSMA): current benefits and future value. Semin Surg Oncol. 2000;18:10–16. doi: 10.1002/(sici)1098-2388(200001/02)18:1<10::aid-ssu3>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh A, Heston WD. Tumor target prostate specific membrane antigen (PSMA) and its regulation in prostate cancer. J Cell Biochem. 2004;91:528–539. doi: 10.1002/jcb.10661. [DOI] [PubMed] [Google Scholar]
- 24.Neale JH, Olszewski RT, Gehl LM, Wroblewska B, Bzdega T. The neurotransmitter N-acetylaspartylglutamate in models of pain, ALS, diabetic neuropathy, CNS injury and schizophrenia. Trends Pharmacol Sci. 2005;26:477–484. doi: 10.1016/j.tips.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Conway RE, Petrovic N, Li Z, Heston W, Wu D, Shapiro LH. Prostate-specific membrane antigen regulates angiogenesis by modulating integrin signal transduction. Mol Cell Biol. 2006;26:5310–5324. doi: 10.1128/MCB.00084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu T, Jabbes M, Nedrow-Byers JR, Wu LY, Bryan JN, Berkman CE. Detection of prostate-specific membrane antigen on HUVECs in response to breast tumor-conditioned medium. Int J Oncol. 2011;38:1349–1355. doi: 10.3892/ijo.2011.946. [DOI] [PubMed] [Google Scholar]
- 27.Hlouchova K, Barinka C, Klusak V, Sacha P, Mlcochova P, Majer P, Rulisek L, Konvalinka J. Biochemical characterization of human glutamate carboxypeptidase III. J Neurochem. 2007;101:682–696. doi: 10.1111/j.1471-4159.2006.04341.x. [DOI] [PubMed] [Google Scholar]
- 28.Robinson MB, Blakely RD, Couto R, Coyle JT. Hydrolysis of the brain dipeptide N-acetyl-L-aspartyl-L-glutamate. Identification and characterization of a novel N-acetylated alpha-linked acidic dipeptidase activity from rat brain. J Biol Chem. 1987;262:14498–14506. [PubMed] [Google Scholar]
- 29.Rasband WS. ImageJ. U. S. National Institutes of Health. Bethesda, Maryland, USA: 1997–2008. http://rsb.info.nih.gov/ij/ [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Barinka C, Rovenska M, Mlcochova P, Hlouchova K, Plechanovova A, Majer P, Tsukamoto T, Slusher BS, Konvalinka J, Lubkowski J. Structural insight into the pharmacophore pocket of human glutamate carboxypeptidase II. J Med Chem. 2007;50:3267–3273. doi: 10.1021/jm070133w. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leatherbarrow RJ. GraFit Version 5. Horley, UK: Erithacus Software Limited; 2001. [Google Scholar]
- 36.Woodward C, Henderson JK, Wielgos T. High-speed amino acid analysis (AAA) on 1.8 µm reversed-phase (RP) columns. Agilent Technologies App. Note. 2007:5989–6297. EN. [Google Scholar]
- 37.Sacha P, Zamecnik J, Barinka C, Hlouchova K, Vicha A, Mlcochova P, Hilgert I, Eckschlager T, Konvalinka J. Expression of glutamate carboxypeptidase II in human brain. Neuroscience. 2007;144:1361–1372. doi: 10.1016/j.neuroscience.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 38.Barinka C, Starkova J, Konvalinka J, Lubkowski J. A high-resolution structure of ligand-free human glutamate carboxypeptidase II. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:150–153. doi: 10.1107/S174430910700379X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson PF, Cole DC, Slusher BS, Stetz SL, Ross LE, Donzanti BA, Trainor DA. Design, synthesis, and biological activity of a potent inhibitor of the neuropeptidase N-acetylated alpha-linked acidic dipeptidase. J Med Chem. 1996;39:619–622. doi: 10.1021/jm950801q. [DOI] [PubMed] [Google Scholar]
- 40.Phan J, Zdanov A, Evdokimov AG, Tropea JE, Peters HK, 3rd, Kapust RB, Li M, Wlodawer A, Waugh DS. Structural basis for the substrate specificity of tobacco etch virus protease. J Biol Chem. 2002;277:50564–50572. doi: 10.1074/jbc.M207224200. [DOI] [PubMed] [Google Scholar]
- 41.Mlcochova P, Plechanovova A, Barinka C, Mahadevan D, Saldanha JW, Rulisek L, Konvalinka J. Mapping of the active site of glutamate carboxypeptidase II by site-directed mutagenesis. FEBS J. 2007;274:4731–4741. doi: 10.1111/j.1742-4658.2007.06021.x. [DOI] [PubMed] [Google Scholar]
- 42.Bucher MH, Evdokimov AG, Waugh DS. Differential effects of short affinity tags on the crystallization of Pyrococcus furiosus maltodextrin-binding protein. Acta Crystallogr D Biol Crystallogr. 2002;58:392–397. doi: 10.1107/s0907444901021187. [DOI] [PubMed] [Google Scholar]
- 43.Schrodinger, LLC, The PyMOL Molecular Graphics System. Version 1.3r1. 2010 [Google Scholar]