Abstract
Plantaricin NC8 (PLNC8), a coculture-inducible two-peptide bacteriocin from Lactobacillus plantarum NC8, has recently been purified and genetically characterized. Analysis of an 8.1-kb NC8 DNA region downstream of the PLNC8 operon revealed the presence of at least four operons involved in bacteriocin production, showing high homology to the plantaricin cluster in L. plantarum C11. However, we found a three-component regulatory operon involving a quorum-sensing mechanism. Two of these components, the induction factor (PLNC8IF) and the histidine kinase, are novel, while the response regulator is identical to PlnD from C11. Homologous expression of plNC8IF in NC8 allowed constitutive bacteriocin production. Heterologous expression of this gene in Lactococcus lactis MG1363 produced supernatants which promoted bacteriocin production in NC8. Reverse transcription-PCR studies indicated that cocultivation of NC8 with inducing cells promoted transcription of the bacteriocin and regulatory operons in NC8. An identical result was obtained after addition of an external source of PLNC8IF. We propose that the presence of specific bacteria could act as an environmental signal that is able to switch on bacteriocin production in L. plantarum NC8 via a quorum-sensing mechanism mediated by PLNC8IF.
Lactic acid bacteria (LAB) can produce a wide range of substances with antimicrobial activity (24, 27, 48). Among these, the proteinaceous compounds called bacteriocins have been a focus of research because of their potential use as natural food preservatives (7, 8). Bacteriocins are peptides or proteins with antimicrobial activity directed against related species (24, 27, 48). Bacteriocins produced by LAB can be classified into four main groups according to their biochemical and genetic properties (17, 27, 35). Class II bacteriocins are small, heat-stable, cationic, and hydrophobic peptides, which are synthesized as precursor molecules containing, in most cases, a leader peptide of the so-called double-glycine type (17, 35). This leader peptide is recognized and cleaved by a dedicated ABC transporter, resulting in translocation of mature, active bacteriocin to the medium (21). The class IIb subgroup contains bacteriocins whose antimicrobial activity depends on the complementary action of two different peptides (17, 35).
Production and export of class II bacteriocins require several genes, which are usually organized into two or more operons (17, 35). The bacteriocin synthesis operon is composed of one or two (in class IIb) structural genes followed by a gene encoding the immunity protein. A second operon encoding the machinery necessary for the processing, transport, and secretion of the bacteriocins is usually located in the vicinity of the bacteriocin genes. This operon is formed by two or more genes encoding an ABC transporter and its accessory protein (17, 35).
Production of several class II bacteriocins is regulated by a so-called three component regulatory system (17, 28, 29, 35, 36). In these cases, a regulatory operon has been found to be involved in bacteriocin production. This operon is composed of a gene encoding an induction factor (IF) or peptide pheromone (Pph) followed by two genes encoding a histidine kinase protein (HK) and a response regulator (RR). Such an IF acts as an indicator of the cell density, which is sensed by the corresponding HK, resulting in activation of the RR, which then activates expression of all operons necessary for bacteriocin synthesis, transport, and regulation. This quorum-sensing or autoinduction mechanism mediated by inducer peptides has been found in Carnobacterium piscicola (3, 30, 40, 45), Lactobacillus plantarum (4, 11), Lactobacillus sakei (4, 12), and Enterococcus faecium (37, 38). In most of these systems, however, bacteriocin production is an unstable phenotype (10, 12, 16, 37, 44). This instability has been attributed to reduced synthesis of the IF under experimental conditions (28, 36). Therefore, it has been suggested that environmental factors may play an important role in the regulation of bacteriocin production, by means of increasing the basal levels of the IFs (4, 36, 42). Thus, sakacin A production by L. sakei Lb706 and Lactobacillus curvatus LTH1174 was found to be a temperature-sensitive process (12). Nevertheless, how the environment interacts with the regulation of bacteriocin production is still poorly understood.
Recently, a novel class IIb bacteriocin, plantaricin NC8 (PLNC8), has been biochemically and genetically characterized (33). Interestingly, this bacteriocin was produced by L. plantarum NC8 only after cocultivation with specific gram-positive strains or the addition of heat-killed cells from some of the inducing strains (33, 34). However, no cell-free supernatants (CFS) from any of the inducing strains had any effect (34). Along with the induced bacteriocin PLNC8, an autoinducing activity was also produced after exposure of NC8 cultures to the inducing conditions. In this case, the CFS from a previously induced (bac+) NC8 culture was able to induce bacteriocin production in a fresh NC8 single culture (34). This observation was confirmed by sequence analysis of the promoter region of the plNC8BAC operon (33), which maintains the consensus of promoters of class II bacteriocin operons whose expression is dependent on an autoinduction mechanism (36, 42).
In this study we have gained insight on the autoinduction phenomenon observed after coculture induction of bacteriocin production in L. plantarum NC8. Thus, molecular analysis of an 8.1-kb DNA sequence downstream of the PLNC8 operon has revealed the presence of at least four operons involved in bacteriocin synthesis, transport, and regulation. Although these operons are very similar to those for bacteriocin production in L. plantarum C11, a new regulatory operon of the three-component type has been found. Homologous and heterologous expression of the novel inducer peptide (PLNC8IF) of this operon supports our previous observations in the sense that an autoinduction mechanism is activated in NC8 as a response to the presence of specific bacteria in its own environment (34). This observation has been confirmed by reverse transcription-PCR (RT-PCR) analysis of the expression of the bacteriocin and regulatory operons found in NC8. To our knowledge, this is the first report on a quorum-sensing mechanism mediating induction of bacteriocin production after cocultivation.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
L. plantarum NC8, kindly provided by Lars Axelsson (MATFORSK, Norwegian Food Research Institute, Osloveien, Norway), has been described recently as producing an inducible two-peptide bacteriocin named PLNC8 in response to cocultivation with specific gram-positive bacteria (33, 34). It was propagated in MRS broth (Oxoid, Basingstoke, Hampshire, England) at 30°C without shaking. Lactococccus lactis MG1363 was grown in M17 broth (Oxoid) plus 1% (wt/vol) glucose (GM17) at 30°C without shaking. Where appropriate, erythromycin (Fluka Chemie GmbH, Buchs, Switzerland) was added to the culture medium at 10 μg/ml (final concentration).
Escherichia coli DH5α was grown in Luria-Bertani broth (43) at 37°C with vigorous agitation. E. coli DH5α transformant cells harboring the recombinant plasmid pBluescript II KS(+) (Stratagene Europe, Amsterdam, The Netherlands) or pSIG306 (this work) were selected on Luria-Bertani agar plates supplemented with 150 μg of ampicillin (Fluka) per ml or 200 μg of erythromycin per ml (final concentration), respectively, 16 μl of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 50 mg/ml; Promega Co., Madison, Wis.) per plate, and 4 μl of isopropyl β-d-thiogalactoside (IPTG; 200 mg/ml; Gibco BRL, Basel, Switzerland) per plate.
DNA isolation and transformation procedures.
Total genomic DNA from L. plantarum NC8 was isolated by the method of Cathcart (5). Plasmid DNAs from L. plantarum and L. lactis were isolated by the method of Anderson and McKay (1). Plasmid DNA from E. coli was extracted as described previously (43). L. plantarum NC8 and L. lactis MG1363 were electroporated according to the methods of Aukrust and Blom (2) and Holo and Nes (22), respectively. E. coli DH5α was electroporated by the method of Dower et al. (15).
Molecular cloning and Southern and colony hybridizations.
Restriction enzymes, T4 DNA ligase, and other DNA-modifying enzymes were used as recommended by the manufacturer (Boehringer Mannheim, Indianapolis, Ind.).
Chromosomal DNA from L. plantarum NC8 was digested with several restriction enzymes, and the resulting fragments were separated on a 0.7% agarose gel and then blotted onto a nylon filter (Amersham Biosciences Europe GmbH, Freiburg, Germany). After double digestion with AccI and ClaI of a previously cloned 2.6-kb AccI fragment containing the plNC8BAC operon (33), an 898-bp double-stranded DNA fragment was generated (Fig. 1). This fragment was labeled with fluorescein 11-dUTP by using the Gene Images Random Prime kit (Amersham) and subsequently used as a probe in Southern hybridization experiments. Labeling, hybridization, washing, and detection were performed as recommended by the manufacturer (Amersham).
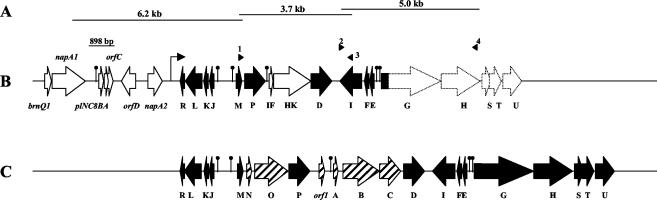
FIG. 1.
Genetic map of the PLNC8 gene cluster. (A) DNA fragments that were either cloned or PCR amplified and then subjected to total or partial DNA sequencing. (B) Genetic map of the PLNC8 gene cluster, showing the genes and ORFs that have been identified. Solid arrows, NC8 genes that share a high degree of homology with, or are virtually identical to, genes found in L. plantarum C11 (see Table 3). Open arrows, L. plantarum NC8 genes that have not been described elsewhere. Dotted arrows, genes that have been identified in NC8 by PCR amplification with specific primers. Numbered arrowheads represent primers used to amplify subsequently sequenced DNA fragments, as follows: 1, PlnM-for; 2, NC8-17; 3, PlnI-rev; 4, PlnH-rev (see Table 2). The bent arrow indicates the start point of the new sequence described in this work. Lollipops indicate the positions of putative promoter sequences. The 898-bp AccI/ClaI DNA fragment used as a probe is indicated. (C) Genetic map of the plantaricin gene cluster in L. plantarum C11, showing the genes that have been described previously (9, 11). Hatched arrows, genes that have not been found in L. plantarum NC8.
In order to extend the known sequence downstream of the plNC8BAC operon, total genomic DNA from L. plantarum NC8 was digested with BclI and separated by size on 0.7% agarose gels, and DNA fragments ranging from 5 to 7 kb were ligated to the dephosphorylated pBluescript II KS(+) cloning vector digested with the same enzyme. The ligation mixture was used to transform E. coli DH5α. The genomic minilibrary generated in this way was screened with the double-stranded AccI/ClaI DNA probe described above by the colony blot technique (43). Hybridization, washing, and detection were performed as described above for the Southern blot. Positive clones were identified as carrying the plasmid construct pSIG303 (Table 1), their plasmid DNA was extracted, and the BclI insert was sequenced as described below.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristics | Source or reference(s) |
|---|---|---|
| pUC18 | E. coli cloning vector; 2.7 kb; Apr | Stratagene |
| pIL252 | Low-copy-number gram-positive cloning vector; 4.8 kb; Emr | 46 |
| pVE3514a | E. coli-Streptococcus pneumoniae shuttle vector pJDC9 containing the L. lactis promoter P59; 7 kb; Emr | 6, 49 |
| pSIG303 | pUC18 containing a 6.2-kb BclI insert from L. plantarum NC8 including the PLNC8 operon; Apr | This work |
| pSIG306 | pVE3514 with a 178-bp KpnI-EcoRI PCR product insert from L. plantarum NC8 containing the plNC8IF gene; Emr | This work |
| pSIG308 | pIL252 containing a 301-bp BamHI-EcoRI insert from pSIG306 which contains the P59::plNC8IF gene fusion; Emr | This work |
This plasmid was previously referred to as pJDC9::P59 (32), and it was kindly supplied by P. Langella, INRA-URLGA, Jouy-en-Josas, France.
PCR and DNA sequencing.
All primers used in PCRs (Table 2) were synthesized by MWG Biotech (Ebersberg, Germany). Primers NC8-7 and NC8-10 (33) were used to amplify an L. plantarum NC8 DNA fragment containing the plNC8A gene plus part of the plNC8B gene, encoding PLNC8. Primer PlnM-for was designed from the end part of the cloned 6.2-kb BclI fragment of NC8 chromosomal DNA (Fig. 1). Primer NC8-17 was designed from the end part of the 3.7-kb PlnM-for-PlnI-rev PCR-amplified fragment from NC8 (Fig. 1). Primers KpnNC8IF-for and EcoNC8IF-rev were used to amplify a 189-bp NC8 DNA fragment corresponding to the plNC8IF structural gene including a putative ribosome-binding site (RBS) (5′-GGAG-3′) located 12 bp upstream of that gene. To facilitate subsequent cloning, KpnI and EcoRI sites were introduced at the 5′ and 3′ ends of these primers, respectively. Primers PlnA-for, PlnA-rev, PlnH-rev, PlnI-rev, and PlnU-rev were designed based on the published DNA sequence of the plnABCDEFGHIJKLMNOP locus of L. plantarum C11 (EMBL accession number X94434) (9, 11).
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| Designed from the L. plantarum NC8 DNA sequence | |
| NC8-7 | 5′-GGTCTGCGTATAAGCATCGC-3′ |
| NC8-10 | 5′-AAATTGAACATATGGGTGCTTTAAATTCC-3′ |
| NC8-17 | 5′-ACTTCCAATGCTTAGAGGATAGG-3′ |
| NC8-22 | 5′-GCGACTTTCTAAACGAAGCC-3′ |
| PlnE-rev | 5′-ATGCTACAGTTTGAGAAGTTACA-3′ |
| PlnF-for | 5′-CTATCCGTGGATGAATCCTC-3′ |
| PlnJ-rev | 5′-ATGACTGTGAACAAAATGATTAAGG-3′ |
| PlnK-for | 5′-TCACTTATTATAATCCCTTGAACC-3′ |
| PlnM-for | 5′-TAAACAGGTAAAGCAGGTTGG-3′ |
| KpnNC8IF-for | 5′-CGCGGGTACCAGTATTTGGAGGGATTAATATG-3′ |
| EcoNC8IF-rev | 5′-CGCGGAATTCTAATGATGGCCTCCAAG-3′ |
| IFNC8-for | 5′-ATGAAAAACATTAATAAGTACACTGAAC-3′ |
| Designed from the L. plantarum C11 DNA sequence | |
| PlnA-for | 5′-ATGAAAATTCAAATTAAAGGTATGAAGC-3′ |
| PlnA-rev | 5′-TTACCATCCCCATTTTTTAAACAGTTTC-3′ |
| PlnH-rev | 5′-TCAATTATTATCCAGCACCTTATC-3′ |
| PlnI-rev | 5′-CCCAACTCAATCACCCATTAAC-3′ |
| PlnU-rev | 5′-TGTGACCAGCCAGTTCACC-3′ |
For amplification of DNA fragments up to 3 kb, 100-μl reaction mixtures containing 2.5 mM MgCl2, 1× reaction buffer, 100 μM each deoxynucleoside triphosphate (dNTP), 100 pmol of each primer, 5 U of Taq DNA polymerase (Promega), and 250 ng of genomic L. plantarum NC8 DNA as the template were used with a GeneAmp PCR system 2400 thermal cycler (Perkin-Elmer Corporation, Norwalk, Conn.). Amplification included denaturation at 94°C for 4 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 1 min, and polymerization at 72°C for 1 min. For amplification of DNA fragments larger than 3 kb, the Expand Long Template PCR system (Roche Applied Science, Barcelona, Spain) was used according to the manufacturer's instructions. PCR amplifications used for sequencing, cloning, or gene expression were performed by using the High-Fidelity PCR system (Roche) under the conditions recommended by the manufacturer.
DNA sequencing was performed by the Servicio de Secuenciación Automática de DNA, Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Cientificas (CSIC), Madrid, Spain, with an ABI PRISM 377 DNA sequencer (Perkin-Elmer Applied Biosystems).
Plasmid construction and gene expression.
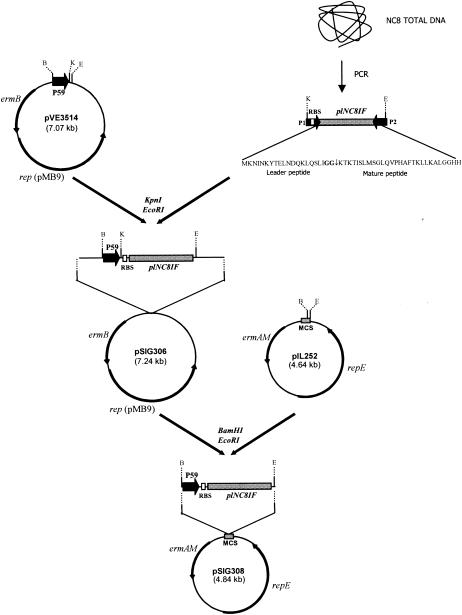
Construction of pSIG303 is described above. The plNC8IF structural gene, including its putative RBS but not its putative promoter, was amplified by PCR as described above. The corresponding 189-bp amplified product was digested with KpnI and EcoRI and ligated to KpnI-EcoRI-digested pVE3514 (Table 1), just downstream of the lactococcal promoter P59 (49), to yield pSIG306 (Fig. 2), which was electroporated into E. coli DH5α. A 301-bp P59::plNC8IF cassette was obtained by double digestion of pSIG306 with BamHI and EcoRI, and this cassette was then cloned into BamHI-EcoRI-digested pIL252 to yield pSIG308 (Fig. 2). This plasmid was transformed into L. plantarum NC8 and L. lactis MG1363 by electroporation as described above.
FIG. 2.
Diagrammatic representation of the construction of recombinant plasmids pSIG306 and pSIG308. Restriction enzymes used to digest parental molecules prior to ligation are indicated. Restriction sites of selected DNA regions are indicated: B, BamHI; E, EcoRI; K, KpnI. P1 and P2 stand for primers KpnNC8IF-for and EcoNC8IF-rev, respectively (see Table 2). The PLNC8IF protein sequence is shown, with the double-glycine motif (boldfaced) and the cleavage site in the native peptide (vertical arrow) indicated. The position of the lactococcal promoter P59 is indicated. MCS, multicloning site.
Induction, autoinduction, and bacteriocin assays.
Induction of bacteriocin production in L. plantarum NC8 can be accomplished either (i) by coculturing with specific gram-positive bacteria (such as L. lactis MG1363), (ii) by addition of heat-killed cells from some of these inducing bacteria, or (iii) by addition of CFS showing inducing activity (33, 34).
Bacteriocin production was induced by cocultivation as follows: a 1% inoculum of L. plantarum NC8 was cultured with a 0.5% inoculum of the inducer strain L. lactis MG1363 in MRS broth, the mixed culture was held at 30°C for 6 to 8 h, and then the inhibitory activity in the CFS was assayed. Bacteriocin induction by heat-killed cells was performed by adding 0.5% of an autoclaved overnight culture of L. lactis MG1363 to a culture of L. plantarum NC8 that had just been started with a 1% inoculum of an overnight culture of this strain. The culture was incubated at 30°C for 6 to 8 h, and then the bacteriocin activity in the CFS was quantitatively tested. To check for inducing activity of CFS, samples for testing, typically 10 to 50 μl, were added to 1 ml of MRS broth containing ca. 108 cells of an overnight culture of L. plantarum NC8, and the mixture was incubated for 6 to 8 h at 30°C; then the resulting CFS were examined for bacteriocin activity. For a quantitative assay of the bacteriocin activity in the CFS, we used the microtiter plate system as described previously (19), with L. plantarum 128/2 as the indicator strain (26).
A quantitative assay of the inducing (autoinducing) activity of a CFS was carried out as described previously (34). In all induction experiments, L. plantarum NC8 cultures were used as negative controls for both bacteriocin and inducer activities.
For gene expression studies, L. plantarum NC8 was induced as described above with either living or heat-killed cells of L. lactis MG1363 (100-ml cultures) or by addition of CFS from L. lactis MG1363(pSIG308) cultures. For the latter purpose, 300 μl of a 10-fold-concentrated CFS of MG1363(pSIG308) (see below) was added to a 100-ml MRS broth culture of L. plantarum NC8 (optical density at 600 nm, 0.1; ca. 108 CFU/ml) and incubated at 30°C for 8 h. As a control, L. plantarum NC8 cultures without any addition (uninduced) were carried out in parallel. Samples from both induced and uninduced NC8 cultures were collected at different points along the growth curve, and the respective CFS were used for induction and bacteriocin assays as described above, while the cells were used to isolate total RNA for use in RT-PCR experiments.
The 10-fold-concentrated CFS of L. lactis MG1363(pSIG308) was obtained from a 110-ml GM17 overnight culture. Briefly, the CFS was treated with solid ammonium sulfate (476 g of culture supernatant per liter; 75% saturation) at 4°C for 16 h with gentle agitation. The protein precipitates were collected by centrifugation at 10,000 × g for 15 min at 4°C, and the resulting pellets were solubilized in 7.5 ml of citrate-phosphate buffer (50 mM; pH 5.0). These 7.5-ml samples were desalted through PD10 gel filtration columns (Amersham) equilibrated with the citrate-phosphate buffer (final volume of the samples, 10.5 ml). The same protocol was used in parallel to obtain the corresponding CFS for control experiments involving pure cultures of L. plantarum NC8 or L. lactis MG1363.
Partial purification of PLNC8IF from CFS of constitutive transformant strains.
In order to verify that the autoinducer peptide PLNC8IF is secreted into the culture media of both L. plantarum NC8(pSIG308) and L. lactis MG1363(pSIG308), partial purification and mass spectrometric analysis of the secreted peptide were carried out. For this purpose, 2-liter cultures of each transformant strain were processed by a protocol similar to that used to purify bacteriocins from LAB (18, 25, 33), but selecting fractions that exhibited induction of bacteriocin production in NC8. After several runs on a C2-C18 reverse-phase column (Pharmacia Biotech, Uppsala, Sweden), samples were analyzed by matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry by F. Canals, Institut de Biologia Fonamental Vicent Villar Palasí, University of Barcelona, Barcelona, Spain.
RNA isolation and RT-PCR.
Samples from L. plantarum or L. lactis cultures were collected at different times along the growth curve, and RNA was extracted from the cell pellets by an adaptation of the method of Raya et al. (41). For reverse transcription, total RNA was treated with RNase-free DNase I (Amersham) as recommended by the manufacturer. cDNA synthesis was carried out in a 20-μl reaction volume containing 1 μg of total RNA, 12.5 U of avian myeloblastosis virus reverse transcriptase (AMV-RT; Roche, Mannheim, Germany), 1× AMV-RT reaction buffer, 10 pmol of antisense primer, 2 mM each dNTP, and 5 U of RNAguard (Amersham) at 42°C for 1 h. For first-strand DNA synthesis of the plNC8BA, plnEF, plnJK, and plNC8IF-plNC8HK-plnD operons, the antisense primers used were NC8-10, PlnF-for, PlnK-for, and NC8-22, respectively (Table 2). After inactivation of the enzyme at 95°C for 5 min, 1 μl of each cDNA was used for PCR analysis using the primer pairs NC8-7-NC8-10, PlnF-for-PlnE-rev, PlnK-for-PlnJ-rev, and IFNC8-for-EcoIFNC8 to detect the plNC8BA, plnEF, plnJK, and plNC8IF-plNC8HK-plnD operons, respectively. The annealing temperature was 58°C for all PCRs. The resulting amplified DNA fragments were resolved on 10% acrylamide gels (43). Control reactions using the same primer pairs prior to reverse transcription were used to check for the absence of contaminating chromosomal DNA.
Nucleotide sequence accession number.
The contiguous 8,107-bp DNA sequence downstream of the PLNC8 operon has been assigned GenBank accession number AF522077.
RESULTS
DNA sequence and genetic analysis of the PLNC8 locus in L. plantarum NC8.
To extend the known DNA sequence downstream of the PLNC8 operon (33), we cloned and sequenced a 6.2-kb BclI L. plantarum NC8 chromosomal DNA fragment (Fig. 1) that hybridized with a labeled probe containing the plnc8BAC operon, thus obtaining as much as 2.84 kb of novel sequence. Analysis of this sequence revealed five open reading frames (ORFs), three of which were identical to the plnR, plnK, and plnJ genes from the plantaricin locus of L. plantarum C11, while the other two showed high homology, respectively, with the plnL and plnM genes from the same locus (Table 3). On the basis of this knowledge, several primers were designed either from the novel NC8 DNA sequence or from the published C11 sequence (Table 2). Hence, to scan for possible correspondences between the plantaricin C11 and PLNC8 loci, primer plnM-for was used in PCRs in combination with other primers designed by us from the DNA sequence of the plantaricin locus (plnABCDEFGHIJKLMNOP) of L. plantarum C11. As a result, a 3.7-kb DNA fragment, which overlapped with the end of the BclI fragment, was amplified with primer pair plnM-for-plnI-rev and entirely sequenced (Fig. 1). On the basis of this new sequence, we designed the NC8-17 primer (Table 2), which was combined with primer plnH-rev in new PCRs. These PCRs resulted in an amplified DNA fragment of 5 kb (Fig. 1), from which 1,681 bp was sequenced. In summary, we have obtained a contiguous 8,107-bp DNA sequence downstream of the PLNC8 operon which has been assigned GenBank accession number AF522077. Furthermore, several other PCRs were carried out to check for correspondence with the C11 operon. From these PCRs we concluded that the plnG, plnH, plnS, plnT, and plnU genes from C11 were also present in NC8, as shown in Fig. 1.
TABLE 3.
Homology to the data banks of the putative proteins coded by the ORFs found in the bacteriocin cluster of L. plantarum NC8
| NC8 ORF (no. of amino acids) | Homologue
|
||
|---|---|---|---|
| C11 gene (no. of amino acids)a | Identity (%) | Feature(s) (reference) | |
| plnR (50) | plnR (50) | 100 | Unknown function (11) |
| plnL (200) | plnL (138) | 99 (in 138 amino acids) | Putative immunity protein for plantaricin J/K (11) |
| plnK (57) | plnK (57) | 100 | Plantaricin J/K component (11) |
| plnJ (55) | plnJ (55) | 100 | Plantaricin J/K component (11) |
| plnM (66) | plnM (66) | 93 (in 44 amino acids) | Putative immunity protein (11) |
| plnP (248) | plnP (247) | 98 | Putative immunity protein (11) |
| plNC8IF (28) | — | — | |
| plNC8HK (446) | plnB (442) | 34 | Histidine kinase, plantaricin A system (9) |
| plnD (247) | plnD (247) | 98 | Response regulator, plantaricin A system (9) |
| plnI (257) | plnI (257) | 99 | Putative immunity protein for plantaricin E/F (11) |
| plnF (52) | plnF (52) | 100 | Plantaricin E/F component (11) |
| plnE (56) | plnE (56) | 100 | Plantaricin E/F component (11) |
| plnG (76)b | plnG (457) | 99 | ABC transporter, plantaricin A system (11) |
A similar plantaricin cluster has been described in L. plantarum WCFS1 (31). —, no homology detected.
Number of amino acids corresponding to the ORF actually sequenced in L. plantarum NC8.
The extended L. plantarum NC8 DNA sequence of the PLNC8 operon revealed the presence of 12 ORFs which seem to be organized into four putative operons (Fig. 1). Despite the high degree of homology of most of these ORFs with genes in the plantaricin locus in L. plantarum C11, two major differences between the bacteriocin operons in C11 and NC8 are noteworthy: (i) a 1.6-kb deletion in L. plantarum NC8 between plnM and plnP and (ii) a 3.5-kb deletion in NC8 between plnP and plnD (Fig. 1). Consequently, plnN, plnO, plnA, plnB, plnC, and orf1 from C11 are not present in NC8. On the other hand, two novel ORFs between plnP and plnD were found in NC8 and were designated plNC8IF and plNC8HK (Fig. 1). These two genes plus plnD (Fig. 1) seem to be organized as a three-component regulatory operon equivalent to the plnABCD regulatory operon in C11 (9), as discussed below.
The regulatory operon in L. plantarum NC8.
In NC8, the regulatory operon for bacteriocin production starts with plNC8IF, which encodes a putative peptide of 49 amino acid residues, showing all the features described for inducer peptides (36). As with bacteriocin-like peptides, the product of plNC8IF possesses a leader sequence of the double-glycine type that, once processed, gives rise to a mature peptide of 28 amino acid residues, with a theoretical pI of 13.0 and a molecular weight (MW) of 3,015. No homology with any known protein in the databases was found for the mature PLNC8IF.
Just 14 bp downstream of plNC8IF we found plNC8HK, which encodes a putative protein of 446 amino acid residues with a predicted pI of 5.34 and an MW of 51,053, showing significant homology with the family of the HKs (20). Actually, the highest similarity for PLNC8HK (34% identity) was obtained with PlnB, the HK of the plantaricin locus from L. plantarum C11 (9). Immediately downstream of plNC8HK, we found plnD. The putative protein encoded by plnD was 98% identical to the RR PlnD of L. plantarum C11 (9). Therefore, based on homology and relative position, the plNC8IF-plNC8HK-plnD gene cluster seems to form a regulatory operon of the so-called three-component type, comprising an autoinducer peptide (PLNC8IF), an HK (PLNC8HK), and an RR (PLND). Interestingly, this regulatory operon is located between plnP and plnI in L. plantarum NC8, in a position analogous to that of the regulatory operon plnABCD in L. plantarum C11 (Fig. 1). In L. plantarum NC8, however, the RR PlnC is not present.
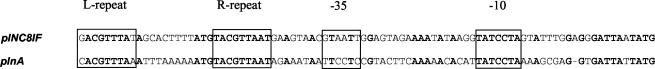
Analysis of the DNA region upstream of plNC8IF revealed the presence of a putative promoter that resembles the regulated promoter of plnA (Fig. 3) as well as other bacteriocin promoters from LAB whose expression is regulated via a three-component regulatory system (11, 36). Therefore, certain DNA motifs are conserved: the L and R repeats are virtually identical to those of plnA, as is the −10 region (Fig. 3). Downstream of plnD, a rho-independent transcription terminator was found, suggesting that the plNC8IF, plNC8HK, and plnD genes would be cotranscribed, as reported below.
FIG. 3.
Alignment of the promoter sequences of plNC8IF and plnA, showing conserved nucleotides (boldfaced) and significant features (L and R repeats; −35 and −10 sites) (boxes).
Homologous and heterologous expression of plNC8IF.
In order to determine whether the product of plNC8IF is in fact an IF for bacteriocin production in L. plantarum NC8, homologous and heterologous expression of this gene was performed. For this purpose, we fused the plNC8IF gene (including its putative RBS) to the constitutive lactococcal promoter P59, producing plasmid pSIG306 (Fig. 2). This fusion was subsequently transferred into pIL252 to yield pSIG308 (Fig. 2), which was then established in L. plantarum NC8. When NC8 transformant cells were tested for bacteriocin production in broth cultures, all of them produced bacteriocin constitutively (Table 4). In contrast, in control experiments, L. plantarum NC8 with or without pIL252 did not produce bacteriocin at all under the same conditions (Table 4). These findings suggested that the product of plNC8IF induces bacteriocin production in NC8 and that its expression must be regulated. MALDI-TOF mass spectrometry analysis of partially purified PLNC8IF from L. plantarum NC8(pSIG308) CFS showed a peak corresponding to a peptide of 3,029.66 Da, i.e., 16 Da more than the theoretical molecular size of the processed peptide PLNC8IF (3,013.71 Da). This difference can be explained by an oxidized methionine in the mature PLNC8IF. These results, together with the fact that no signal corresponding to the unprocessed PLNC8IF peptide was detected in MALDI-TOF mass spectrometry analysis (theoretical molecular size, 5,401.0 Da), strongly indicate that PLNC8IF is effectively exported out of the pSIG308-transformed NC8 cells in its mature form after cleavage at the double-glycine motif.
TABLE 4.
Bacteriocin and inducing activities of different CFS
| CFS source | Bacteriocin activitya (BU/ml) | Inducing activityb (AIU/ml) |
|---|---|---|
| L. plantarum NC8 | 0 | 0 |
| L. lactis MG1363 | 0 | 0 |
| L. plantarum NC8 + L. lactis MG1363 | 1,280 | 640 |
| L. plantarum NC8(pSIG308) | 640 | 80 |
| L. lactis MG1363(pSIG308) | 0 | 160 |
| L. plantarum NC8(pIL252) | 0 | 0 |
| L. lactis MG1363(pIL252) | 0 | 0 |
L. plantarum 128/2 was used as the sensitive strain.
Twenty microliters of each CFS was added to 1 ml of MRS medium inoculated with ca. 108 saline-washed cells of an overnight culture of L. plantarum NC8. This mixture was incubated for 8 h at 30°C, and bacteriocin activity was tested by using L. plantarum 128/2 as the sensitive strain. AIU, autoinducing units.
Transformants of L. lactis MG1363 carrying pSIG308 were analyzed for their ability to produce and export PLNC8IF to the culture medium. Addition of CFS from L. lactis MG1363(pSIG308) to L. plantarum NC8 cultures resulted in induction of bacteriocin production (Table 4), indicating that PLNC8IF was being produced and exported. In order to corroborate the presence of PLNC8IF in the L. lactis MG1363(pSIG308) CFS and to check whether the product of the plNC8IF gene had been processed in such a heterologous strain, MALDI-TOF mass spectrometry analysis of fractions exhibiting induction of bacteriocin production in NC8 was carried out. Two peaks corresponding to peptides of 3,013.70 and 3,029.70 Da were detected, corresponding to the PLNC8IF mature peptide and to the same peptide with an oxidized methionine, respectively. This finding indicates that L. lactis MG1363 was able to process and export mature PLNC8IF to the culture medium, as was the original NC8 strain. No bacteriocin activity was detected in L. lactis MG1363(pSIG308) CFS (Table 4). Finally, CFS from MG1363 or MG1363(pIL252) cultures did not induce bacteriocin production in L. plantarum NC8 or show any bacteriocin activity (Table 4).
Coculture with inducing cells induces transcription of the bacteriocin and regulatory operons in L. plantarum NC8.
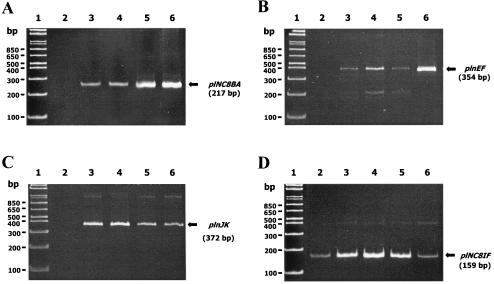
To confirm at the molecular level that exposure of NC8 to inducing cells results in expression of its bacteriocin genes and of the PLNC8IF (regulatory) operon, we studied the expression of all these genes. Total RNA was isolated from samples collected at different points of the growth phase from NC8 cultures which had been cocultured with the inducer strain L. lactis MG1363. As shown in Fig. 4, cocultivation with MG1363 induced in NC8 the expression of the structural genes for the three two-peptide bacteriocins detected in the NC8 chromosome—PLNC8 (plNC8A and plNC8B [Fig. 4A]), plantaricin EF (plnE and plnF [Fig. 4B]), and plantaricin JK (plnJ and plnK [Fig. 4C])-at the early-exponential phase of growth. Bacteriocin activity was detected in the CFS of all these samples, with titers ranging from 640 to 1,280 bacteriocin units (BU)/ml. Control experiments involving pure cultures of NC8 showed no transcription of these genes (Fig. 4), and no bacteriocin activity was found in their CFS at any time. On the other hand, cocultivation with MG1363 resulted in induction of the putative regulatory operon plNC8IF-plNC8HK-plnD in L. plantarum NC8 (Fig. 4D). In contrast to the bacteriocin operons, a basal level of transcription for this regulatory operon was detected in some samples of NC8 growing as a pure culture (Fig. 4D).
FIG. 4.
RT-PCR analysis of expression of the bacteriocin and regulatory operons in L. plantarum NC8 at the early-exponential phase of growth in MRS medium. Expression of the plNC8BA (A), plnEF (B), plnJK (C), and plNC8IF-plNC8HK-plnD (D) operons was analyzed. Lanes: 1, MW marker (1 kb plus DNA ladder; Gibco BRL); 2, pure culture of L. plantarum NC8; 3, pure culture of L. plantarum NC8 that had been induced with a PLNC8IF-rich, 10-fold-concentrated CFS from L. lactis MG1363(pSIG308); 4, L. plantarum NC8 culture that had been induced with heat-killed cells of L. lactis MG1363; 5, L. plantarum NC8 that had been cocultivated with L. lactis MG1363; 6, control PCR using total DNA from L. plantarum NC8 as the template. MWs of relevant bands of the marker are given on the left. Designations and sizes of the expected products are given on the right.
The same transcriptional analysis was carried out using autoclaved L. lactis MG1363 cells to induce bacteriocin production in NC8, and virtually the same transcriptional pattern as that for induction with living MG1363 cultures was detected (Fig. 4).
Finally, PCR experiments involving relevant primer pairs failed to detect in L. lactis MG1363 DNA any of those operons expressed in L. plantarum NC8.
PLNC8IF induces transcription of all the bacteriocin operons in NC8 as well as its own (regulatory) operon.
The role of PLNC8IF as an inducer of bacteriocin production in NC8 as well as an autoinducer of its own synthesis was shown at the molecular level by studying the expression of the relevant operons when exposed to an external source of PLNC8IF. For this purpose, total RNAs were isolated from samples collected at different points of the growth phase from NC8 cultures that had been induced by addition of a 10-fold-concentrated CFS from an L. lactis MG1363(pSIG308) culture, containing PLNC8IF at 10,240 IU/ml. As shown in Fig. 4, addition of PLNC8IF to NC8 cultures results in induction of the expression of all the bacteriocin structural genes in NC8 (plNC8A and plNC8B [Fig. 4A], plnE and plnF [Fig. 4B], and plnJ and plnK [Fig. 4C]) during the exponential phase of growth, as expected from the induction by cocultivation described above. Moreover, a PLNC8IF-rich CFS was able to induce the expression of the plNC8IF-plNC8HK-plnD operon, i.e., its own synthesis, in NC8 cultures. These findings constitute evidence that the product of plNC8IF is an autoinducer molecule.
DISCUSSION
With this work we have gained a better knowledge of the regulation of bacteriocin production genes whose expression is dependent on an external stimulus, such as cocultivation with other strains. Thus, for L. plantarum NC8, a quorum-sensing mechanism is predicted to be the actual mediator between the external stimulus provided by the presence of specific bacterial strains in the same culture medium and the production of bacteriocins.
According to our results, the presence of either living or heat-killed inducing cells promotes expression of the three bacteriocin operons in NC8 (plNC8BA, plnEF, and plnJK) (Fig. 4A to C). This is readily detected by the bacteriocin activity found in the corresponding CFS. Actually, from induced NC8 CFS, we purified and determined the amino acid sequence of plantaricin F as well as MALDI-TOF chromatographic peaks corresponding to peptides with MWs identical to those of the mature proteins encoded by plnE, plnJ, and plnK (data not shown). Therefore, to our knowledge, NC8 is the strain of LAB naturally producing the largest number of different bacteriocin peptides (PLNC8α, PLNC8β, PlnE, PlnF, PlnJ, and PlnK) described to date.
Concomitantly with bacteriocin production, the same stimulus (the inducing cells) is able to activate the expression of an operon (plNC8IF-plNC8HK-plnD) that exhibits all the features of a three-component regulatory system operating via a quorum-sensing mechanism. Actually, a similar regulatory operon (plnABCD) has been described for the bacteriocin cluster in L. plantarum C11, whose expression is dependent on the inoculum size (10, 11). Interestingly, though, the two regulatory operons possess completely different IFs (the products of plnA and plNC8IF for C11 and NC8, respectively), which have in common only certain conserved motifs at the leader sequences and conserved regions at the promoters: L and R repeats (Fig. 3). In fact, these direct repeats have been shown to play an essential role in the activation-repression of the regulatory promoter itself by specific RRs encoded in the same operon: plnC and plnD in C11 (13, 14). However, only plnD is present in NC8. Overexpression of plnD in its homologous host C11 had a repressive effect on both bacteriocin production and its own (plnABCD) transcription (14). However, this repressive effect could be quite different on a different genetic background, in a nonoverexpressed dose, and in the absence of the related gene plnC, as is the case in L. plantarum NC8. On the other hand, it is known that in order to obtain the (auto)induction phenomenon, the IF interacts with its specific HK, which subsequently activates its cognate RR (4, 17, 36).
The putative gene for HK in NC8 (plNC8HK) shows quite different homologies at the amino- and carboxy-terminal sequences: while the N terminus shows just 21% identity with the HKs from other bacteriocin regulatory operons (plnB for plantaricin C11 [9] and sppK for sakacin P [23]), the carboxy terminus shows up to 47% identity with other bacteriocin HKs and kinases from the HPK10 group (20). Prediction models for the topology of the HKs suggest that they possess two structural domains (36). The external domain (located at the N-terminal part) would be responsible for recognizing the specific IF, while the internal domain (C-terminal part) would be able to activate the RR by phosphorylation. Hence, it is not surprising that the N-terminal part of the protein encoded by plNC8HK is unrelated, because it has to recognize a specific molecule, its own autoinducer (PLNC8IF). On the other hand, its C-terminal part is quite conserved, because it has a common function: to phosphorylate an RR (PlnD). In turn, this RR has to recognize specific sequences at the regulated promoters, including the promoter governing its own synthesis (14, 39, 42, 47). Accordingly, conserved L and R repeats can be found at the promoter regions of all the bacteriocin and regulatory operons in NC8. This constitutes evidence that those genes are regulated by the expression of plnD. In summary, this is the first report of interchangeable IFs (PLNC8IF and PlnA) acting through the same RR and bacteriocin genes.
Constitutive expression of PLNC8IF by L. plantarum NC8 or addition of a PLNC8IF-containing CFS to an NC8 culture results in bacteriocin and autoinducer production by this strain, in agreement with the model that PLNC8IF mediates the induction provided by the inducing cells. This observation was confirmed by analysis of the RT-PCR experiments. Thus, addition of purified PLNC8IF promoted the expression of the bacteriocin and regulatory operons in NC8 in the same way as the presence of either living or heat-killed inducer cells does. A basal level of transcription was detected for the plNC8IF-plNC8HK-plnD regulatory operon, in agreement with previous observations regarding the need for maintenance of a minimum level of regulatory proteins so that the cell can sense the external IF concentration and respond to it at once (3).
On the other hand, as predicted by the DNA sequence analysis, mature PLNC8IF results after cleavage at the double-glycine motif in L. plantarum NC8, as well as in the heterologous host L. lactis MG1363, according to the MALDI-TOF analyses. This observation corroborates our previous results, for during amino acid sequencing of PLNC8 by the Edman degradation method, some of the samples assayed showed sequences corresponding to mature PLNC8IF. In addition, it has been demonstrated that PLNC8IF does not exhibit any bacteriocin activity by itself.
In conclusion, we propose that the presence of specific bacteria acts as an environmental signal to switch bacteriocin production on in L. plantarum NC8. A quorum-sensing mechanism mediated by PLNC8IF appears to be involved in this process. In addition, L. plantarum NC8 offered the exceptional opportunity of studying how a particular IF (i.e., PLNC8IF) is able to autoinduce a regulatory system as well as to regulate the expression of a series of bacteriocin operons which had previously been described as being governed by a different autoinducer (plantaricin A, in C11). Both PLNC8IF and PlnA need specific HKs (encoded by plNC8HK and plnK in NC8 and C11, respectively) due to their high ligand specificities, but they both act through the same RR (plnD), provided that consensus motifs (L and R repeats) are present at the promoter sequences of the operons to be regulated.
Acknowledgments
This work was supported by the Spanish Government through MCYT project AGL2000-1611-CO3-01. A.M. was the recipient of a grant from MCYT, Madrid, Spain.
We thank Belén Caballero-Guerrero for excellent technical assistance.
REFERENCES
- 1.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aukrust, T., and H. Blom. 1992. Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res. Int. 25:253-261. [Google Scholar]
- 3.Axelsson, L., and A. Holck. 1995. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J. Bacteriol. 177:2125-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brurberg, M. B., I. F. Nes, and V. G. H. Eijsink. 1997. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol. Microbiol. 26:347-360. [DOI] [PubMed] [Google Scholar]
- 5.Cathcart, D. P. 1995. Purification, characterization and molecular analysis of plantaricin S, a two-peptide bacteriocin from olive fermenting Lactobacillus plantarum strains. Ph.D. thesis. Cranfield University, Bedford, United Kingdom.
- 6.Chen, J.-D., and D. A. Morrison. 1988. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64:155-164. [DOI] [PubMed] [Google Scholar]
- 7.Daeschel, M. A. 1993. Applications and interactions of bacteriocins from lactic acid bacteria in foods and beverages, p. 63-91. In D. G. Hoover and L. R. Steenson (ed.), Bacteriocins of lactic acid bacteria. Academic Press, Inc., New York, N.Y.
- 8.de Vuyst, L., and E. J. Vandamme. 1994. Antimicrobial potential of lactic acid bacteria, p. 91-142. In L. de Vuyst and E. J. Vandamme (ed.), Bacteriocins of lactic acid bacteria: microbiology, genetics and applications. Blackie Academic & Professional, London, United Kingdom.
- 9.Diep, D. B., L. S. Håvarstein, J. Nissen-Meyer, and I. F. Nes. 1994. The gene encoding plantaricin A, a bacteriocin from Lactobacillus plantarum C11, is located on the same transcription unit as an agr-like regulatory system. Appl. Environ. Microbiol. 60:160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diep, D. B., L. S. Håvarstein, and I. F. Nes. 1995. A bacteriocin-like peptide induces bacteriocin synthesis in Lactobacillus plantarum C11. Mol. Microbiol. 18:631-639. [DOI] [PubMed] [Google Scholar]
- 11.Diep, D. B., L. S. Håvarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178:4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diep, D. B., L. Axelsson, C. Grefsli, and I. F. Nes. 2000. The synthesis of the bacteriocin sakacin A is a temperature-sensitive process regulated by a pheromone peptide through a three-component regulatory system. Microbiology 146:2155-2160. [DOI] [PubMed] [Google Scholar]
- 13.Diep, D. B., O. Johnsborg, P. A. Risøen, and I. F. Nes. 2001. Evidence for dual functionality of the operon plnABCD in the regulation of bacteriocin production in Lactobacillus plantarum. Mol. Microbiol. 41:633-644. [DOI] [PubMed] [Google Scholar]
- 14.Diep, D. B., R. Myhre, O. Johnsborg, Å. Aakra, and I. F. Nes. 2003. Inducible bacteriocin production in Lactobacillus is regulated by differential expression of the pln operons and by two antagonizing response regulators, the activity of which is enhanced upon phosphorylation. Mol. Microbiol. 47:483-494. [DOI] [PubMed] [Google Scholar]
- 15.Dower, W. J., F. J. Miller, and W. C. Ragsdale. 1998. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eijsink, V. G. H., M. B. Brurberg, P. H. Middelhoven, and I. F. Nes. 1996. Induction of bacteriocin production in Lactobacillus sake by a secreted peptide. J. Bacteriol. 178:2232-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eijsink, V. G. H., L. Axelsson, D. B. Diep, L. S. Håvarstein, H. Holo, and I. F. Nes. 2002. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie Leeuwenhoek 81:639-654. [DOI] [PubMed] [Google Scholar]
- 18.Floriano, B., J. L. Ruiz-Barba, and R. Jiménez-Díaz. 1998. Purification and genetic characterization of enterocin I from Enterococcus faecium 6T1a, a novel antilisterial plasmid-encoded bacteriocin which does not belong to the pediocin family of bacteriocins. Appl. Environ. Microbiol. 64:4883-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geis, A., J. Singh, and M. Teuber. 1983. Potential of lactic streptococci to produce bacteriocin. Appl. Environ. Microbiol. 45:205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41:139-227. [DOI] [PubMed] [Google Scholar]
- 21.Håvarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 22.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 23.Hühne, K., A. Holck, L. Axelsson, and L. Krockel. 1996. Analysis of the sakacin P gene cluster from Lactobacillus sake Lb674 and its expression in sakacin-negative Lb. sake strains. Microbiology 142:1437-1448. [DOI] [PubMed] [Google Scholar]
- 24.Jack, R., J. R. Tagg, and B. Ray. 1995. Bacteriocins of Gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiménez-Díaz, R., R. M. Rios-Sánchez, M. Desmazeaud, J. L. Ruiz-Barba, and J. C. Piard. 1993. Plantaricin S and T, two new bacteriocins produced by Lactobacillus plantarum LPCO10 isolated from a green olive fermentation. Appl. Environ. Microbiol. 59:1416-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiménez-Díaz, R., J. L. Ruiz-Barba, D. P. Cathcart, H. Holo, I. F. Nes, K. H. Sletten, and P. J. Warner. 1995. Purification and partial amino acid sequence of plantaricin S, a bacteriocin produced by Lactobacillus plantarum LPCO10, the activity of which depends on the complementary action of two peptides. Appl. Environ. Microbiol. 61:4459-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-86. [DOI] [PubMed] [Google Scholar]
- 28.Kleerebezem, M., L. E. N. Quadri, O. P. Kuipers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 29.Kleerebezem, M., and L. E. N. Quadri. 2001. Peptide pheromone-dependent regulation of antimicrobial peptide production in Gram-positive bacteria: a case of multicellular behavior. Peptides 22:1579-1596. [DOI] [PubMed] [Google Scholar]
- 30.Kleerebezem, M., O. P. Kuipers, W. M. de Vos, M. E. Stiles, and L. E. N. Quadri. 2001. A two-component signal transduction cascade in Carnobacterium piscicola LV17B: two signaling peptides and one sensor-transmitter. Peptides 22:1597-1601. [DOI] [PubMed] [Google Scholar]
- 31.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Loir, Y., A. Gruss, S. D. Ehrlich, and P. Langella. 1998. A nine-residue synthetic propeptide enhances secretion efficiency of heterologous proteins in Lactococcus lactis. J. Bacteriol. 180:1895-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maldonado, A., J. L. Ruiz-Barba, and R. Jiménez-Díaz. 2003. Purification and genetic characterization of plantaricin NC8, a novel coculture-inducible two-peptide bacteriocin from Lactobacillus plantarum NC8. Appl. Environ. Microbiol. 69:383-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maldonado, A., R. Jiménez-Díaz, and J. L. Ruiz-Barba. 2004. Production of plantaricin NC8 by Lactobacillus plantarum NC8 is induced in the presence of different types of Gram-positive bacteria. Arch. Microbiol. 181:8-16. [DOI] [PubMed]
- 35.Nes, I. F., D. B. Diep, L. S. Håvarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 36.Nes, I. F., and V. G. H. Eijsink. 1999. Regulation of group II peptide bacteriocin synthesis by quorum sensing mechanisms, p. 175-192. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 37.Nilsen, T., I. F. Nes, and H. Holo. 1998. An exported inducer peptide regulates bacteriocin production in Enterococcus faecium CTC492. J. Bacteriol. 180:1848-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Keeffe, T., C. Hill, and R. P. Ross. 1999. Characterization and heterologous expression of the genes encoding enterocin A production, immunity, and regulation in Enterococcus faecium DPC1146. Appl. Environ. Microbiol. 65:1506-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkinson, J. S. 1993. Signal transduction schemes in bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 40.Quadri, L. E. N., M. Kleerebezem, O. P. Kuipers, W. M. De Vos, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1997. Characterization of a locus from Carnobacterium piscicola LV17B involved in bacteriocin production and immunity: evidence for global inducer-mediated transcriptional regulation. J. Bacteriol. 179:6163-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raya, R., J. Bardowski, P. S. Andersen, S. D. Ehrlich, and A. Chopin. 1998. Multiple transcriptional control of the Lactococcus lactis trp operon. J. Bacteriol. 180:3174-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Risøen, P. A., M. B. Brurberg, V. G. H. Eijsink, and I. F. Nes. 2000. Functional analysis of promoters involved in quorum sensing-based regulation of bacteriocin production in Lactobacillus. Mol. Microbiol. 37:619-628. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 44.Saucier, L., A. Poon, and M. E. Stiles. 1995. Induction of bacteriocin in Carnobacterium piscicola LV17. J. Appl. Bacteriol. 78:684-690. [Google Scholar]
- 45.Saucier, L., A. S. Paradkar, L. S. Frost, S. E. Jensen, and M. E. Stiles. 1997. Transcriptional analysis and regulation of carnobacteriocin production in Carnobacterium piscicola LV17. Gene 188:271-277. [DOI] [PubMed] [Google Scholar]
- 46.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 47.Stock, J. B., A. M. Stock, and J. M. Mottonen. 1990. Signal transduction in bacteria. Nature 334:395-400. [DOI] [PubMed] [Google Scholar]
- 48.Tagg, J. R., A. S. Dajani, and L. W. Wannamaker. 1976. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40:722-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Vossen, J. M. B. M., D. van der Lelie, and G. Venema. 1987. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 53:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]