Abstract
A mammalian A-type cyclin, cyclin A1, is highly expressed in testes of both human and mouse and targeted mutagenesis in the mouse has revealed the unique requirement for cyclin A1 in the progression of male germ cells through the meiotic cell cycle. While very low levels of cyclin A1 have been reported in the human hematopoietic system and brain, the sites of elevated levels of expression of human cyclin A1 were several leukemia cell lines and blood samples from patients with hematopoietic malignances, notably acute myeloid leukemia. To evaluate whether cyclin A1 is directly involved with the development of myeloid leukemia, mouse cyclin A1 protein was overexpressed in the myeloid lineage of transgenic mice under the direction of the human cathepsin G (hCG) promoter. The resulting transgenic mice exhibited an increased proportion of immature myeloid cells in the peripheral blood, bone marrow, and spleen. The abnormal myelopoiesis developed within the first few months after birth and progressed to overt acute myeloid leukemia at a low frequency (≈15%) over the course of 7–14 months. Both the abnormalities in myelopoiesis and the leukemic state could be transplanted to irradiated SCID (severe combined immunodeficient) mice. The observations suggest that cyclin A1 overexpression results in abnormal myelopoiesis and is necessary, but not sufficient in the cooperative events inducing the transformed phenotype. The data further support an important role of cyclin A1 in hematopoiesis and the etiology of myeloid leukemia.
In the process of blood formation or hematopoiesis, stringent control of the cell cycle is required for hematopoietic cells to ensure the replicative potential needed for self-renewal, as well as the differentiation into appropriate numbers of the various lineages (1). The cyclins and cyclin-dependent kinases (Cdks) are key components of the cell cycle machinery that is responsible for the progression through the G1/S and G2/M phases, as well as for the exit from the cell cycle to a quiescent G0 state (2). Several lines of evidence suggest that many blood disorders, including acute leukemia and aplastic anemia, are derived from alterations in the cell cycle control of hematopoietic stem cells (1). Differential expression of cyclins and Cdks was observed between normal and tumor cells in a murine leukemia model that was generated by injection of clonogenic Wehi-3b cells into BALB/c mice (3). In this model, the G1 cyclins and Cdks were significantly increased in tumor cells when compared with normal cells. Elevated levels of cyclin E have been observed in patients with acute myeloid leukemia (AML; ref. 4) and acute lymphoblastic leukemias (ALL; ref. 5). The combination of cyclin D1 and Cdk4 expression has been shown to be an important prognostic factor in ALL: there was a significant correlation between expression of cyclin D1 and frequency of disease recurrence in children with ALL (6).
We have previously reported (7, 8) the identification of a mammalian A-type cyclin, mouse cyclin A1, that is expressed at highest levels, if not exclusively, in the male germ line. An absolute requirement of cyclin A1 for progression through the meiotic cell cycle in spermatocytes, but not oocytes, was demonstrated by gene targeting (9). The presence of two A-type cyclins is a general feature of other higher eukaryotes: human cyclin A1 has also been identified and shown to be highly expressed in the testis and at very low levels in only the brain (10) and peripheral blood (11, 12). Of particular interest to the present study was the observation of elevated levels of cyclin A1 in several leukemia cell lines (10) and in patients with leukemia at the promyelocyte and myeloblast stages (11, 12). The aim of this report was therefore to test whether the altered expression of cyclin A1 is a cause of malignancy of myeloid cells in an animal model. Because cyclin A1 overexpression was observed in several subsets of myeloid leukemias, especially acute promyelocytic leukemia (12), we speculated that the deregulation of cyclin A1 might directly contribute to the development of myeloid leukemia. To test our hypothesis, we selectively expressed cyclin A1 cDNA in the early myeloid lineage, using a transgenic mouse model. Two types of abnormalities were observed in the transgenic mice overexpressing cyclin A1 under the direction of human cathepsin G (hCG). A low frequency of the transgenic mice developed two kinds of vascular tumors, hemangioma and angiosarcoma, which will be described elsewhere. In the present report, we describe the profound perturbation of myelopoiesis in the transgenic mice and the development of acute myeloid leukemia.
Materials and Methods
Generation of Transgenic Mice.
A 1.8-kb mouse cyclin A1 cDNA was cloned into a vector generously provided by Timothy Ley (Washington University Medical School, St. Louis), which contains hCG regulatory sequences and a portion of the coding sequence (13, 14). This construct has been used previously to drive expression of reporter constructs to the myeloid lineage and of itself does not produce any abnormalities (13, 14). Transgenic mice were generated following standard procedures used routinely in our laboratory (15, 16). Transgenic founders and their progeny were identified by Southern blotting, using sequences from cyclin A1 cDNA and hCG gene as probes. F1 progeny were obtained by breeding founder animals with B6CBAF1/J mice.
Reverse Transcription (RT)–PCR.
Total cellular RNA was extracted from bone marrow cells by using the TRI REAGENT kit following the manufacturer's instructions (Molecular Research Center, Cincinnati). Total RNA (1 μg) was analyzed by RT-PCR according to the manufacturer's instructions (CLONTECH), using primers specific for hCG and cyclin A1 cDNA or for the endogenous cyclin A1 mRNA. The primer sequences were: 5′-TAATCGCCCAGACAAGAAGAAC-3′, derived from the upstream region of cyclin A1 cDNA, and 5′-ATGATCTCCCCTGCCTCAGC-3′, from exon 1 and exon 2 junctions for the hCG-cyclin A1 transgene. For the endogenous cyclin A1 mRNA, the forward primer was the same as the upstream primer used above. The downstream primer sequence was: 5′-CCTGCTGATGTGGCCAATGAG-3′. A set of primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also used in each reaction as positive controls. The primer sequences were: 5′-TTGTCAGCAATGCATCCTGC-3′ and 5′-GCTGACAATCTTGAGTGAGTTG-3′. The PCR products were analyzed by Southern blotting using cyclin A1 cDNA or GAPDH cDNA as a probe.
Immunoblotting.
Lysates were prepared from bone marrow cells isolated from transgenic and nontransgenic mice by using TRI REAGENT according to the manufacturer's instructions. Equal amounts of lysates (200 μg) were run on a 7.5% protein gel and transferred to nitrocellulose as described (8). Cyclin A1 proteins were detected by using a rabbit anti-cyclin A1 antibody diluted 1:500, as described (9, 17).
Hematologic Measurements.
White blood cell (WBC), hemoglobin and platelet counts in peripheral blood (PB) were measured by using an automated Coulter Counter. Differential counts of PB and bone marrow (BM) were performed microscopically on Wright-Giemsa-stained smears. A minimum of 100 cells for PB and 200–400 cells for BM were examined and the percentage of blast cells was counted.
Immunohistochemistry and Immunofluorescence.
Slides of bone marrow cells were prepared by using a cytospin apparatus and fixed in methanol at −20°C for 15 min. The slides were incubated with anti-cyclin A1 antibody diluted 1:100 overnight at 4°C and were then stained with the Vectastain ABC kit (Vector Laboratories). Diaminobenzidine (DAB)-stained slides were counterstained with hematoxylin. For immunofluorescence, cytospin preparations were made as described above and blocked in PBS with 4% normal goat serum or 4% horse serum for 1 h. Primary antibodies included rabbit anti-cyclin A1 (see above) and a rat monoclonal anti-mouse 7/4 (Serotec), both diluted 1:100. The slides were incubated for 1 h at room temperature and secondarily incubated in the dark for 1 h with rhodamine-tagged goat anti-rabbit IgG (Molecular Probes) and FITC-tagged anti-rat IgG (Serotec), both diluted 1:200. Finally, the slides were incubated with 1 μM DAPI (4′,6-diamidino-2-phenylindole; Molecular Probes) for 30 min.
Fluorescence-Activated Cell Sorting (FACS)/Cell Cycle Analysis.
For flow cytometry, single cell suspensions of 1 × 105 cells prepared as above were incubated with phycoerythrin and FITC-conjugated antibodies, C-kit, Gr-1, and Mac-1 (PharMingen) for 15 min at 4°C. Cells were washed three times in 1× PBS containing 2% FCS and were applied onto a FACSCalibur flow cytometer equipped with MAC CELLQUEST software (Becton Dickinson). For cell cycle analysis, 5 × 105 cells were fixed and permeabilized. Cells were then washed twice with 1× PBS containing 10% FCS, resuspended and incubated in 1× PBS buffer containing 5 mg/ml propidium iodide (Sigma), 100 mM sodium citrate, pH 7.3, and 0.05 mg RNase A (Sigma) for 30 min at 37°C. The cell fluorescence was measured in a FACSCalibur flow cytometer.
Myeloperoxidase Staining.
Spleens from transgenic and nontransgenic mice were weighed, and frozen sections were prepared and histochemically stained with the leukocyte peroxidase (myeloperoxidase) kit according to the manufacturer's instructions (Sigma).
Assays for Hematopoietic Colony Formation.
Single cell suspensions were obtained by dispersing bone marrow cells in 1× PBS containing 2% FCS. Cells (1 × 105) were plated in duplicate in 35-mm tissue culture dishes containing Methocult 3430 methylcellulose medium (StemCell Technologies, Vancouver), and incubated at 37°C in a humidified incubator with 5% CO2. Colonies were scored after 7–8 days of incubation.
Bone Marrow Transplantation.
Seven- to twelve-week old C3H/SCID and NOD/SCID mice (The Jackson Laboratory) were exposed to a 137Cs source totaling 6.6 Gy, with a focal skin distance of 75 cm, delivered in two equal fractions 3 h apart at a dose rate of 1 Gy/min. Bone marrow cells (1 × 107) isolated from a single donor transgenic or control mouse were suspended in 1× PBS + 2% FCS. The cells were divided for i.v. tail injection into five irradiated recipient mice. Peripheral blood counts of recipient mice were monitored starting 2 weeks after transplantation.
Results
Generation of Transgenic Mice Expressing Cyclin A1.
To examine the effects of overexpression of cyclin A1 on myeloid development, we generated transgenic mice expressing mouse cyclin A1 cDNA under the direction of regulatory sequences of the human CG gene (Fig. 1A) that have been shown to drive expression of reporter constructs to promyelocytes and monocytes in vivo (13, 14). Four transgenic founders were identified, one of which was an infertile male. The other three fertile founders were used to establish three independent transgenic lines, designated 6, 10, and 19, which were then used to generate F1 progeny.
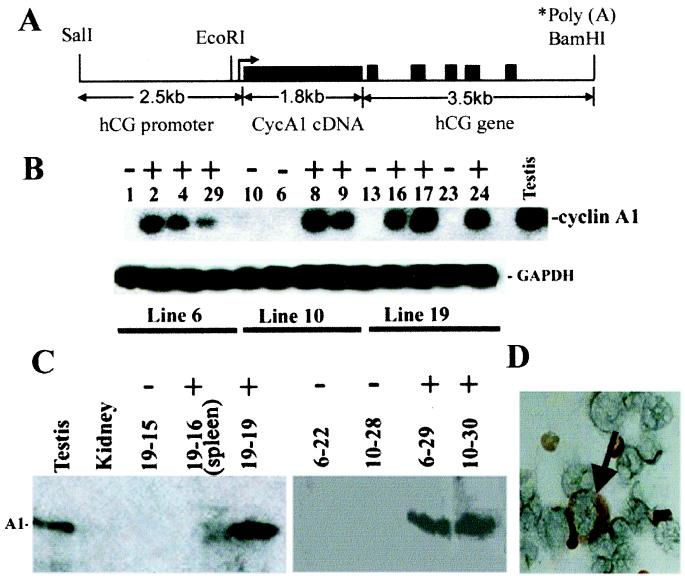
Figure 1.
(A) Diagram of the hCG/cyclin A1 transgene. The transcription start site is indicated with an arrow. The hCG sequences containing the promoter region extend upstream ≈2.4 kb from the start site. The large solid black box represents cyclin A1 cDNA and the small boxes represent the five exons of the hCG gene (≈3.5 kb, as noted); the polyadenylation signal is provided by the hCG gene. (B) RT-PCR analysis of bone marrow RNA from the indicated F1 transgenic mice (+) and nontransgenic controls (−) from three founder lines indicated below. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for RNA quality. (C) Immunoblot. Same amount of protein (≈200 μg) isolated from bone marrow and one sample from spleen (line 19, #16) of F1 transgenic (+) and nontransgenic littermate controls (−), as well as lysates from testis and kidney were loaded into each lane (founder line-mouse number). (D) Immunohistochemistry. Bone marrow cells from a 3-month-old transgenic mouse were incubated with antiserum specific for cyclin A1. The arrow indicates intensive staining of cytoplasm of an early myeloid cell.
To assess the expression of the hCG/cyclin A1 transgene, total cellular RNA was isolated from bone marrow cells of mice at 3 months of age and analyzed by RT-PCR (Fig. 1B), using primers designed to detect expression of the hCG/cyclin A1 fusion mRNA. PCR products amplified from RNA from adult mouse testes, which express high levels of cyclin A1 (8), were used as a positive control, using primers to detect the endogenous cyclin A1 mRNA (Fig. 1B). Signals were detected in the RT-PCR products from the bone marrow RNA from the majority of the transgenic mice, but none were detected in the nontransgenic littermates (Fig. 1B). RT-PCR of RNA isolated from bone marrow of the nontransgenic littermates, using primers to detect any endogenous cyclin A1 expression, failed to detect signals (data not shown). Expression of cyclin A1 was also monitored by immunoblotting analysis (Fig. 1C). Cyclin A1 protein was readily detected in the transgenic bone marrows, at levels even higher than testis controls on a mass basis (Fig. 1C). Cyclin A1 protein was also detected in the spleen of transgenic mice, but not in the spleen of a young and apparently healthy transgenic mouse from line 19, whose spleen, upon histological analysis, was noted to not have been infiltrated with early myeloid cells (Fig. 1C and data not shown). These results suggested that high levels of the transgenic cyclin A1 expression were indeed restricted to the bone marrow of transgenic mice, which histological analysis revealed to have a high proportion of myeloid cells (see below).
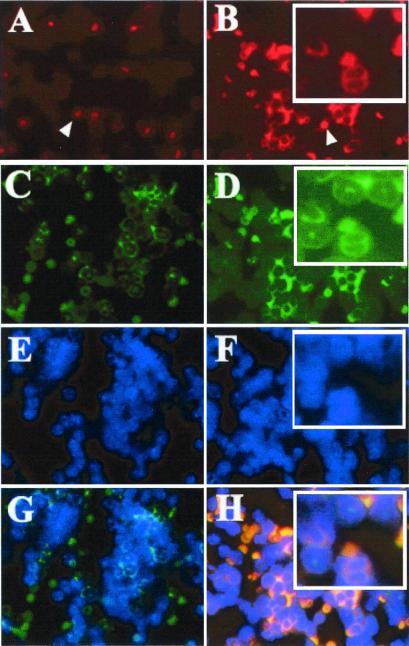
Immunohistochemistry was then used to identify the types of cells expressing cyclin A1 in the bone marrow. Cyclin A1 was detected in bone marrow cells with an early myeloid morphology from transgenic (Fig. 1D) but not normal mice (data not shown). Correct targeting of the transgene to the myeloid lineage was further confirmed by immunofluorescence analysis in which bone marrow cells were simultaneously stained with anti-cyclin A1 antibodies and a rat anti-mouse 7/4 antibody, which detects a murine myeloid-specific protein (13), as well as DAPI (4′,6-diamidino-2-phenylindole) to highlight the distinctive nuclei (Fig. 2). No signal was detected with cyclin A1 antibody in the cells from nontransgenic littermate (Fig. 2A). The colocalization of cyclin A1 (red) and 7/4 (green) produced a yellow signal in the merged images in Fig. 2 G and H. The yellow signal was detected in the cytoplasm of cells with large, doughnut-shaped nuclei, characteristic of myeloid precursor cells, which stained blue by DAPI (Fig. 2 E–H). Although staining for cyclin A1 and antigen 7/4 colocalized in myeloid precursor cells, the most intense signal for cyclin A1 was present in what appear to be immature myeloid precursor cells, whereas the most intense localization of 7/4 was in neutrophils (Fig. 2B Inset). The results of the mRNA and protein expression studies thus showed that the transgene was expressed in all three transgenic lines and that its expression was restricted to the myeloid lineage. The immunolocalization also revealed that the subcellular distribution was apparently cytoplasmic, as compared with the almost exclusive nuclear localization seen for cyclin A1 in spermatocytes (7, 9).
Figure 2.
Immunofluorescence to detect cell type and subcellular localization. Bone marrow cells from transgenic mouse (B, D, F, and H) and the nontransgenic littermate control (A, C, E, and G) were prepared by using a cytospin apparatus, and incubated simultaneously with antibody against cyclin A1 (A and B) and antibody against antigen 7/4 (C and D), as well as DAPI (4′,6-diamidino-2-phenylindole; E and F). Red blood cells also showed signals due to their nonspecific cross-reaction with dyes (indicated by white arrowhead in A and B). Note that cyclin A1 antibody strongly stained with the cells with myeloid precursors (B). The colocalization of cyclin A1 and 7/4 was detected in myeloid precursors of the transgenic line (H). Although cells with a neutrophilic morphology weakly expressed cyclin A1 (red staining in cells at top of Inset in B), the green staining for antigen 7/4 was significantly more intense (Inset in D). These data suggested that although both cyclin A1 and antigen 7/4 are expressed in myeloid cells, cyclin A1 appears to be more abundant in earlier stages of differentiation than in later stages. It is also noted that we did not detect cyclin A1 protein in myeloid cells in the nontransgenic littermate BM (A and G).
Altered Myelopoiesis in hCG-Cyclin A1 Transgenic Mice.
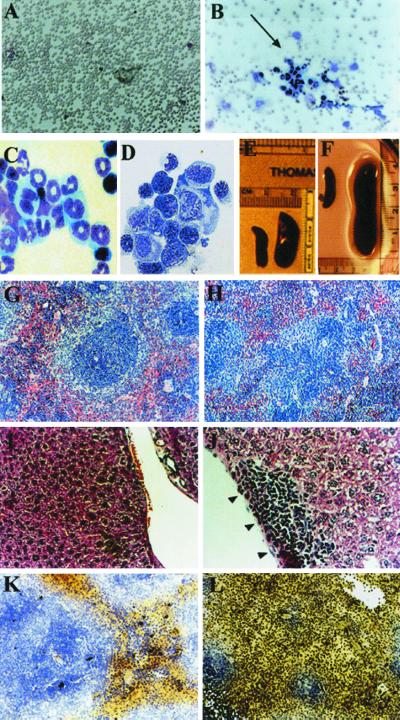
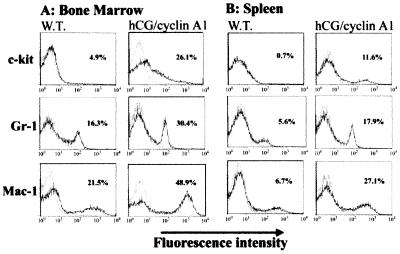
The immunohistochemical analysis also revealed the presence of elevated numbers of myeloid precursor cells in the bone marrow of the transgenic mice as compared with their control littermates. Although the transgenic mice had nearly normal peripheral blood counts up to about 6 months of age (Table 1), histological analysis of their spleen and bone marrow at 2-month intervals revealed increased numbers of early myeloid cells in the bone marrow and the splenic red pulp in all animals examined. Light microscopic examination revealed that the bone marrow of the transgenic mice was packed with myeloid cells at all stages of maturation (data not shown). The spleens and livers of the transgenic mice were also abnormal in comparison to littermate controls. The spleens were much enlarged (Fig. 3E) and the follicular architecture was disrupted (Fig. 3 G and H). The livers exhibited a disrupted morphology and an infiltration with immature myeloid cells (Fig. 3 I and J), an abnormality also observed in transgenic mice at later ages. To determine whether the population of premature hematopoietic cells was indeed increased and myelopoiesis perturbed in the transgenic mice, we performed FACS analysis of isolated bone marrow and spleen cells by using markers for myeloid maturation (18, 19). The transgenic samples displayed a markedly increased population of c-kit positive hematopoietic precursors, as well as increased numbers of cells that were positive for Gr-1 (differentiated granulocytes) and Mac-1 (macrophage) cell surface markers (Fig. 4), relative to the control. Abnormal myelopoiesis was further confirmed by using a methylcellulose colony-formation assay (18, 19). After 7–8 days of methylcellulose culture, both bone marrow cells and splenocytes from transgenic mice gave rise to significantly more colonies (4-fold increase) than the controls, although the colony sizes were not different between the transgenic mice and wild-type controls. The maturation process of the myeloid cells is not completely disrupted, as is the case in overt myeloid leukemia. As described above, the PB profile remains essentially normal in nonleukemic transgenic mice, which is not the case in overt myeloid leukemia (see Table 1). However, the BM, spleen, and liver were observed to progressively accumulate myeloid precursors.
Table 1.
Hematologic parameters from hCG/cyclin A1 transgenic and control mice
| Mice | White blood cells (103/μl) | Hb (g/dl) | Plt (103/μl) | % blasts |
|---|---|---|---|---|
| Nontransgenic (n = 22) | 7.8 ± 2.524 | 15.59 ± 2.153 | 1132.8 ± 176.3 | 0 |
| Nonleukemic transgenic (n = 22) | 8.8 ± 2.923 | 14.06 ± 1.866 | 1000.4 ± 197.7 | 0 |
| Leukemic transgenic (n = 9) | 110.3 ± 8.68 | 9.44 ± 2.542 | 570.0 ± 113.5 | 12.3 ± 4.19 |
Hb, hemoglobin; Plt, platelet; Kruskal–Wallis variance analysis used (P < 0.0001).
Figure 3.
Phenotype of hCG/cyclin A1 transgenic mice. Morphology and pathology of PB, BM, spleen, and liver. PB (A and B) or BM (C and D) preparations were stained with Wright's Giemsa. (A) PB from a nontransgenic littermate control. (B) PB from a leukemic mouse (line 19). Note that the PB shown in B contains clusters of premature myeloid cells (indicated by arrow). BM preparations are from nontransgenic littermates (C) and leukemic line 19 (D). (E–H) Morphology and pathology of spleen. Spleens on the left in E and F are from nontransgenic littermates and those on the right of transgenic mice from nonleukemic transgenic line 6 (E) and leukemic line 19 (F) mice. (G and H) Histological sections of spleens from 7-month-old nontransgenic littermate (G) and transgenic (H) from line 6 and reveal highly disrupted follicular structures. (I and J) Sections from the same mice in G and H, showing infiltration of liver with immature myeloid cells (black arrowheads in J). The sections in G–J were stained with Hematoxylin and Eosin. (K and L) Histochemical staining for myeloperoxidase activity in a leukemic mouse (line 19, #45; L) and the littermate control (K). A majority of cells stain positive for myeloperoxidase and also reveal the highly disrupted follicles.
Figure 4.
FACS analysis of the cells isolated from the bone marrow and spleen of the transgenic mice and the littermate controls. Cells in suspension were stained with C-kit, Gr-1, and Mac-1, conjugated with fluorescein isothiocyanate (Gr-1) or R-phycoerythrin (C-kit and Mac-1), and are depicted by the solid lines. The control antibodies of the same isotype are displayed as dotted lines. The values in each histogram indicate the percent of cells in the population positive for the respective antibodies. The vertical axes indicate the relative cell counts (from 0 to 200) and the horizontal axes indicate the log fluorescence intensity.
Development of Myeloid Leukemia in hCG-Cyclin A1 Transgenic Mice.
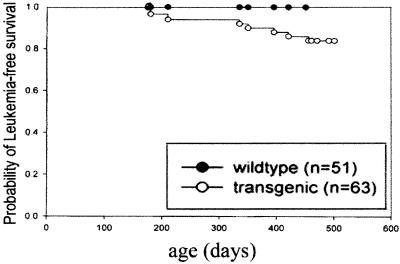
The three founder lines were expanded for a 14-month follow up study. The Kaplan–Meyer plot in Fig. 5 depicts the long latency and low penetrance of development of leukemia. Approximately 15% of the transgenic mice (14% from line 6, 13% from line 10, 16% from line 19) developed the leukemic phenotype, and 85% of mice greater than 7 months of age are at risk. The diagnosis of leukemia was based on several parameters, including hematological changes, the presence of high levels of myeloid precursors in the BM and PB, and pathological characteristics of infiltration of organs by the myeloid blast population. Blood counts of the leukemic transgenic mice showed striking differences from the controls or nonleukemic transgenics, with anemia and elevated white blood cell count (Table 1). The leukemic mice exhibited a severe leukocytosis accompanied by anemia and thrombocytopenia and a pale bone marrow that was associated with hepatosplenomegaly (Fig. 3F and data not shown). Microscopic examination of their peripheral blood and bone marrow cells revealed a high percentage of immature myeloid cells, particularly blast cells in the bone marrow (Fig. 3 B and D). Some of the leukemic cells exhibit marked nuclear irregularity, with folding or lobulation (Fig. 3D). A great increase in the number of cells that stained positive for myeloperoxidase was observed in the spleen of the transgenic mice (Fig. 3 K and L). The architecture of the spleen and liver was completely disrupted by a massive invasion of leukemic cells. Predictably, FACS analysis of cells from the leukemic bone marrow again showed a dramatic accumulation of hematopoietic cells (c-Kit positive) and increased myeloid populations (Gr-1 and Mac-1) (data not shown), confirming the early myeloid origin of the leukemia. Methylcellulose culture of leukemic bone marrow cells produced abundant colonies, and myeloid blasts and promyelocytes were the predominant cell types in cytospins of cells from these cultures (data not shown).
Figure 5.
Survival of hCG/cyclin A1 transgenic mice from founder lines 6, 10, and 19 compared with that of nontransgenic mice determined with Kaplan–Meyer plot. Survival at various time points was calculated on the basis of the size of each group of mice when an animal died or became terminally ill. The plot was generated by using Sigma Plot 2000 (SPSS Science, Chicago). The numbers (n) of transgenic and nontransgenic mice used for the analysis are shown on the right part of the graph.
Finally, we performed preliminary analysis of the cell cycle profiles of bone marrow cells in normal, nonleukemic transgenic, and leukemic mice. The bone marrow of both nonleukemic and leukemic transgenic mice appeared to have fewer cells in the G2 and M phases of the cell cycle (data not shown).
Acute Myeloid Leukemia and Altered Myelopoiesis Are Transplantable.
To determine whether tumor cells from the primary leukemic mice can initiate leukemia in nontransgenic recipients, bone marrow cells or splenocytes isolated from leukemic or overtly healthy transgenic versus normal control animals were transplanted to irradiated C3H-SCID and NOD-SCID mice by i.v. injection. Peripheral blood profiles were monitored at 2-week intervals. Among the 15 recipient mice transplanted with BM from the leukemic hCG/cyclin A1 transgenic mice, four mice died 3–6 weeks posttransplantation, but were not suitable for postmortem analysis. Of the 11 remaining tested recipients, all showed abnormal blood profiles. Two developed overt leukemia within 4–6 weeks and were killed. Three additional recipients were also killed within 4–6 weeks and shown to have markedly increased myeloid precursors in BM, spleen, and liver, typical of leukemic animals (see above). Further evidence for the leukemogenesis caused by overexpression of cyclin A1 was obtained by transplanting bone marrow cells from young, apparently healthy transgenic mice into irradiated SCID mice (15 recipients). By 4–8 weeks posttransplantation, 3 recipient mice died (without postmortem analysis), and 5 of the remaining 12 recipient mice examined displayed abnormalities in the BM, spleen, and liver. Mice transplanted with wild-type BM cells (11 recipients) did not develop any evidence of abnormalities.
Discussion
In this report, we have described the effects of targeted overexpression of cyclin A1 in promyelocytes and monocytes on myelopoiesis in a transgenic mouse model. Disorders in myelopoiesis were observed in all animals examined, notably the accumulation of early myeloid cells in the bone marrow, which presumably led to the invasion of the same type of cells in the spleen and liver. A low frequency (≈15%) of the transgenic mice progressed to overt leukemia after a latency period of 7–12 months. The disease was readily transferable to nontransgenic recipients. A similar delayed onset of disease and low penetrance of leukemia and subsequent transferability was observed in the PML/RARα fusion transgenic mice generated by several groups (14, 18, 19) and in the AML1/MDS1/EV1 fusion transgenics (20). As suggested in these other studies as well, the delayed onset and subsequent rapid onset following transfer of the diseased bone marrow may reflect the acquisition of additional genetic abnormalities during the development of the acute myeloid leukemia in the transgenic animals; that is, elevated levels of cyclin A1 alone may not be sufficient for leukemogenesis, although our data demonstrate a clear correlation with the development of the disease. However, the fact that the leukemic phenotype could be transferred to SCID mice suggested that the abnormality in the myeloid lineage of the transgenic mice was caused by the overexpression of cyclin A1 protein. Even nonleukemic transgenic bone marrow from the cyclin A1 transgenic mice transplanted into SCID mice retained the abnormal characteristics in myeloid lineage and the susceptibility to develop the leukemia.
Various cyclins and Cdks have been shown to exhibit cell cycle phase-specificity of expression in hematopoietic stem cells (21), indicating their essential role in regulating proliferation and differentiation, as well as self-renewal. In normal human hematopoietic cells, only a very low level of expression of cyclin A1 has been detected by RT-PCR (11, 22). To date, we have been unable to detect mouse cyclin A1 in normal total bone marrow cells of B6CBAF1/J mice by RT-PCR, nor have we observed positively stained cells in nontransgenic bone marrow when using anti-cyclin A1 antibodies. Whether this represents a true difference in distribution of expression between species of this extremely tightly regulated cell cycle gene or a difference in sensitivity of detection remains to be determined. It is of interest to note that we have not observed any obvious abnormalities in the hematopoietic system of our cyclin A1 deficient mice; the only phenotype observed to date is the arrest of spermatogenesis (9). It is also of interest that, although the mouse and human cyclin A1 proteins are highly conserved, with 87% sequence identity, they do differ at the amino terminus: the human cyclin A1 has 44 extra amino acids that are not found in the mouse protein (8, 10).
Elevated levels of human cyclin A1 expression were shown to be predominantly expressed in hematological malignancies with myeloid differentiation (11) and the highest frequency of cyclin A1 overexpression was observed in AML (12). In our transgenic mice, the leukemic phenotype resembles human AML. Similar to human, anemia and thrombocytopenia were consistently observed in the leukemic mice. There was also a high percentage of myeloblasts in bone marrow and severe infiltration of spleen and liver with immature myeloid cells.
The predominant site of cyclin A1 localization in the early myeloid cells in the transgene-expressing mice was cytoplasmic, as shown by immunohistochemistry and immunofluorescence. This is in the contrast to the predominantly nuclear localization of cyclin A1 in spermatocytes in the normal adult testis (7). The loss of its normal nuclear localization may imply that in these cells, cyclin A1 has a function distinct from its proposed role in the activation of MPF and progression into M-phase. For example, the altered subcellular distribution of Cdk1 in oral carcinomas has been suggested to affect its normal function in binding to cyclins (23). Previous studies have shown that the functional status of each population of the hematopoietic stem cells reflects cell cycle profiles that are tightly regulated by the cell cycle control system (1). The abnormal expression and/or cytoplasmic localization of cyclin A1 in the early myeloid cells may perturb the cell cycle progression at the onset of G1-S transition and therefore perturb the differentiation of early myeloid cells into mature cells.
Our initial FACS/cell cycle analysis has suggested that, in nonleukemic transgenic mice and leukemic hCG/cyclin A1 transgenic mice, bone marrow cells appeared to have fewer cells in the G2/M phases of the cell cycle, when compared with their littermate controls. This may reflect a loss of normal growth characteristics in hematopoietic cells in the transgenic mice. Although we are not aware of comparable analyses of AML patients, this observation is analogous to previous observations in patients with ALL; that is, most of the ALL blast samples examined had few or no cells in the S or G2/M phases of the cell cycle, despite exhibiting overexpression of cyclin E protein that correlated with the accumulation of early hematopoietic cells (5). The attenuation of cell differentiation could be due to a block of cell cycle progression. A-type cyclins are known to function in regulating G1-S phase and G2-M phase transition in the association with Cdk1 and Cdk2. The expression of cyclin A2 peaks at the onset of G1 to S phase transition, as well as G2-M phase transition. Cyclin A2 expression was shown to be regulated in a cell-cycle-dependent manner in the human myeloid cell line HL-60, with the peak expression of cyclin A2 observed at S-G2 (21).
Specific chromosomal translocations have been observed in many types of leukemia (24). The expression of the fusion gene(s) derived from the translocations has been postulated to inhibit differentiation and maturation of hematopoietic progenitor cells (25). Translocations in APL create a fusion gene between RARα and several genes—including PML, PLZF, NPM, NuMA, and Stat5b (25–28). The fusion gene encoding the chimeric PML-RARα transcription factor yields a protein with altered transactivation properties compared with the normal PML and RARα proteins. Recent studies have shown that PML affects cell cycle progression by mediating expression of several key proteins, including pRb, cyclin E, Cdk2, and p27, that normally control cell cycle progression (29). Interestingly, very recent transfection experiments expressing PML-RARα or PLZF-RARα fusion proteins in cultured cell lines suggested that cyclin A1 may be a direct target of these oncogenic fusion proteins (30). It will be interesting to determine whether cyclin A1 is a target of PML-RARα or PLZF-RARα in the induction of acute myeloid leukemia in vivo.
The strong correlations between cyclin A1 and the murine myeloid leukemia described here suggest that cyclin A1 acts as an oncogene that is involved in the etiology of acute myeloid leukemia. Although the pathways affected by the overexpression of cyclin A1 in the myeloid lineage in transgenic mice that result in the development of AML are not fully understood, these transgenic mice provide a valuable tool with which to unravel the mechanisms underlying the role of cell cycle regulatory gene cyclin A1 in leukemogenesis.
Acknowledgments
We thank Dr. Timothy Ley for the hCG vector and Dr. Stefan Karlsson and colleagues, Dr. Guy Sauvageau, and Dr. Marja Ekblom for helpful advice. We also thank Dr. Riccardo Della-Favera and Jovenal Quintana for use of their cytospin apparatuses, Dr. Jolanda Grudzien for performing the blood counts, Dr. Robert Grossman for teaching us the murine bone marrow transplantation, members of Dr. Pandolfi's lab, especially Dr. Eduardo Rego, for advice and helpful discussion, Chris Marshall for help in organizing the mouse colony, and Erika Laurion and YuJing Xie for help in preparing the figures. This work was supported in part by grants from the National Institutes of Health (HD34915 to D.J.W. and CA 08748 and CA 74031 to P.P.P.), The Adler Foundation (to D.J.W.), and the Wenner-Gren Foundation and The Swedish Foundation for International Cooperation in Research and Higher Education (STINT) (to C.L.).
Abbreviations
- AML
acute myeloid leukemia
- ALL
acute lymphoblastic leukemia
- RT
reverse transcription
- hCG
human cathepsin G
- FACS
fluorescence-activated cell sorting
- Cdk
cyclin-dependent kinase
- SCID
severe combined immunodeficient
- PB
peripheral blood
- BM
bone marrow
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Furukawa Y. Hum Cell. 1998;11:81–92. [PubMed] [Google Scholar]
- 2.Sherr C J. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 3.Yerly-Motta V, Contassot E, Pavy J J, Tiberghien P, Herve P. Cell Mol Biol (Noisy-le-grand) 1999;45:1217–1228. [PubMed] [Google Scholar]
- 4.Iida H, Towatari M, Tanimoto M, Morishita Y, Kodera Y, Saito H. Blood. 1997;90:3707–3713. [PubMed] [Google Scholar]
- 5.Scuderi R, Palucka K A, Pokrovskaja K, Bjorkholm M, Wiman K G, Pisa P. Blood. 1996;87:3360–3367. [PubMed] [Google Scholar]
- 6.Volm M, Koomagi R, Stammler G, Rittgen W, Zintl F, Sauerbrey A. Int J Cancer. 1997;74:508–512. doi: 10.1002/(sici)1097-0215(19971021)74:5<508::aid-ijc5>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 7.Ravnik S E, Wolgemuth D J. Dev Biol. 1999;207:408–418. doi: 10.1006/dbio.1998.9156. [DOI] [PubMed] [Google Scholar]
- 8.Sweeney C, Murphy M, Kubelka M, Ravnik S E, Hawkins C F, Wolgemuth D J, Carrington M. Development. 1996;122:53–64. doi: 10.1242/dev.122.1.53. [DOI] [PubMed] [Google Scholar]
- 9.Liu D, Matzuk M M, Sung W K, Guo Q, Wang P, Wolgemuth D J. Nat Genet. 1998;20:377–380. doi: 10.1038/3855. [DOI] [PubMed] [Google Scholar]
- 10.Yang R, Morosetti R, Koeffler H P. Cancer Res. 1997;57:913–920. [PubMed] [Google Scholar]
- 11.Kramer A, Hochhaus A, Saussele S, Reichert A, Willer A, Hehlmann R. Leukemia. 1998;12:893–898. doi: 10.1038/sj.leu.2401051. [DOI] [PubMed] [Google Scholar]
- 12.Yang R, Nakamaki T, Lubbert M, Said J, Sakashita A, Freyaldenhoven B S, Spira S, Huynh V, Muller C, Koeffler H P. Blood. 1999;93:2067–2074. [PubMed] [Google Scholar]
- 13.Grisolano J L, Sclar G M, Ley T J. Proc Natl Acad Sci USA. 1994;91:8989–8993. doi: 10.1073/pnas.91.19.8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grisolano J L, Wesselschmidt R L, Pelicci P G, Ley T J. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- 15.Behringer R R, Crotty D A, Tennyson V M, Brinster R L, Palmiter R D, Wolgemuth D J. Development. 1993;117:823–833. doi: 10.1242/dev.117.3.823. [DOI] [PubMed] [Google Scholar]
- 16.Packer A I, Crotty D A, Elwell V A, Wolgemuth D J. Development. 1998;125:1991–1998. doi: 10.1242/dev.125.11.1991. [DOI] [PubMed] [Google Scholar]
- 17.Liu D, Liao C, Wolgemuth D J. Dev Biol. 2000;224:388–400. doi: 10.1006/dbio.2000.9776. [DOI] [PubMed] [Google Scholar]
- 18.He L Z, Tribioli C, Rivi R, Peruzzi D, Pelicci P G, Soares V, Cattoretti G, Pandolfi P P. Proc Natl Acad Sci USA. 1997;94:5302–5307. doi: 10.1073/pnas.94.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown D, Kogan S, Lagasse E, Weissman I, Alcalay M, Pelicci P G, Atwater S, Bishop J M. Proc Natl Acad Sci USA. 1997;94:2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuenco G M, Nucifora G, Ren R. Proc Natl Acad Sci USA. 2000;97:1760–1765. doi: 10.1073/pnas.030421197. . (First Published February 4, 2000; 10.1073/pnas.030421197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burger C, Wick M, Muller R. J Cell Sci. 1994;107:2047–2054. doi: 10.1242/jcs.107.7.2047. [DOI] [PubMed] [Google Scholar]
- 22.Yong A S, Goldman J M. Bone Marrow Transplant. 1999;23:827–828. doi: 10.1038/sj.bmt.1701729. [DOI] [PubMed] [Google Scholar]
- 23.Goodger N M, Gannon J, Hunt T, Morgan P R. J Pathol. 1996;178:422–428. doi: 10.1002/(SICI)1096-9896(199604)178:4<422::AID-PATH497>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Look A T. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 25.Merghoub T, Gurrieri C, Piazza F, Pandolfi P P. Blood Cells Mol Dis. 2001;27:231–248. doi: 10.1006/bcmd.2001.0385. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Brand N J, Chen A, Chen S J, Tong J H, Wang Z Y, Waxman S, Zelent A. EMBO J. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Redner R L, Rush E A, Faas S, Rudert W A, Corey S J. Blood. 1996;87:882–886. [PubMed] [Google Scholar]
- 28.Wells R A, Catzavelos C, Kamel-Reid S. Nat Genet. 1997;17:109–113. doi: 10.1038/ng0997-109. [DOI] [PubMed] [Google Scholar]
- 29.Mu Z M, Le X F, Vallian S, Glassman A B, Chang K S. Carcinogenesis. 1997;18:2063–2069. doi: 10.1093/carcin/18.11.2063. [DOI] [PubMed] [Google Scholar]
- 30.Muller C, Yang R, Park D J, Serve H, Berdel W E, Koeffler H P. Blood. 2000;96:3894–3899. [PubMed] [Google Scholar]