Abstract
Despite the wide use of plant regeneration for biotechnological purposes, the signals that allow cells to become competent to assume different fates remain largely unknown. Here, it is demonstrated that the Regeneration1 (Rg1) allele, a natural genetic variation from the tomato wild relative Solanum peruvianum, increases the capacity to form both roots and shoots in vitro; and that the gibberellin constitutive mutant procera (pro) presented the opposite phenotype, reducing organogenesis on either root-inducing medium (RIM) or shoot-inducing medium (SIM). Mutants showing alterations in the formation of specific organs in vitro were the auxin low-sensitivity diageotropica (dgt), the lateral suppresser (ls), and the KNOX-overexpressing Mouse ears (Me). dgt failed to form roots on RIM, Me increased shoot formation on SIM, and the high capacity for in vitro shoot formation of ls contrasted with its recalcitrance to form axillary meristems. Interestingly, Rg1 rescued the in vitro organ formation capacity in proRg1 and dgtRg1 double mutants and the ex vitro low lateral shoot formation in pro and ls. Such epistatic interactions were also confirmed in gene expression and histological analyses conducted in the single and double mutants. Although Me phenocopied the high shoot formation of Rg1 on SIM, it failed to increase rooting on RIM and to rescue the non-branching phenotype of ls. Taken together, these results suggest REGENERATION1 and the DELLA mutant PROCERA as controlling a common competence to assume distinct cell fates, rather than the specific induction of adventitious roots or shoots, which is controlled by DIAGEOTROPICA and MOUSE EARS, respectively.
Key words: Cell fate, competence, determination, hormonal mutants, Micro-Tom, plant development, regeneration
Introduction
A remarkable and intriguing aspect of plant development is the capacity to form new and adventitious organs during the whole life cycle. This capacity, which has both evolutionary (Fosket, 1994; Sugimoto et al., 2011) and ecological significance (Kauffman, 1991), confers the plasticity necessary for sessile organisms to face the changing environment. It also has a practical importance in agriculture, explored since the domestication of vegetatively propagated crops (Harlan, 1992). Early studies in the field of mineral nutrition (White, 1934) and plant hormones (Miller et al., 1955) allowed the further sophistication of cloning practices (revised in Vasil, 2008), which were attempted to be extended to almost all agriculturally important crops as a prerequisite for modern genetic manipulations in vitro. Despite the importance of flexible plant development for agriculture, its in vitro manipulation remains largely empirical, and sometimes unsuccessful, since its molecular basis has only recently started to be unravelled (revised in Duclercq et al., 2011).
An important step in the process of formation of novel and ectopic organs is the acquisition of competence of a given cell or tissue to assume a new developmental fate. For instance, plant propagation by stem cuttings, although a rather simple horticultural technique, involves complex changes that culminate in the formation of cells with a new root identity at the base of the shoot cuttings. As proposed by Christianson and Warnick (1988) for in vitro propagation, the acquisition of competence precedes the phase of induction of different organs. Upon induction, a given cell or tissue become determined (committed) to form the induced organ, and this last developmental step can be interpreted as opposite to the initial non-committed state of competent cells or tissues (Wareing, 1982). Thanks to the seminal work of Skoog and Miller (1957), it is currently known that the induction of organs, such as roots and shoots, depends on the balance between the plant hormones auxin and cytokinin, rather than their absolute levels. This implies that any given exogenous or endogenous event that alters the levels of these two hormones, or the capacity to respond to them, will probably influence the capacity for organ formation. Thus, it is conceivable that some plants overproducing cytokinin become prone to form shoots (Estruch et al., 1991; Peres and Kerbauy, 1999; Catterou et al., 2002) and that enhanced auxin sensitivity usually increases root formation capacity (Visser et al., 1996; Lima et al., 2009). Additionally, it has been shown that the expression of genes controlling cytokinin response or shoot meristem identity, such as ARABIDOPSIS RESPONSE REGULATOR5 (ARR5), SHOOTMERISTEMLESS (STM), and WUSCHEL (WUS), correlates with shoot induction, and these may also serve as markers for this event (Cary et al., 2002; Gallois et al., 2002; Che et al., 2006, 2007). However, a similar level of knowledge on the control of the phase of acquisition of competence is not yet available, despite its importance as a key step in organ regeneration (Christianson and Warnick, 1988).
Some transcription factors identified as important molecules in the shoot induction step in Arabidopsis (Cary et al., 2002; Gallois et al., 2002; Che et al., 2007) are homeoboxes (e.g. STM and WUS), whose homologues in maize and tomato were also proposed to regulate the switch from determinate to indeterminate cell fates in the control of developmental processes, such as leaf architecture (Sinha et al., 1993). Considering that adventitious organ formation is recognized to be dependent on the presence of indeterminate (non-committed) stem cells (Sugimoto et al., 2011), one may hypothesize that the acquisition of competence could be related to the action of homeobox genes, such as STM and WUS. However, contrary to the early hypothesis of a specific competence for each kind of organ to be formed (see Christianson and Warnick, 1985), there is recent evidence suggesting that the beginning of organogenesis follows a common and general pathway (Atta et al., 2009; Sugimoto et al., 2010). This pathway is then channelled toward the induction of roots or shoots under the influence of specific hormonal balances (Skoog and Miller, 1957). Thus, it may be that genes controlling competence should have an impact in the capacity to form both roots and shoots, which seems not to be the case for the shoot identity-associated homeobox genes studied thus far (Smith et al., 1997; Cary et al., 2002; Gallois et al., 2002; Che et al., 2007).
Moreover, it is generally assumed that the pre-incubation on auxin-rich ‘callus-inducing medium’ (CIM) helps the acquisition of competence to generate the respective organ in ‘shoot-inducing medium’ (SIM) or ‘root-inducing medium’ (RIM) (Christianson and Warnick, 1988; Valvekens et al., 1988). This assumption suggests auxin as a possible molecular player in the acquisition of competence, a knowledge that was also used in the identification of the genes ENHANCER OF SHOOT REGENERATION 1 (ESR1), ARR15, POLYGALCTURONASE INHIBITING PROTEIN 2 (PGIP2), and WUS as requiring CIM pre-incubation for up-regulation on SIM (Banno et al., 2001; Che et al., 2007). These findings indicate that genes and molecules controlling acquisition of competence may be upstream to the above-mentioned genes in a common cellular signalling pathway prior to the specification of the kind of organ to be formed. Direct approaches to find such genes would be the identification of genes expressed before the commitment to organogenesis (Santos et al., 2009) or the screening for genotypes (induced mutants and natural genetic variation) with higher or lower in vitro organ formation concomitantly on SIM or RIM. The identification of key genes/molecules controlling the acquisition of competence, at either cellular or organismal levels, is relevant to understand the molecular basis of the plant development plasticity.
In the present work, a collection of tomato (Solanum lycopersicum L.) mutants (www.esalq.usp.br/tomato) introgressed into the cv Micro-Tom (MT) genetic background (Scott and Harbaugh, 1989; Meissner et al., 1997) were used to look for genetic variation associated with in vitro regeneration capacity. The Regeneration1 (Rg1) allele, a natural genetic variation originating from S. peruvianum (Koornneef et al., 1993), and the gibberellin (GA) constitutive mutant procera (pro), a loss-of-function in the DELLA-like protein (Bassel et al., 2008; Jasinski et al., 2008), were characterized as affecting the competence phase, since they alter both shoot and root in vitro formation and influence a series of apparently unrelated processes that are linked to the capacity to assume different cell fates.
Materials and methods
Plant material
Tomato (S. lycopersicum L.) cv. MT and the near-isogenic lines (NILs) (Table 1) harbouring the alleles Rg1, pro, and diageotropica (dgt) were obtained as described in previous studies (Pino et al., 2010; Carvalho et al., 2011). Similarly, the mutants Mouse ears (Me) and lateral suppresser (ls) were introgressed into MT as described for the Rg1 allele (Pino et al., 2010), and pro and dgt mutants into MT (Carvalho et al., 2011). MT seeds carrying the DR5::GUS gene (Ulmasov et al., 1997) were kindly provided by Dr José Luiz Garcia-Martinez from Universidad Politécnica de Valencia, Spain. The crosses and phenotypical screening procedures used to obtain the double mutants dgtRg1, proRg1, lsRg1 Mels, and Rg1DR5::GUS were as described previously (Lima et al., 2009). All the genotypes used here are maintained in the tomato mutant collection of the Escola Superior de Agricultura ‘Luiz de Queiroz’ (ESALQ), Universidade de São Paulo (USP), Brazil (http://www.esalq.usp.br/tomato/).
Table 1.
Tomato genotypes in the cv Micro-Tom background used here
| Genotype | Effect/gene function | Origin | Reference |
|---|---|---|---|
| diageotropica (dgt) | Low sensitivity to auxin. Defect in a cyclophilin biosynthesis gene (a putative auxin signal transduction component) | LA1529 cv unknown | Oh et al. (2006) |
| procera (pro) | Constitutive response to gibberellin. Contains a point mutation that convert the VHVID putative DNA-binding domain in the tomato DELLA gene to VHEID | LA0565 cv Condine Red | Bassel et al. (2008) |
| lateral suppresser (ls) | No initiation of lateral branches. Mutated in the VHIID domain of a gene from the GRAS family, which includes the DELLA gene | LA0329 hybrid | Schumacher et al. (1999) |
| Mouse ears (Me) | Highly dissected leaves. Overexpression of a tomato KNOX gene (TKn2/LeT6) | LA0715 cv unknown | Parnis et al. (1997) |
| DR5::GUS | Plants present enzymatic staining in sites where auxin accumulates. Synthetic auxin-responsive promoter fused to the reporter gene uid GUS (encoding a β-glucuronidase) | Micro-Tom | Martí et al. (2010) |
| Regeneration1 (Rg1) | High organ formation capacity in different explants, including roots. Unknown gene function | LA4136 hybrid | Koornneef et al. (1993) |
Plant cultivation
Plants were grown in 150ml plastic pots (MT) containing a 1:1 mixture of commercial substrate (Plantmax HT, Eucatex, São Paulo, Brazil) and expanded vermiculite, supplemented with 1g of NPK 10:10:10 l–1 substrate and 4g of dolomite limestone (MgCO3+CaCO3) l–1 substrate. Plants were kept in a greenhouse under automatic irrigation (four times a day), at an average mean temperature of 28 °C; 11.5/13h (winter/summer) photoperiod, and 250–350 µmol m–2 s–1 photosynthetically active radiation (PAR) by natural radiation reduction with a reflecting mesh (Aluminet-Polysack Industrias Ltda, Leme, SP, Brazil). At the flowering stage (~35 d after sowing), plants were supplemented with NPK (~0.2g per 150ml pot). About 40 d after each crossing, mature fruits were harvested and the seed pulp was removed by fermentation for 12h using commercial baker’s yeast (Saccharomyces cerevisae, Fermix, São Paulo, Brazil). Seeds were subsequently washed, air-dried, and stored at 10 ºC for further use.
Grafting and branch analyses
Grafting procedures were conducted as previously described (Peres et al., 2005) with some modifications. Briefly, 7-day-old seedlings cultivated in 150ml pots were used as both scion and rootstock. Scions were prepared by cutting the stem with a razor blade below the second or third leaf from the apex into a wedge shape. Rootstocks were prepared by cutting transversely with a razor blade ~5cm above soil level, followed by inserting the scion into a ‘V’-shaped incision in the stock. A small peg was used to fasten the scion and rootstock together. Grafted plants were then covered with transparent polyethylene terephthalate (PET) bottles to provide a humid environment. After 1 week, the cover and peg were removed and the plants were maintained in the greenhouse as described above. The branching index, the ratio between the total length of lateral ramification and the main axis length (Morris et al., 2001), was estimated 33 d after grafting.
In vitro culture
Seeds were surface-sterilized by shaking in 100ml of 30% (v/v) commercial bleach (2.7% sodium hypochloride) plus two drops of commercial detergent for 15min, followed by three rinses with sterile water. The seeds were then germinated on media containing half-strength MS salts (Murashige and Skoog, 1962) and B5 vitamins (Gamborg et al., 1968); 15g l–1 sucrose; and 6g l–1 agar (Merck, Darmstadt, Germany). Medium pH was adjusted to 5.8 before autoclaving. Approximately 40 seeds were sown per flask containing 30ml of medium. Cultures were sealed with polyvinyl chloride (PVC) and incubated at 25±1 °C in the dark for 4 d, followed by 4 d or 8 d under a 16h photoperiod provided by a 40W cool white fluorescent tube (~45 µmol m–2 s–1 PAR). Cotyledons were then isolated from 8- or 12-day-old (after sowing) seedlings. The distal and proximal tips were removed, and the cotyledons were divided transversally in two or three pieces. Explants were placed with the abaxial side down immediately after isolation onto semi-solid SIM, composed of MS salts, B5 vitamins, 30g l–1 sucrose, 6g l–1 agar, and 5 µM 6-benzyl adenine (BA) (Sigma, St Louis, MO, USA), or RIM, which has the same composition as SIM, except that BA is replaced with 0.4 µM naphthalene acetic acid (NAA) (Sigma). The CIM has the same salt, sucrose, and vitamin composition as SIM and RIM, plus 0.5 µM BA and 1.0 µM 2,4-dichlorophenoxyacetic acid (2,4-D). During explanting, a Petri dish containing potassium permanganate salts was kept inside the laminar flow hood to avoid ethylene accumulation, which can reduce tomato regeneration afterwards (Lima et al., 2009). Twenty cotyledonary explants were cultured per sterile polystyrene Petri dish (90×15mm), with six plates per treatment. Plates were sealed with PVC and maintained under a 16h photoperiod at 25±1 °C for 3 weeks.
Histological analysis
For light microscopy analysis, five samples of cotyledons from 8-day-old seedlings grown in vitro or three samples of the fourth leaf of 33-day-old plants grown in the greenhouse were collected. Transverse sections (30–60 µm thick) of the base of the petiole were hand cut with a razor blade. Petiole sections were placed in a sodium hypochlorite solution (20%) for bleaching, and then washed with distilled water until total removal of sodium hypochlorite, as evidenced by the loss of the characteristic odour. Petiole sections were stained with 1% aqueous iodine green for 2min, and then washed with distilled water to remove excess dye. Subsequently, the petiole sections were stained with Congo red for 30 s and then washed twice with distilled water. The petiole sections were then mounted on glass slides, adding a small amount of liquid glycerin gelatin (heated to 40 °C), before covering with a cover slip. Images from petiole sections were digitally captured using a Leica (Leica™, Wetzlar, Germany) DMLB microscope with a camera connected to a computer, and the IM50 software (Leica™) was used for image analysis. Cotyledons were fixed in Karnovsky solution (Karnovsky, 1965) for 24h at 8 ºC. Dehydration was performed with a graded ethanol series 10–100%, followed by embedding in synthetic 2-hydroxyethylmethacrylate resin (Leica Historesin embedding kit™), according to the manufacturer’s recommendations. Sections (5 µm) of cotyledons were obtained on a rotary microtome, and stained with 0.05% toluidine blue in phosphate buffer and citric acid pH 4.5 (Sakai, 1973). Slides were prepared with synthetic permanent resin (Entellan™). The images from cotyledon sections were obtained using a Zeiss Axiophot photomicroscope with a digital camera attached, and analysed by image acquisition software Axiovision 4.6 (Carl Zeiss™, Oberkochen, Germany).
DR5::GUS reporter assay
Histochemical GUS (β-glucuronidase) staining was performed in DR5::GUS transgenic MT and MT-Rg1, as described by Jefferson et al. (1987). Roots from 14-day-old seedlings or cotyledons from 8-day-old seedlings grown in vitro in basal MS medium were used. The cotyledons were incubated on RIM for 1 d before staining. Roots and cotyledons were incubated at 37 °C overnight in GUS staining solution [80mM sodium phosphate buffer, pH 7.0; 8mM EDTA; 0.4mM potassium ferrocyanide; 0.05% Triton X-100; 0.8mg ml–1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc); 20% methanol]. Following GUS staining, the reaction was stopped with 70% ethanol. The root tips and cotyledons were prepared and observed in a Trinocular Leica DM LB microscope. Representative phenotypes were photographed (Leica DC 300 F) at ×200 magnification.
RNA analysis and semi-quantitative RT-PCR
Briefly, total RNA was isolated from young leaves and shoot apexes using Trizol (Invitrogen). RNA was quantified in an Ultrospec 1100 pro spectrophotometer (Amersham Biosciences, Piscataway, NJ, USA) and RNA integrity was examined by gel electrophoresis. A 1 µg aliquot of DNase I-treated RNA was used to perform cDNA synthesis using ImProm-II Reverse Transcriptase (Promega). Reverse transcription-PCRs (RT-PCRs) were normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal control. Mature microRNA164 (miR164) was detected using stem–loop RT-PCR, a standard technique to quantify miRNAs accurately (Chen et al., 2005). Primers used for RT-PCR are described in Supplementary Table S1 available at JXB online.
Results
Characterization of Rg1, a natural genetic variation improving both shoot and root formation in vitro
Given that Rg1 was originally characterized as improving shoot formation in vitro from root (Koornneef et al., 1993) and hypocotyl explants (Lima et al., 2004), whether it also improves root formation in adequate medium was tested here. Cotyledonary explants harbouring the Rg1 allele not only formed significantly more shoots on SIM (Figs 1A, 2A), but also more roots on RIM (Figs 1B, 2B), when compared with explants from the NIL MT. Besides displaying more explants undergoing de novo shoot and root formation (Fig. 1), the number of both organs formed per explant also increased in MT-Rg1 (Fig. 2). As previously reported by Koornneef et al. (1993), Rg1 maintained the capacity for shoot regeneration in SIM after long-term pre-incubation on CIM, a capacity not present in most tomato cultivars (Koornneef et al., 1987, 1993), such as the cv MT used here (Fig. 1C).
Fig. 1.
In vitro regeneration ability of Micro-Tom (MT) and MT-Rg1. (A) Shoot formation in cotyledonary explants from 12-day-old (after in vitro sowing) seedlings cultivated on SIM (5.0 µM BA). (B) Root formation in cotyledonary explants from 12-day-old seedlings cultivated on RIM (0.4 µM NAA). (C) Shoot formation in callus explants cultivated on SIM. Cotyledonary explants from 8-day-old seedlings received no, one, or two pre-incubations of 21 d on CIM (0.5 µM BA+1.0 µM 2,4-D) before transfer to SIM. Error bars represent the mean ± SE, n = 5 Petri dishes each containing 20 cotyledons. *P < 0.05 and **P < 0.01, according to Student’s t-test. The measurements were taken 10 d and 21 d after explant inoculation on RIM or SIM, respectively.
Fig. 2.
In vitro regeneration of 8-day-old cotyledonary explants of MT (left) and MT-Rg1 (right) cultivated on SIM (A) or RIM (B). Bar = 1cm. (This figure is available in colour at JXB online.)
A phenotypic characterization of plants harbouring the Rg1 allele was conducted comparing them with the NIL MT. The high in vitro shoot and root formation capacity of Rg1 seemed to parallel an improved shoot (branching) and root system in Rg1 plants growing in the greenhouse (Fig. 3A, 3C). Rg1 seedlings also tended to form supernumerary cotyledons (Fig. 3B), with an average number of seedlings forming three or bifurcated cotyledons, statistically different from MT (Table 2). It was also noticed that MT-Rg1 plants presented thicker petiole bases (Fig. 3D) with a diameter significantly larger than that of MT (Table 2). Histological analysis of MT-Rg1 petiole bases showed that they presented more parenchymatic cell layers, with smaller cells, when compared with MT (Fig. 3E). Although MT-Rg1 tended to display increased fresh and dry mass of the roots and shoots, paralleling its capacity to form these organs in vitro, it did not differ statistically from MT (Table 2).
Fig. 3.
The MT-Rg1 phenotype. (A) Branching phenotype of MT-Rg1. (B) MT-Rg1 seedlings showing two (1), three (2), and bifurcated cotyledons (3). (C) Root system of 85-day-old MT and MT-Rg1 plants. (D) MT and MT-Rg1 leaves. Note the thicker base of the MT-Rg1 petiole (arrow). (E) Transversal section of an MT and MT-Rg1 petiole. Bar = 1cm (A–D) and 100 µm (E). (This figure is available in colour at JXB online.)
Table 2.
Phenotypical data of MT and MT-Rg1
Means followed by the same letters are not significantly different according to Student’s t-test (P < 0.05). For cotyledons per seedlings, n = 4 trays, each containing 70 seedlings. For germination, n = 3 germinating boxes, each containing 50 seeds. For petiole diameter and organ weight, n = 10 plants. Shoot and root dry mass were determined on 60-day-old plants (n = 10) after drying in an oven at 60 °C.
| Cotyledons per seedling (%) | MT | MT-Rg1 |
|---|---|---|
| Two cotyledons | 99.72±0.28 a | 90.45±1.59 b |
| Three cotyledons | 0.28±0.28 b | 3.71±1.39 a |
| Bifurcated cotyledons | 0.00±0.00 b | 5.84±1.77 a |
| Germination (%) | 97.33±1.76 a | 96.67±1.76 a |
| Petiole diameter (mm) | 3.54±0.09 b | 3.97±0.06 a |
| Dry and fresh mass (g) | ||
| Root fresh mass | 2.81±0.22 a | 3.35±0.25 a |
| Root dry mass | 0.26±0.02 a | 0.28±0.02 a |
| Shoot fresh mass | 16.29±1.17 a | 16.81±0.78 a |
| Shoot dry mass | 1.64±0.16 a | 1.75±0.08 a |
In vitro shoot and root formation capacity of MT single mutants and MT-Rg1 double mutants
Tomato mutants were searched based on their effects on branching, the main ex vitro phenotype presented by Rg1, and also based on their gene functions as indicative of a possible capacity to control cell fates (Table 1). Mutants (dgt, pro, ls, and Me) introgressed into the same MT genetic background were tested for their capacity to form organs in SIM or RIM. Double mutants were then produced, combining the selected mutants with MT-Rg1, and tested for their regeneration capacity (Fig. 4).
Fig. 4.
Shoot (A) and root (B) regeneration in vitro, as well as the number of roots formed (C) in cotyledonary explants of single and double mutants. The explants were obtained from 8-day-old seedlings. SIM and RIM were used for shoot and root induction, respectively. Measurements were taken 21 d (A) and 8 d (B and C) after explant inoculation. Error bars represent the mean ±SE, n = 6 Petri dishes each containing 20 explants. Different letters indicate significant differences at P ≤ 0.01 (unpaired Student’s t-test). The mutants utilized are described in Table 1.
The pro mutant displayed a reduced capacity to form both roots and shoots in vitro (Fig. 4A, 4B), whereas dgt exhibited a reduced capacity to form roots (Fig. 4B). Although Me phenocopied the high shoot formation of Rg1 on SIM, it failed to increase root formation in RIM (Fig. 4). It is noteworthy that Rg1 rescued the low organ formation capacity of pro in the double mutant proRg1 and the reduced root formation capacity of dgt in dgtRg1, appearing as epistatic to both mutations. On the other hand, the presence of ls in a double mutant with Rg1dramatically reduced the shoot formation capacity, while ls alone did not present a reduction in organ formation in vitro. This epistatic reduction in shoot formation presented by the double mutant lsRg1 was not observed in Mels.
Phenotypes of MT single mutants and MT-Rg1 double mutants
Histological analysis of cotyledons from 8-day-old seedlings, which was the tissue used as explant in both RIM and SIM, was performed in both single and double mutants (Fig. 5). The Rg1 cotyledons had more spongy parenchyma. This high proportion of spongy parenchyma was also present in Me cotyledons, which also seem to have increased layers of palisade parenchyma. On the other hand, a clear reduction in the number of spongy parenchyma layers was observed for dgt cotyledons. Although the reduced parenchyma proliferation in dgt cotyledons was not reversed in the double mutant dgtRg1, Rg1 increased the number of parenchyma cells in pro and ls double mutants. The increased number of parenchyma cells in the double mutant lsRg1 was not observed in Mels, which indeed had fewer parenchyma cells than Me (Fig. 5).
Fig. 5.
Transversal sections of 8-day-old cotyledons of single and double mutants. Note the presence of the middle vein (vascular bundle) and part of the mesophyll constituted by palisade and spongy parenchyma in the upper (adaxial) and lower (abaxial) sides, respectively. Bar = 100 µm.
Greenhouse-grown double mutant plants (Fig. 6) consistently presented phenotypes confirming the epistatic interactions observed in vitro, but also showing some additive ex vitro phenotypes. The double dgtRg1 mutant showed increased root formation, as compared with the single mutant dgt (Fig. 6A), as well as a trend for a horizontal orientation in the first lateral roots (compare Fig. 6A with Fig. 3C). The lsRg1 plants combined the absence of petals, a pleitropic effect of ls (Fig. 6B), with axillary bud formation (Fig. 6C) and vigorous branching of Rg1 (Fig. 6D). Although the vigorous branching of lsRg1 plants does not parallel the reduction of shoot formation observed in vitro for this genotype (Fig. 4A), these results confirm the epistatic interactions between Rg1 and ls.
Fig. 6.
Phenotype of mutants and double mutants. (A) Increased root formation and growth in the double mutant dgtRg1, as compared with dgt. (B) Absence of petals in ls flowers. (C) Lack of axillary bud formation in ls and the reversion of this phenotype in the double mutant lsRg1. (D) Branching formation in the double lsRg1 mutant (right) and its absence in ls (left). Note that the double lsRg1 mutant combines branch outgrowth with flowers lacking the corolla (inset). (E) GUS staining of DR5::GUS root tips in the MT (left) and the Rg1 (right) background. (F) Increased lobing in leaflets of the double proRg1 mutant (right), as compared with pro (left). (G) Aspect of an Me leaf showing high dissection. Bar = 1cm (A, B, D, F, and G), 5mm (C), and 1mm (E). (This figure is available in colour at JXB online.)
Although the epistasis of Rg1 over the auxin low sensitivity mutant dgt might suggest that Rg1 increased auxin sensitivity, it does not seem to be the case, as indicated by the activity of the synthetic auxin promoter DR5 driving GUS expression in root tips (Fig. 6E). On the contrary, the staining of Rg1 suggests less sensitivity to auxin, which is reinforced by the GUS staining of Rg1 cotyledons incubated on RIM (Supplementary Fig. S1 at JXB online). As for the double proRg1 mutant, it was possible to observe a trend to revert the smooth leaflet margins of pro (Fig. 6F; Jasinski et al., 2008). However, it should be noted that Rg1 plants do not display intensely dissected leaves (Fig. 3D), as the knotted-like homeobox mutant Me does (Fig. 6G).
The double proRg1 mutant also showed that the presence of the Rg1 allele was able to reverse the characteristic non-branching phenotype of pro plants (Bassel et al., 2008), as evidenced in the analysis of the branching index (Fig. 7A). As mentioned, the double lsRg1 mutant proved that Rg1 can rescue the lack of axillary meristem in ls (Fig. 6C), increasing the branching index (Fig. 7A). Conversely, the high branching phenotype of Me was reversed when combined with ls (Fig. 7A). While Rg1 was able to improve the root system of dgt (Fig. 6A), its effect was less evident in shoots of the double mutant dgtRg1, as the plants showed significantly less branching than Rg1, but more branching than dgt (Fig. 7A).
Fig. 7.
Branching phenotype of MT-Rg1 and double mutants. (A) Branching index of single and double mutants measured in 80-day-old plants. The mutants utilized are described in Table 1. (B) Branching index of reciprocal grafting measured in 50-day-old plants. Genotypes are expressed as scion/rootstock. Error bars represent the mean ±SE, n = 7 (A) or n = 12 (B) plants. Different letters indicate significant differences at P ≤ 0.01 (unpaired Student’s t-test). The branching index represents the sum of the length of lateral branches divided by the main stem length (Morris et al., 2001).
One important question is whether the branching phenotype of Rg1 is grafting transmissible. Reciprocal graftings between Rg1 and MT were performed. A high branching phenotype was only observed when Rg1 was the scion (Fig. 7B), suggesting that Rg1 acts locally, probably modulating the capacity of the tissue to respond to the branching stimulus. This tissue-autonomous effect is consistent with the action of Rg1 improving in vitro shoot and root formation in isolated explants (Fig. 1).
Gene expression analysis of different tomato mutants and their double mutants with Rg1
Semi-quantitative RT-PCR analysis was conducted of genes associated with meristem formation, as well as auxin and GA signalling in Rg1 and double mutant plants. The aim of this analysis was to confirm, at the transcriptional level, the epistatic interactions observed in the in vitro and ex vitro phenotypes of the double mutants (Figs 4–7), as well as to gain more insight into the classes of genes that could be differentially regulated in Rg1.
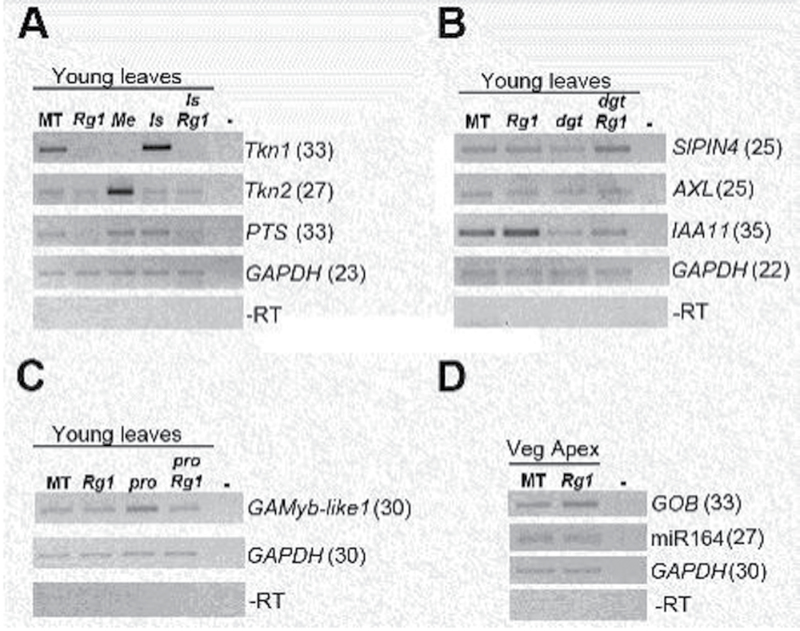
One class of regulatory genes analysed was the tomato Knotted-like homeobox (KNOX) genes Knotted1 (TKn1), Knotted2 (TKn2), and PETROSELINUM (PTS). In tomato, which is a compound-leaf species, this class of genes is expressed in young leaves (Koltai and Bird, 2000; Kim et al., 2003), contrasting with the expression restricted to the shoot meristem in the simple-leaf species Arabidopsis thaliana (Lincoln et al., 1994; Barton, 2001). In general, Rg1 presented a slight reduction in transcript accumulation of all KNOX genes analysed (TKn1, TKn2, and PTS; Fig. 8A). An increased transcript accumulation of TKn1 in the ls mutant was noticeable, which was reverted in the double lsRg1 mutant, thus resembling the epistasis presented by Rg1 over ls in the analysis of branching phenotype (Figs 6, 67). The Me mutation is a gene fusion that causes the overexpression of TKn2 (Chen et al., 1997; Parnis et al., 1997), and leaf tissues of Me plants consistently presented high transcript accumulation of this gene, when compared with the other genetic backgrounds, as expected (Fig. 8A).
Fig. 8.
Expression pattern of genes associated with meristem formation as well as auxin and gibberellin signalling in tomato cv Micro-Tom by semi-quantitative RT-PCR using GAPDH as the reference gene. (A) RT-PCR of tomato Knotted1 (TKn1), tomato Knotted2 (TKn2), and PETROSELINUM (PTS) in young leaves of the given genetic backgrounds: Micro-Tom, MT; Rg1; Mouse ears, Me; lateral suppresser, ls, and the lsRg1 double mutant. (B) RT-PCR analysis of the auxin pathway-associated genes S. lycopersicum PIN-FORMED4 (SlPIN4), AXR1-like (AXL), and AUX/IAA11 (IAA11) in young leaves of MT, Rg1, diageotropica (dgt), and the dgtRg1 double mutant. (C) RT-PCR analysis of the gibberellin pathway-associated gene GAMyb-like1 in young leaves of MT, Rg1, procera (pro), and the proRg1 double mutant. (D) Stem-loop RT-PCR analysis of the microRNA164 (miR164) and its target the NAC-domain GOBLET (GOB) in MT and Rg1 vegetative apexes. Tomato GAPDH was used as the reference gene. PCRs without reverse transcriptase (RT–) or without cDNA (–) are shown as negative controls. Numbers in parentheses represent PCR cycles for each amplicon. Two biological replicates were used for each genetic background with at least two technical PCR replicates.
For auxin-related genes, there was an increase in transcript accumulation of SlPIN4 in the double dgtRg1 mutant, when compared with dgt (Fig. 8B), suggesting that this gene, which is the most highly expressed PIN gene in tomato (Pattison and Catalá, 2012), may be one of the components of the epistatic interaction seen in vitro and ex vitro between dgt and Rg1 (Figs 4–7). On the other hand, the low transcript accumulation of IAA11 in dgtRg1 when compared with Rg1 and MT indicated an epistatic effect of dgt, which presented a low transcript accumulation of this gene, as previously reported (Nebenfuhr et al., 2000).
The same type of epistatic interaction at the transcript accumulation level was also observed for the GA pathway-associated gene GAMyb-like1 (Fig. 8C). High GAMyb-like1 transcript levels in leaf tissues of the GA-constitutive mutant pro (Fig. 8C) (see Bassel et al., 2008) was no longer observed in the double mutant proRg1, which correlated well with the epistasis of Rg1 over pro observed in vitro and ex vitro (Figs 4–7). No obvious alterations in transcript accumulation of the miR164 and its target gene the NAC-domain GOBLET (GOB; Berger et al., 2009) were observed in Rg1 vegetative apexes, when compared with MT (Fig. 8D).
Discussion
Organogenic capacity is not necessarily linked to alterations of hormonal status
In the present work, the characterization of the natural genetic variation Rg1 was extended by demonstrating its additional capacity to improve root formation in vitro, besides shoot formation. This allele is also able to reverse the low levels of in vitro root and shoot formation in the DELLA mutant pro, as well as the low in vitro root formation of the cyclophilin mutant dgt. These results may suggest increased auxin and reduced GA signalling in Rg1 to overcome the low auxin sensitivity of dgt and the constitutive GA signalling in pro, respectively. However, analysis of transcript accumulation from key genes associated with the auxin signalling pathway (Fig. 8B), combined with the analysis of the activity of the synthetic DR5 auxin response promoter (Fig. 6E; Supplementary Fig. S1 at JXB online), did not support an increase in auxin sensitivity in Rg1. Moreover, tomato mutants with increased auxin sensitivity would present a reduced complexity of leaf architecture (e.g. leaves with fewer leaflets and less dentated leaflets), as has been described for the AUX/IAA9 tomato mutant entire (Wang et al., 2005; Zhang et al., 2007). This seems not to be the case for Rg1 leaves (Fig. 3D), despite the fact that the RT-PCR analysis suggested that Rg1 is expressed in leaves, since it affects the expression of a number of genes in young leaves (Fig. 8A–C). Similarly, a reduction in GA sensitivity in Rg1 would result in plant dwarfism and low seed germination, as described for various GA-deficient tomato mutants at different intensities (Koornneef et al., 1990). Neither a reduction in seed germination (Table 2) nor dwarfism were observed in Rg1 (Fig. 3A), which is consistent with the lack of alteration in levels of GAMyb-like1 transcripts in MT-Rg1 when compared with MT (Fig. 8C). Although the possibility that the Rg1 locus affects distinct auxin- and GA-related pathways not tested here cannot be ruled out, Rg1 is most probably not affecting hormonal homeostasis, but rather developmental processes also affected by the auxin and GA signalling pathway.
The relationship between in vitro organogenic capacity and shoot branching
A remarkable unexpected result was that the Rg1 allele was able to reverse the absence of axillary meristems in the ls mutant, rescuing the non-branching phenotype. There is considerable evidence that the flux of auxin from lateral buds, which can be influenced by cytokinins and the novel hormone strigolactone, controls branching (Leyser, 2009). The role of GA in controlling branching is also evident (Fig. 7A; Bassel et al., 2008), although less considered in the current signalling pathway. Additionally, before bud outgrowth, the presence of pre-existing axillary meristems and lateral shoot primordia is required, and these are promoted by the GRAS protein LATERAL SUPPRESSER (LS). This predicts that the loss-of-function ls mutant is probably epistatic to most mutations affecting the main components controlling lateral bud outgrowth. In this context, Rg1, which is epistatic to ls, should be situated at least at the same genetic hierarchical level as LS, acting locally to promote the presence of cells capable of undergoing the formation of axillary meristem and bud primordium. This is consistent with the fact that Rg1 is not graft transmissible (Fig. 7B), and that Rg1 is also epistatic to the non-branching phenotype of the GA-constitutive mutant pro.
Competence as the capacity to improve different types of organs in vitro and ex vitro
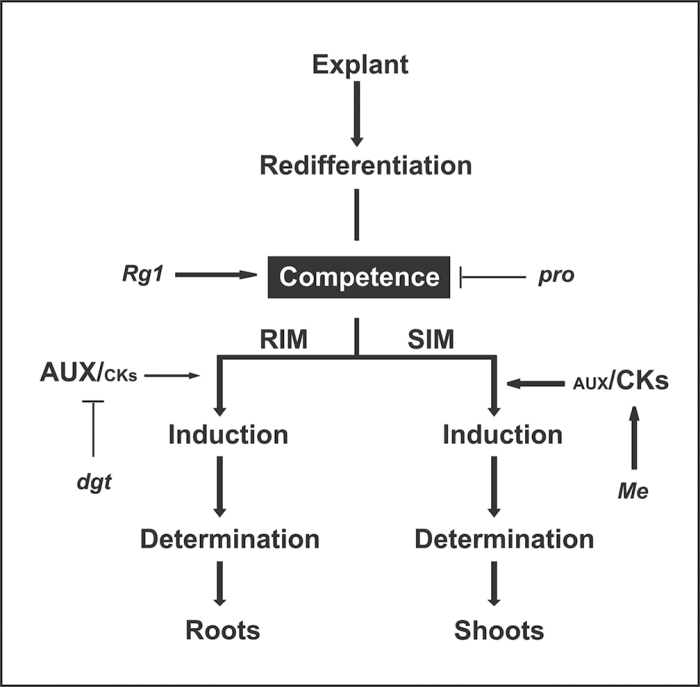
The fact that Rg1 can be epistatic to distinct and apparently unrelated non-branching mutants, together with its capacity to improve different types of organs in vitro, indicate thats Rg1 is controlling a common requirement necessary for all these processes. This requirement would probably be the production of competent cells able to undergo further differentiation and to form roots and shoots in vitro, as well as axillary meristems ex vitro. The present results, together with both classical (Bonnet and Torrey, 1966) and recent studies (Atta et al., 2009; Sugimoto et al., 2010) suggesting a common initial developmental pathway for different organs, led to the reinterpretation of the Christianson and Warnick (1988) model, adding that the stage of ‘acquisition of competence’ is probably a general process, necessary for both shoot and root formation. This initial common competence would contrast with the stage of ‘induction’, which requires specific auxin-to-cytokinin balance (Skoog and Miller, 1957), leading to subsequent ‘determination’ to form shoots or roots, but not both organs. Based on these concepts, it is proposed here that Rg1 and pro mutants are probably positive and negative regulators, respectively, of the stage of ‘acquisition of competence’, since they have a large influence on both root and shoot formation. On the other hand, the specific high shoot formation in Me (Fig. 4A) and the low root formation in dgt (Fig. 4B) suggest that these mutants are affecting the stage of ‘induction’ of roots and shoots (Fig. 9). It is reasonable to assume that the low sensitivity of dgt to auxin (Kelly and Bradford, 1986), and the known effect of KNOX genes, such as Me, improving cytokinin levels (Hay et al., 2004), probably represent disturbances in the auxin-to-cytokinin balance, or the endogenous response to these hormones, disfavouring root induction in dgt and favouring shoot induction in Me (Fig. 9). Different from Me and dgt, Rg1 seems to be controlling competence to form both shoots and roots, instead of the specific induction of these organs. If so, Rg1 should not present conspicuous alterations in the auxin-to-cytokinin balance, which, otherwise, would improve the induction of one type of organ to the detriment of the other (Skoog and Miller, 1957; Li et al., 1992). This is supported by the evidence presented here that Rg1 has no significant alterations in auxin, together with hormonal measurements made by Boiten et al. (2004), which showed that Rg1 does not confer an increase in the endogenous levels of cytokinin. Moreover, it appears that the high competence of Rg1 is able not only to revert the lack of competence of pro, consequently improving both root and shoot formation in the double mutant proRg1, but also to compensate the poor root induction in dgt, improving root formation in the double mutant dgtRg1.
Fig. 9.
A working hypothesis for the contribution of the mutations studied here (see Fig. 4) in the proposed phases controlling in vitro regeneration (Christianson and Warnick, 1988). The pro mutant and the Rg1 allele have negative and positive effects, respectively, in the phase of acquisition of competence, which are reflected in their capacity to form both root and shoot. The dgt mutant probably affects the induction phase, since its low auxin sensitivity represents an altered response to the auxin-to-cytokinin balance (AUX/CKs) necessary for root induction. The known effect of KNOX genes (represented here by the Me mutant) increasing cytokinin (Hay et al., 2004) may cause an indirect effect, through alterations in the auxin-to-cytokinin balance or a direct effect on the shoot induction phase.
The more determined the less competent
One important corollary of the model proposed by Christianson and Warnick (1988) is that determination can also be interpreted as opposite to competence, since an explant highly committed (‘determined’) to a particular developmental pathway (Tran Thanh Van, 1973) will probably be more recalcitrant (‘non-competent’) to assume a different fate. These considerations may help further elucidation of the molecular basis of competence, since genes positively affecting this process, such as Rg1, may be arresting the specification of cell fates, or maintaining the population of indeterminate cells (stem cells) in a given explant (Sugimoto et al., 2011). It is noteworthy that Rg1 was initially selected by its capacity to maintain shoot regeneration after long-term in vitro callus culture, which is lost much more quickly for other tomato genotypes under the same circumstances (Koornneef et al., 1987). This behaviour of Rg1 was also confirmed here by comparing the shoot formation capacity of MT and MT-Rg1 calli induced on CIM and then transferred to SIM (Fig. 1C). Similarly, in a previous study, it had been shown that the capacity for shoot formation in cotyledon explants of MT decreases proportionally with age, but it is maintained in the near isogenic Rg1 line (see fig. 2B in Pino et al., 2010). Rg1 also presented an increased number of cells in transversal sections of petioles and cotyledons (Fig. 5). However, the higher regeneration capacity does not seem to be linked to this higher number of cells phenotype, since the double mutant dgtRg1, which presented enhanced regenerations of both roots and shoots, displays a reduced number of cells in the explants. Further, despite the fact that it is being considered here that Me is only affecting the phase of induction of shoots, but not the competence to form both roots and shoots, it presented an enhanced number of cells similar to that observed in Rg1.
The genetic basis of organogenic competence
Among the genes that maintain indeterminate cell fates are specific members of homeoboxes, such as the maize KNOTTED1 (Sinha et al., 1993). However, if KNOTTED-like genes in general can improve the capacity to assume different developmental fates, it would be expected that an improved root formation on RIM in the TKn2 overexpresser mutant Me studied here would be obtained, which was not the case (Fig. 4B). Considering the model plant Arabidopsis, another important requisite of a gene controlling competence is probably the capacity to be expressed in roots, since these organs are the preferred source for explants (Valvekens et al., 1988). Cultivated tomato does not exhibit the capacity to regenerate shoots from root explants (Koornneef et al., 1993; Peres et al., 2001), but this capacity is gained by the presence of the Rg1 allele (Koornneef et al., 1993; Lima et al., 2004). When tomato cotyledons or true leaves are used as explants, an initial down-regulation of shoot-specific homeobox genes might be necessary. This may explain the observed reduction of transcript accumulation of TKn1 and, to a lesser extent, TKn2 in Rg1 young leaves (Fig. 8A), which are tomato explants with the same regeneration capacity as cotyledons (Kut and Evans, 1982). Conversely, a surprising result was the observed high transcript accumulation of TKn1 in the mutant ls. It is noteworthy that ls presented a high in vitro formation of shoots from cotyledon explants (Fig. 4A), which suggests that the developmental programme controlling ex vitro axillary meristem formation is distinct from that controlling in vitro adventitious meristem formation. Whether the homeobox TKn1 is relevant for such developmental programmes remains to be determined. Curiously, in Arabidopsis, elevated expression of KNAT1, homologous to TKn1 (Hay ande Tsiantis, 2010), alters leaf architecture (Chuck et al., 1996), a phenotype not observed in the ls mutant.
In summary, the characterization of tomato mutants indicated that the acquisition of competence for organogenesis is a common pathway controlled by genes that are most probably upstream to those controlling the process of induction of specific organs. In the case of shoot induction in Arabidopsis, there are indications that some of the downstream genes may be the ENHANCER OF SHOOT REGENERATION 1 (ESR1), ARR15, POLYGALCTURONASE INHIBITING PROTEIN 2 (PGIP2) (Banno et al., 2001; Che et al., 2007) and the homeobox genes WUS and STM (Cary et al., 2002; Gallois et al., 2002; Che et al., 2007), whose expression usually occurs after transfer to SIM. Currently, very few genes are known to act upstream of the aforementioned genes in the control of organ formation. However, a recent study (Motte et al., 2011) demonstrated that CUP SHAPED COTYLEDON2 (CUC2), which is expressed upstream to the homeoboxes WUS and STM during shoot induction in cultured root explants (Gordon et al., 2007), marks the site of both shoot and lateral root primordium formation in Arabidopsis. It is noteworthy that the tomato mutant Goblet-4d, which has a gain of function in the CUC2 gene (Berger et al., 2009), presents a high frequency of supernumerary cotyledons, a phenotype also shown by Rg1 and other mutants affecting in vitro regeneration capacity (Chaudhury et al., 1993; Chandler, 2008). Therefore, it is possible that the expression of CUC2 during acquisition of competence is necessary for further action of the homeobox genes in the determination of shoot meristems. The present data suggested that the action of the DELLA protein PROCERA is also required during acquisition of competence, which is consistent with previous studies in Arabidopsis (Ezura and Harberd, 1995) showing that reduced levels or sensitivity to GA, implying that the DELLA proteins are active as a growth repressor (Harberd et al., 2009), are associated with enhanced in vitro organ formation. It is interesting to note that PROCERA was also considered as modulating competence to respond to KNOX-dependent signals that direct leaflet formation (Jasinski et al., 2008). Besides PROCERA and the NAC transcription factor CUC2, there would be more genes controlling competence. One of those key genes is likely to be RG1, whose future cloning and identity will provide more insights into this developmental pathway, and the understanding of the intriguing capacity of plants to form organs during their life cycle.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. GUS staining of DR5::GUS cotyledons in MT (left) and Rg1 genetic background.
Table S1. Primers used in the RT-PCR analysis.
Supplementary Material
Acknowledgements
This work was partially funded by ‘Fundação de Amparo à Pesquisa do Estado de São Paulo’ (FAPESP-07/07175-0). We thank ‘Conselho Nacional de Desenvolvimento Científico e Tecnológico’ (CNPq) for fellowships (AF, BA da G, and LEPP), and ‘Coordenação de Aperfeiçoamento de Pessoal de Nível Superior’ (CAPES) (SL-C) and FAPESP (MSA, GFFS, and LEP) for scholarships granted. We thank Dr Roger Chetelat (Tomato Genetics Resource Center, Davis, USA) and Dr Maarten Koornneef (Max Plank Institute for Plant Breeding Research, Cologne, Germany) for the donation of tomato seeds in their original genetic backgrounds. Guilherme P. Oliveira is acknowledged for conducting callus culture in MT. We thank Cassia R.F. Figueiredo for laboratory assistance, NAP/MEPA ESALQ/USP for microscopy facilities, and Francisco Vitti for greenhouse assistance.
Glossary
Abbreviations
- BA
benzyl adenine
- CIM
callus-inducing medium
- 2,4-D
2,4-dichlorophenoxyacetic acid
- GA
gibberellin
- MT
Micro-Tom
- NAA
naphthaleneacetic acid
- PVC
polyvinyl chloride
- RIM
root-inducing medium
- SIM
shoot-inducing medium
This paper is available online free of all access charges (see http://jxb.oxfordjournals.org/open_access.html for further details)
References
- Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc’h A, Carnero E, Giraudat-Pautot V, Rech P, Chriqui D. 2009. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro The Plant Journal 57 626–644 [DOI] [PubMed] [Google Scholar]
- Banno H, Ikeda Y, Niu Q-W, Chua N-H. 2001. Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration Thye Plant Cell 13 2609–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK. 2001. Leaving the meristem behind: regulation of KNOX genes Genome Biology 2 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Mullen RT, Bewley JD. 2008. procera is a putative DELLA mutant in tomato (Solanum lycopersicum): effects on the seed and vegetative plant Journal of Experimental Botany 59 585–593 [DOI] [PubMed] [Google Scholar]
- Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, Zinder M, Samach A, Eshed Y, Ori N. 2009. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves Development 136,823–832 [DOI] [PubMed] [Google Scholar]
- Boiten H, Azmi A, Dillen W, Schepper S, Debergh P, Gerats T, Onckelen H, Prinsen E. 2004. The Rg-1 encoded regeneration capacity of tomato is not related to an altered cytokinin homeostasis New Phytologist 161 761–771 [DOI] [PubMed] [Google Scholar]
- Bonnett HT, Jr, Torrey JG. 1966. Comparative anatomy of endogenous bud and lateral root formation in Convolvulus arvensis roots cultured in vitro American Journal of Botany 53 496–507 [Google Scholar]
- Carvalho RF, Campos ML, Pino LE, Crestana SL, Zsögön A, Lima JE, Benedito VA, Peres LEP. Convergence of developmental mutants into a single tomato model system: ‘Micro-Tom’ as an effective toolkit for plant development research. Plant Methods. 2011;7:18. doi: 10.1186/1746-4811-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary AJ, Che P, Howell SH. 2002. Developmental events and shoot apical meristem gene expression patterns during shoot development in Arabidopsis thaliana The Plant Journal 32 867–877 [DOI] [PubMed] [Google Scholar]
- Catterou M, Dubois F, Smets R, Vaniet S, Kichey T, Van Onckelen H, Sangwan-Norreel BS, Sangwan RS. 2002. hoc: an Arabidopsis mutant overproducing cytokinins and expressing high in vitro organogenic capacity The Plant Journal 30 273–287 [DOI] [PubMed] [Google Scholar]
- Chandler JW. 2008. Cotyledon organogenesis Journal of Experimental Botany 59 2917–2931 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Letham S, Craig S, Dennis ES. 1993. amp1: a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering The Plant Journal 4 907–916 [Google Scholar]
- Che P, Lall S, Howell SH. 2007. Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture Planta 226 1183–1194 [DOI] [PubMed] [Google Scholar]
- Che P, Lall S, Nettleton D, Howell SH. 2006. Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture Plant Physiology 141 620–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Research. 2005;33:179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Janssen BJ, Williams A, Sinha N. 1997. A gene fusion at a homeobox locus: alterations in leaf shape and implications for morphological evolution The Plant Cell 9 1289–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson ML, Warnick DA. 1985. Temporal requirement for phytohormone balance in the control of organogenesis in vitro Developmental Biology 112 494–497 [Google Scholar]
- Christianson ML, Warnick DA. 1988. Organogenesis in vitro as a developmental process HortScience 23 515–519 [Google Scholar]
- Chuck G, Lincoln C, Hake S. 1996. KNAT1 induces lobed leaves with ectopic meristem when overexpressed in Arabidopsis The Plant Cell 8 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclercq J, Sangwan-Norreel B, Catterou M, Sangwan RS. 2011. De novo shoot organogenesis: from art to science Trends in Plant Science 16 597–606 [DOI] [PubMed] [Google Scholar]
- Estruch JJ, Prinsen E, Van Onckelen H, Schell J, Spena A. 1991. Viviparous leaves produced by somatic activation of an inactive cytokinin-synthesizing gene Science 254 1364–1367 [DOI] [PubMed] [Google Scholar]
- Ezura H, Harberd NP. 1995. Endogenous gibberellin levels influence in vitro shoot regeneration in Arabidopsis thaliana (L.) Heynh. Planta 197 301–305 [DOI] [PubMed] [Google Scholar]
- Fosket DE. 1994. Plant growth and development New York: Academic Press; [Google Scholar]
- Gallois JL, Woodward C, Reddy GV, Sablowski R. 2002. Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis Development 129 3207–3217 [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. 1968. Nutrient requirement of suspension cultures of soybean root cells Experimental Cell Research 50 151–158 [DOI] [PubMed] [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM. 2007. Pattern formation during de novo assembly of the Arabidopsis shoot meristem Development 134 3539–3548 [DOI] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y. 2009. The angiosperm gibberellin–GID1–DELLA growth regulatory mechanism: how an ‘inhibitor of an inhibitor’ enables flexible response to fluctuating environments The Plant Cell 21 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JR. 1992. Domestication of vegetatively reproduced crops. In: Harlan JR, ed. Crops and man 2nd edn Madison, WI: American Society of Agronomy and Crop Science Society of America; 130–133 [Google Scholar]
- Hay A, Craft J, Tsiantis M. 2004. Plant hormones and homeoboxes: bridging the gap? BioEssays 26 395–404 [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2010. KNOX genes: versatile regulators of plant development and diversity Development 137 3153–3165 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Tattersall A, Piazza P, Hay A, Martinez-Garcia JF, Schmitz G, Theres K, McCormick S, Tsiantis M. 2008. PROCERA encodes a DELLA protein that mediates control of dissected leaf form in tomato The Plant Journal 56 603–612 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants EMBO Journal 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky MJ. 1965. A formaldehyde–glutaraldehyde fixative of high osmolality for use in electron microscopy Journal of Cell Biology 27 137–138 [Google Scholar]
- Kauffman JB. 1991. Survival by sprouting following fire in tropical forests of the Eastern Amazon Biotropica 23 219–224 [Google Scholar]
- Kelly MO, Bradford KJ. 1986. Insensitivity of the diageotropica tomato mutant to auxin Plant Physiology 82 713–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Pham T, Hamidi A, McCormick S, Kuzoff RK, Sinha N. 2003. Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX gene in generating compound leaves Development 130 4405–4415 [DOI] [PubMed] [Google Scholar]
- Koltai H, Bird DM. 2000. Epistatic repression of PHANTASTICA and class 1 KNOTTED genes is uncoupled in tomato The Plant Journal 22 455–459 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Bade J, Hanhart C, Horsman K, Schel J, Soppe W, Verkerk R, Zabel P. 1993. Characterization and mapping of a gene controlling shoot regeneration in tomato The Plant Journal 3 131–141 [Google Scholar]
- Koornneef M, Bosma T DG, Hanhart CJ, Van der Veen JH, Zeevaart JAD. 1990. The isolation and characterization of gibberellin-deficient mutants in tomato Theoretical and Applied Genetics 80 852–857 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Martinelli L. 1987. A genetic analysis of cell culture traits in tomato Theoretical and Applied Genetics 74 633–641 [DOI] [PubMed] [Google Scholar]
- Kut SA, Evans DA. 1982. Plant regeneration from cultured leaf explants of eight wild tomato species and two related Solanum species In Vitro 18 593–598 [Google Scholar]
- Leyser O. 2009. The control of shoot branching: an example of plant information processing Plant, Cell and Environment 32 694–703 [DOI] [PubMed] [Google Scholar]
- Li Y, Hagen G, Guilfoyle TJ. 1992. Altered morphology in transgenic tobacco plants that overproduce cytokinins in specific tissues and organs Developmental Biology 153 386–395 [DOI] [PubMed] [Google Scholar]
- Lima JE, Benedito VA, Figueira A, Peres LEP. 2009. Callus, shoot and hairy root formation in vitro is affected by the sensitivity to auxin and ethylene in tomato mutants Plant Cell Reports 28 1169–1177 [DOI] [PubMed] [Google Scholar]
- Lima JE, Carvalho RF, Neto AT, Figueira A, Peres LEP. 2004. Micro-MsK: a tomato genotype with miniature size, short life cycle, and improved in vitro shoot regeneration Plant Science 167 753–757 [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. 1994. A knotted -like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants The Plant Cell 6 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí E, Carrera E, Ruiz-Rivero O, Garcia-Martinez JL. 2010. Hormonal regulation of tomato gibberellin 20-oxidase1 expressed in Arabidopsis Journal of Plant Physiology 167 1188–1196 [DOI] [PubMed] [Google Scholar]
- Meissner R, Jacobson Y, Melamed S, Levyatuv S, Shalev G, Ashri A, Elkind Y, Levy A. 1997. A new model system for tomato genetics The Plant Journal 12 1465–1472 [Google Scholar]
- Miller CO Skoog F Von Saltza MH Strong F 1955Kinetin, a cell division factor from deoxyribonucleic acid Journal of the American Chemical Society 77,1392 [Google Scholar]
- Morris SE, Turnbull CGN, Murfet IC, Beveridge CA. 2001. Mutational analysis of branching in pea Evidence that Rms1 and Rms5 regulate the same novel signal Plant Physiology 126 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motte H, Verstraeten I, Werbrouck S, Geelen D. 2011. CUC2 as an early marker for regeneration competence in Arabidopsis root explants. Journal of Plant Physiology 168 1598–1601 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures Physiologia Plantarum 15 473–497 [Google Scholar]
- Nebenfuhr A, White TJ, Lomax T. 2000. The diageotropica mutation alters auxin induction of a subset of the Aux/IAA gene family in tomato Plant Molecular Biology 44 73–84 [DOI] [PubMed] [Google Scholar]
- Oh K, Ivanchenko MG, White TJ, Lomax TL. 2006. The diageotropica gene of tomato encodes a cyclophilin: a novel player in auxin signaling Planta 224 133–144 [DOI] [PubMed] [Google Scholar]
- Parnis A, Cohen O, Gutfinger T, Hareven D, Zamir D, Lifschitz E. 1997The dominant developmental mutants of tomato Mouse-ear and Curl are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene The Plant Cell 9 2143–2158 [Google Scholar]
- Pattison RJ, Catalá C. 2012. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families The Plant Journal 70 585–598 [DOI] [PubMed] [Google Scholar]
- Peres LEP, Carvalho RF, Zsögön A, Bermudez-Zambrano OD, Robles WGR, Tavares S. 2005. Grafting of tomato mutants onto potato rootstocks: an approach to study leaf-derived signaling on tuberization Plant Science 169 680–688 [Google Scholar]
- Peres LEP, Kerbauy GB. 1999. High cytokinin accumulation following root tip excision changes the endogenous auxin to cytokinin ratio during root-to-shoot conversion in Catasetum fimbriatum Lindl. (Orchidaceae) Plant Cell Reports 18 1002–1006 [Google Scholar]
- Peres LEP, Morgante PG, Vechi C, Kraus JE, Van Sluys M-A. 2001. Shoot regeneration capacity from roots and transgenic hairy roots of different tomato cultivars and wild related species Plant Cell, Tissue and Organ Culture 65, 37–44 [Google Scholar]
- Pino LE, Lombardi-Crestana S, Azevedo MS, Scotton DC, Borgo L, Quecini V, Figueira A, Peres LEP. The Rg1 allele as a valuable tool for genetic transformation of the tomato Micro-Tom model system. Plant Methods. 2010;6:23. doi: 10.1186/1746-4811-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai WS. 1973. Simple method for differential staining of paraffin embedded plant material using toluidine blueO. Stain Technology 48 247–249 [DOI] [PubMed] [Google Scholar]
- Santos AM, Oliver MJ, Sánchez AM, Payton PR, Gomes JP, Miguel C, Oliveira MM. 2009. An integrated strategy to identify key genes in almond adventitious shoot regeneration Journal of Experimental Botany 60 4159–4173 [DOI] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K. 1999. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family Proceedings of the National Academy of Sciences, USA 96 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J, Harbaugh B. 1989. Micro-Tom: a miniature dwarf tomato Florida Agricultural Experiment Station Circular 370 1–6 [Google Scholar]
- Sinha NR, Williams RE, Hake S. 1993. Overexpression of the maize homeobox gene, KNOTTED1, causes a switch from determinate to indeterminate cell fates Genes and Development 7 787–795 [DOI] [PubMed] [Google Scholar]
- Skoog F, Miller CO. 1957. Chemical regulation of growth and organ formation in plant tissues cultured in vitro Symposia of the Society for Experimental Biology 11 118–131 [PubMed] [Google Scholar]
- Smith LG, Jackson D, Hake S. 1995. Expression of knotted1 marks shoot meristem formation during maize embryogenesis Developmental Genetics 16 344–348 [Google Scholar]
- Sugimoto K, Gordon SP, Meyerowitz EM. 2011. Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends in Cell Biology 21 212–218 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM. 2010. Arabidopsis regeneration from multiple tissues occurs via a root development pathway Developmental Cell 18 463–471 [DOI] [PubMed] [Google Scholar]
- Tran Thanh Van M. 1973. Direct flower neoformation from superficial tissue of small explants of Nicotiana tabacum L Planta 115 87–92 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux⁄IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements The Plant Cell 9 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. 1988. Agrobacterium tumefaciens mediated transformation of Arabidopsis thaliana root explants by using kanamycin seletion Proceedings of the National Academy of Sciences, USA 85 5536–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil IK. 2008. A history of plant biotechnology: from the Cell Theory of Schleiden and Schwann to biotech crops Plant Cell Reports 27 1423–1440 [DOI] [PubMed] [Google Scholar]
- Visser EJW, Cohen JD, Barendse GWM, Blom CWPM, Voesenek LACJ. 1996. An ethylene-mediated increase in sensitivity to auxin induces adventitious root formation in flooded Rumex palustris Sm. Plant Physiology 112 1687–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latché A, Pech J, Bouzayen M. 2005. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis The Plant Cell 17 2676–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareing PF. 1982. Determination and related aspects of plant development. In: Smith H, Grierson D, eds. The molecular biology of plant development Oxford: Blackwell Scientific Publications; 517–541 [Google Scholar]
- White PR. 1934. Potentially unlimited growth of excised tomato root tips in a liquid medium Plant Physiology 9 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen R, Xiao J, Qian C, Wang T, Li H, Ouyang B, Ye Z. 2007. A single-base deletion mutation in SlIAA9 gene causes tomato (Solanum lycopersicum) entire mutant Journal of Plant Research 120 671–678 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.