Abstract
In 2003, a new fruit fly pest species was recorded for the first time in Kenya and has subsequently been found in 28 countries across tropical Africa. The insect was described as Bactrocera invadens, due to its rapid invasion of the African continent. In this study, the morphometry and DNA Barcoding of different populations of B. invadens distributed across the species range of tropical Africa and a sample from the pest's putative aboriginal home of Sri Lanka was investigated. Morphometry using wing veins and tibia length was used to separate B. invadens populations from other closely related Bactrocera species. The Principal component analysis yielded 15 components which correspond to the 15 morphometric measurements. The first two principal axes contributed to 90.7% of the total variance and showed partial separation of these populations. Canonical discriminant analysis indicated that only the first five canonical variates were statistically significant. The first two canonical variates contributed a total of 80.9% of the total variance clustering B. invadens with other members of the B. dorsalis complex while distinctly separating B. correcta, B. cucurbitae, B. oleae and B. zonata. The largest Mahalanobis squared distance (D2 = 122.9) was found to be between B. cucurbitae and B. zonata, while the lowest was observed between B. invadens populations against B. kandiensis (8.1) and against B. dorsalis s.s (11.4). Evolutionary history inferred by the Neighbor-Joining method clustered the Bactrocera species populations into four clusters. First cluster consisted of the B. dorsalis complex (B. invadens, B. kandiensis and B. dorsalis s. s.), branching from the same node while the second group was paraphyletic clades of B. correcta and B. zonata. The last two are monophyletic clades, consisting of B. cucurbitae and B. oleae, respectively. Principal component analysis using the genetic distances confirmed the clustering inferred by the NJ tree.
Introduction
Globally, Dacine fruit flies of the genus Bactrocera Macquart (Diptera: Tephrtitidae) are among the most important pests of fruits and vegetables [1]. In addition to the polyphagous nature of some species, several are considered highly invasive; aided by globalization of trade and poor quarantine infrastructure in the developing countries. Adults often exhibit a strong tendency for dispersal and the immature stages are readily transported to new areas via fruits movement [2]. In Africa, a member of the genus Bactrocera was detected in 2003 at the Kenyan coast [3] and later described as Bactrocera invadens Drew, Tsuruta & White [4]. The pest is believed to be native to Sri Lanka [5] and has rapidly expanded its geographical range, now reported from 28 African countries including the Indian Ocean archipelago of the Comoros [4], [6], [7], [8], [9], [10], [11], [12]. Bactrocera invadens is an emerging polyphagous fruit fly pest and in Africa it has been reported to attack over 43 fruit species from 23 families with mango being one of the most preferred cultivated host [10], [12], [13], [14]. Direct damage to mango due to B. invadens has been reported to range from 30–80% depending on the cultivar, locality and season [8], [12], [15]. In addition to the direct losses, indirect losses attributed to quarantine restrictions have been enormous. The direct and indirect damage continue to have wide reaching socio-economic implications for millions of rural and urban populations involved in the mango value chain across Africa. The pest has been described as “a devastating quarantine pest” by the Inter-African Phytosanitary Council [6].
Many economically important fruit fly pest species from the family Tephritidae belong to complexes of sibling species, presenting difficulties in identification, even to the experts [16]. Bactrocera invadens is believed to belong to the B. dorsalis (Hendel) complex of tropical fruit flies [4]. This complex comprises of more than 75 species largely endemic to South-East Asia [17] with undescribed species remaining in collections [18]. Indeed, the B. dorsalis complex of fruit fly species appear to be evolving rapidly demanding the need for closer assessment of their taxonomic identity through morphometric and genetic analysis. For example, Drew et al. [4] depicted different thoracic colourations of B. invadens that are morphotypes of the same pest but that has complicated the taxonomic identity of this pest.
Detail review of the B. dorsalis complex by Drew & Hancock [17] has led to considerable debate over species, and a number of published works has aimed at defining the limits of some species populations [19], [20], [21], [1]. A study by Tan et al. [22] compared the profiles of phenylpropanoid metabolites of four Bactrocera species from the B. dorsalis complex, that includes B. dorsalis s.s., B. invadens, B. correcta and B. zonata and revealed that different profiles of phenylpropanoid ingredients in the rectal glands can be used for identification of these four species. Other studies on identification of the B. dorsalis complex by Schutze et al. [23] used geometric morphometric analysis of wing size and shape to discriminate species within this complex. However, recent observations by Drew et al. [5] emphasized the need to continue research on this complex to provide validity or otherwise, for all species in the complex, for both economic reasons and for refining the systematics of the Subfamily Dacinae. Due to the complexity of this group of fruit flies and the arrival of B. invadens into Africa, the need to undertake the inventory of the B. dorsalis complex in Asia and make comparison with what is in Africa becomes important in order to redefine this complex.
Morphometric analyses have been a useful technique in detecting morphological differences among organisms to distinguish closely related species including fruit flies, justify synonymies, demonstrate morphological variation along altitudinal or geographical gradients and propose new species [24], [25], [26], [27], [28], [29]. Indeed, in some frugivorous tephritid fruit fly species, diagnostic morphological characters for the identification of adult flies are now available [29], [30], [31], [5]. However, morphological tools present some limitations, mainly due to high homoplasy in most morphological characters and the existence of cryptic species groups across the family. Thus, the classification of Tephritids to the species level based on adult morphology alone is sometimes unreliable [32], [33], [34]. These limitations have led several taxonomists and quarantine officials alike to seek viable alternative ways of fruit fly identification including the use of molecular markers [35], [32], [36], [37]. Recently, the current molecular tool of choice is DNA Barcoding, which is a system that employs sequence diversity in short, standardized gene regions aiding in identification of species [38]. This standardized method for identifications of species focuses sequencing efforts on one target gene, cytochrome c oxidase subunit I (COI) [39], [40].
The main objective of our study was to establish whether B. invadens individuals collected from Africa could be distinguished from other Asian Bactrocera species using both multivariate morphometric analysis and molecular methods. Because B. invadens belongs to a complex, we believe that information generated from this investigation should help clarify its identity, resolve its placement in the right phylogeny, ease quarantine restrictions and potentially contribute to better management of the pest if sterile insect technique or eradication from particular locality or region becomes an option.
Results
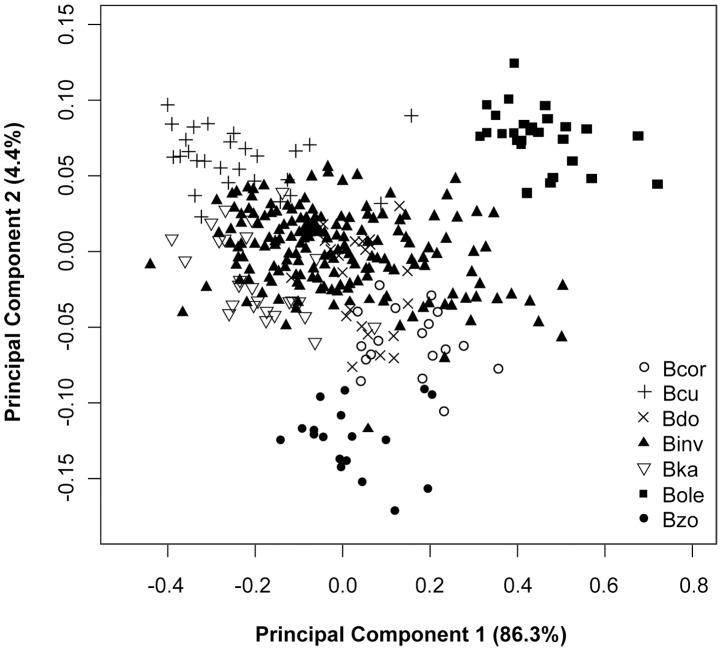
The PCA yielded 15 components which correspond to the 15 morphometric measurements. Bartlett χ tests indicated that only the first 13 components were statistically significant. Projection of the morphometric data of the Bactrocera species on the first two principal axes showed a partial separation of these populations (Figure 1). The first two principal components contributed to 90.7% of the total variance (PC1 = 86.3% and PC2 = 4.4%) (Table 1). The third, fourth and fifth components contributed 2.3%, 2.0% and 1.7%, respectively, but did not improve separation of the populations. Bactrocera invadens populations and the other Bactrocera species belonging to the B. dorsalis complex could not be separated by PCA (Figure 1). However, the first two principal components separated B. cucurbitae, B. oleae and B. zonata into distinct groups (Figure 1). Contributions or loadings of the individual measurements indicate that PC1 represented the overall size, thus all loadings on PC1 are negative and within a small range (−0.34 to −0.15). PC2 is a contrast between vein 3, 4, 5, 6, 14 and tibia length with negative coefficients and the rest of the variables with positive coefficients hence PC2 is associated with wing shape (Table 1).
Figure 1. Projection of the wing and tibia data of Bactrocera invadens compared with the other Bactrocera species on the first two principal components.
Table 1. Eigen values and coefficients (loadings) of the first two principal components (PC1 and PC2) for the log-transformed wing measurements data of the fruit fly populations.
| PC1 | PC2 | |
| Proportion of variance | 86.30% | 4.40% |
| Eigen value | 0.046 0.002 | |
| Variable | Loadings | |
| Vein1(a_m) | −0.24 | 0.08 |
| Vein2 (a_b) | −0.21 | 0.34 |
| Vein3 (b_c) | −0.25 | −0.05 |
| Vein4 (d_e) | −0.34 | −0.73 |
| Vein5 (e_f) | −0.28 | −0.21 |
| Vein6 (f_g) | −0.31 | −0.09 |
| Vein7 (g_h) | −0.23 | 0.04 |
| Vein8 (h_i) | −0.29 | 0.09 |
| Vein9 (i_e) | −0.24 | 0.36 |
| Vein10 (c_j) | −0.29 | 0.27 |
| Vein11 (d_k) | −0.23 | 0.14 |
| Vein12 (i_l) | −0.15 | 0.23 |
| Vein13 (j_k) | −0.24 | 0.03 |
| Vein14 (n_o) | −0.26 | −0.01 |
| Tibia length | −0.26 | −0.04 |
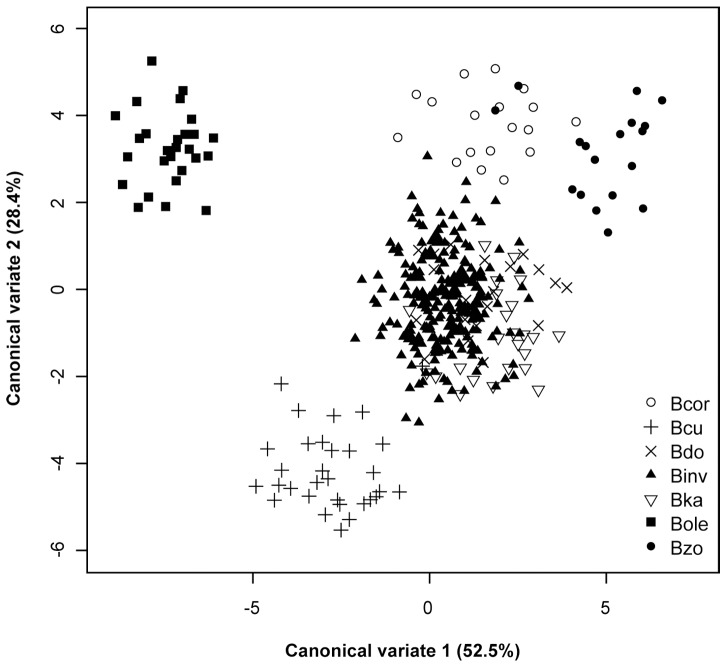
Tests for dimensionality for the canonical discriminant analysis revealed that only the first five canonical variates were statistically significant. Projection of the data on the first two canonical variate axes showed a better pattern of separation (Figure 2) compared with PCA. The first two canonical variates contributed a total of 80.9% (CV1 = 52.5% and CV2 = 28.4%) of the total variance. The third, fourth and fifth canonical variates contributed 9.3%, 5.7% and 2.9% of the total variance, respectively. The standardized canonical coefficients (Table 2) showed that CV1 is strongly dominated by positive correlation with vein4 and tibia length while strongly negatively correlated with vein1. CV2 is dominated by positive correlation with vein 14 and negative correlation with vein 3 and vein 10. Bactrocera invadens populations and the other B. dorsalis species complex clustered together while B. correcta, B. cucurbitae, B. oleae and B. zonata distinctly separated (Figure 2).
Figure 2. Projection of the wing and tibia data of Bactrocera invadens compared with other Bactrocera species on the first two canonical variates.
Table 2. Raw and standardized canonical coefficients for the canonical discriminant analysis on log-transformed wing measurements data for the fruit fly populations.
| Raw coefficients | Standardised coefficients | |||
| Variable | CV1 | CV2 | CV1 | CV2 |
| Vein1(a_m) | −46.32 | 3.75 | −1.71 | 0.14 |
| Vein2 (a_b) | −16.96 | −18.1 | −0.63 | −0.67 |
| Vein3 (b_c) | 14.87 | −38.17 | 0.49 | −1.26 |
| Vein4 (d_e) | 37.84 | 16.95 | 1.89 | 0.85 |
| Vein5 (e_f) | −1.00 | 17.93 | −0.04 | 0.78 |
| Vein6 (f_g) | 8.56 | −19.66 | 0.39 | −0.90 |
| Vein7 (g_h) | 17.82 | 4.65 | 0.68 | 0.18 |
| Vein8 (h_i) | −12.93 | −16.33 | −0.59 | −0.74 |
| Vein9 (i_e) | 2.50 | 18.49 | 0.11 | 0.85 |
| Vein10 (c_j) | −2.01 | −22.88 | −0.10 | −1.13 |
| Vein11 (d_k) | −12.44 | −22.2 | −0.46 | −0.82 |
| Vein12 (i_l) | −22.96 | 27.55 | −0.72 | 0.87 |
| Vein13 (j_k) | −2.47 | −6.96 | −0.11 | −0.32 |
| Vein14 (n_o) | 0.26 | 26.42 | 0.01 | 1.14 |
| Tibia length | 26.65 | 8.56 | 1.11 | 0.36 |
Mahalanobis distances used to compare morphometric divergence among populations group centroids showed a large degree of segregation in populations outside the B. dosrsalis complex and little interpopulation variability within the complex. For example, the largest Mahalanobis squared distance (D2 = 122.9) was found to be between B. cucurbitae and B. zonata, followed by B. oleae and B. zonata (111.8), B. correcta and B. curcubitae (88.4), and B. cucurbiate and B. oleae (68) (Table 3). Comparison of B. invadens populations against B. dorsalis sample gave a square distance of 11.4. The smallest distance was between the B. invadens populations and B. kandiensis (8.1).
Table 3. Mahalanobis Squared Distances (D2) between clusters representing the species/populations of Bactrocera invadens and other Bactrocera species.
| Species | Bcor | Bcu | Bdo | Binvadens | Bka | Bole | Bzo |
| Bcor | - | ||||||
| Bcu | 88.4 | - | |||||
| Bdo | 21.8 | 48.4 | - | ||||
| Binvadens | 22.1 | 40.4 | 11.4 | - | |||
| Bka | 26.5 | 43.1 | 15.9 | 8.1 | - | ||
| Bole | 54.6 | 68.0 | 61.4 | 45.1 | 71.7 | - | |
| Bzo | 26.6 | 122.9 | 39.6 | 43.4 | 36.6 | 111.8 | - |
Bcor – B. correcta, Bcu – B. cucurbitae, Bdo – B. dorsalis, Binvadens – B. invadens, Bka – B. kandiensis, Bole – B. oleae and Bzo – B. zonata.
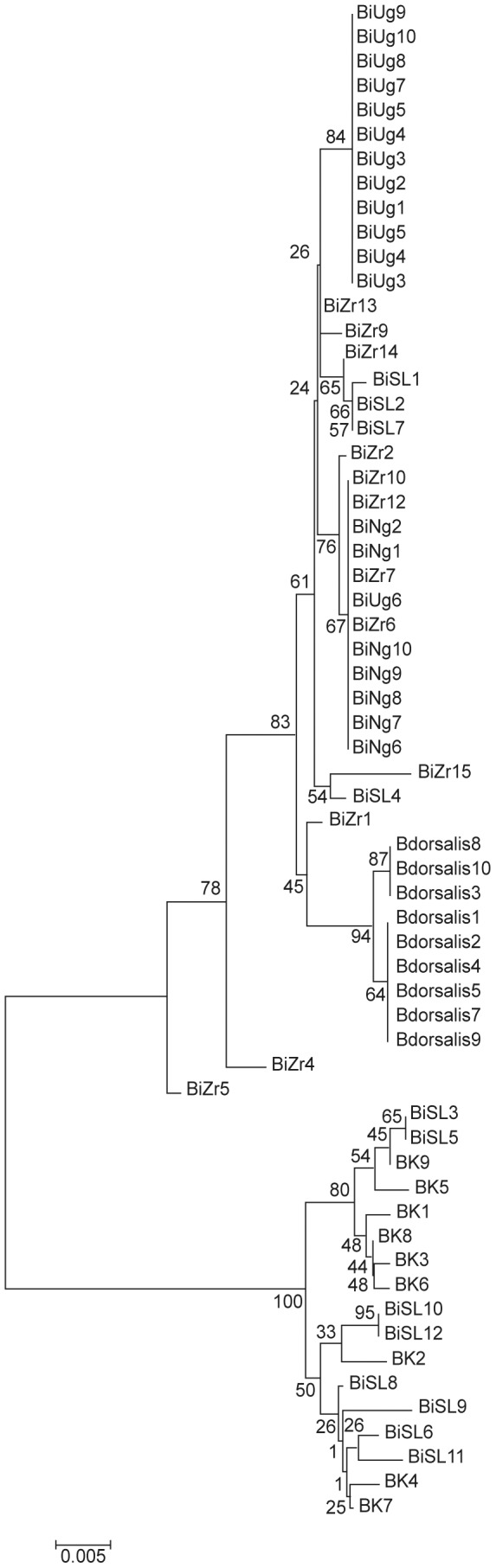
The first phylogenetic tree was derived considering only the species belonging to the Bactrocera dorsalis complex, the B. invadens populations from Kenya, Uganda, Zaria and Sri Lanka, B. dorsalis s.s. and B. kandiensis. The optimal tree with the sum of branch length = 0.14854468 is as shown in Figure 3. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [41]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method [42] and are in the units of the number of base substitutions per site. The analysis involved 62 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 658 positions in the final dataset. The tree separated the B. invadens populations into two clusters (Figure 3). One cluster consisted of B. invadens populations from Kenya, Uganda, Zaria and Sri Lanka with a separate branch consisting of B. dorsalis population. The second cluster was dominated by B. invadens individuals from Sri Lanka that were grouped with B. kandiensis (Figure 3).
Figure 3. Evolutionary relationships between Bactocera invadens populations, B. dorsalis s.s and B. kandiensis as inferred using Neighbour-Joining method by Mega 5 program (Tamura et al., 2011).
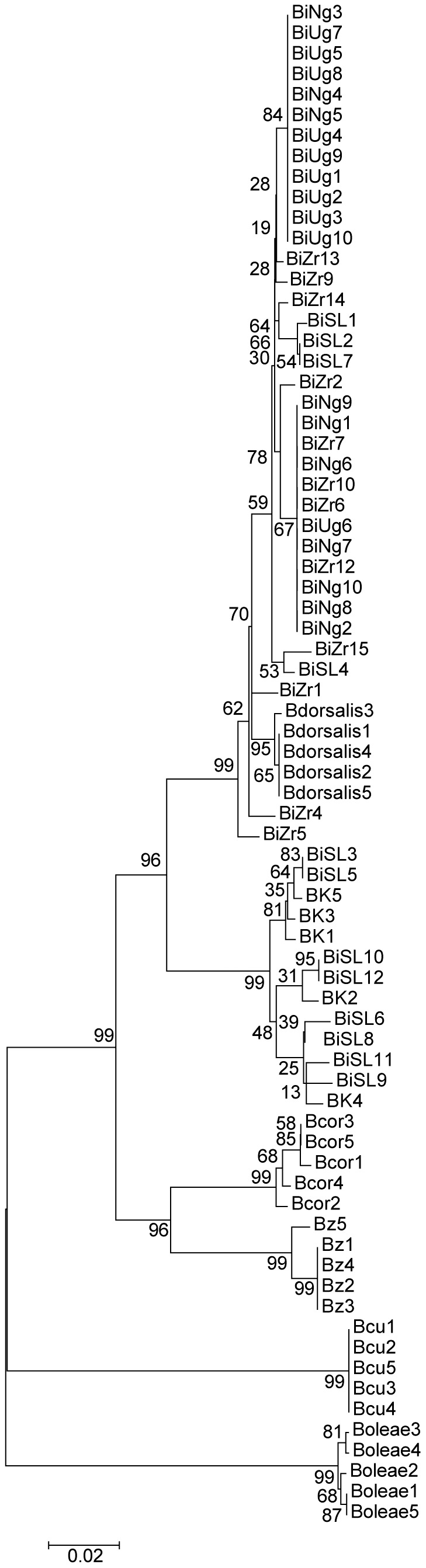
The second tree was constructed for all the Bactrocera species for which DNA barcodes have been generated in this study. The optimal tree with the sum of branch length = 0.49981941 is shown in Figure 4. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [41]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method [42] and are in the units of the number of base substitutions per site. The analysis involved 74 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 658 positions in the final dataset. This analysis clustered the Bactrocera species populations into four groups (Figure 4). The first group had the clustering of the Bactrocera dorsalis species complex (B. invadens, B. kandiensis and B. dorsalis sensu stricto), branching from the same node. The second group consisted of B. correcta and B. zonata branching from the same node. While the last two groups are clades, consisting of B. cucurbitae and B. oleae, respectively (Figure 4).
Figure 4. Evolutionary relationships between B. invadens populations and other Bactrocera species included in the study as inferred using Neighbour-Joining method by Mega 5 program (Tamura et al., 2011).
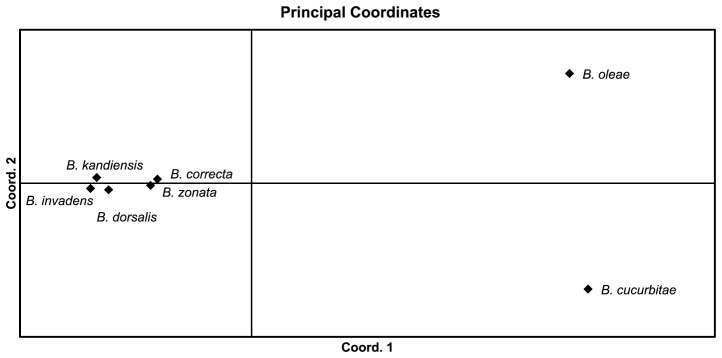
The table of genetic distances (Table 4) was constructed by Mega 5 [43] using the Kimura 2-parameter model. The output was used to generate principal component plots, using GenAlEx 6.41 [44]. In this analysis of the Bactrocera species using principal coordinate analysis (PCA), the first two axes explained 59.38% of the variation (the first axis 34.93%, and the second axis 24.46%) (Figure 5). The PCA separated the seven species into four distinct clusters. A cluster was occupied by the species belonging to the B. dorsalis complex, a cluster consisting of B. correcta and B. zonata, B. cucurbiate on its own cluster and likewise, B. oleae (Figure 5).
Table 4. Estimates of Evolutionary Divergence over Sequence Pairs between Groups generated by Mega 5 program (Tamura et al., 2011).
| Species | Bcu | Bcor | Bzo | Binvadens | Bka | Bdo | Bole |
| Bcu | - | ||||||
| Bcor | 0.181 | - | |||||
| Bzo | 0.183 | 0.076 | - | ||||
| Binvadens | 0.176 | 0.098 | 0.105 | - | |||
| Bka | 0.187 | 0.099 | 0.101 | 0.057 | - | ||
| Bdo | 0.167 | 0.091 | 0.100 | 0.025 | 0.06 | - | |
| Bole | 0.194 | 0.175 | 0.185 | 0.177 | 0.175 | 0.17 | - |
Bcor – B. correcta, Bcu – B. cucurbitae, Bdo – B. dorsalis, Binvadens – B. invadens, Bka – B. kandiensis, Bole – B. oleae and Bzo – B. zonata.
Figure 5. Plots of the principal coordinate analysis (PCA) from the covariance matrix with data standardization calculated using GenAlEx for the Bactrocera species.
Discussion
Among tephritid fruit flies, morphometric analysis has been tested and successfully used for determining differences among species of the B. dorsalis complex and to analyse variability among populations of Anastrepha fraterculus (Wiedemann) (Diptera: Tephritidae) collected from different host plants using aculeus, wing and head (frontal plate) characters [25], [28], [29], [45]. Our results showed that B. invadens can be morphometrically separated from other Bactrocera species (B. correcta, B. cucurbitae, B. oleae and B. zonata) used in this study with respect to wing morphology and the tibia length. However, the degree of morphological variation with respect to B. invadens populations and B. dorsalis s. s. used in this study was extremely low (Mahalanobis distance = 11.4). This is in line with the observations of Drew et al. [4] who classified B. invadens as a member of the B. dorsalis complex in possessing a very narrow coastal band and anal streak in addition to other abdominal and thoracic features. In a later study, Drew et al. [5] using aedeagus length was able to further discriminate between populations of B. invadens from those of B. carambolae, B. dorsalis, B. papayae and B. philippinensis. In our study, though the Sri Lankan populations of B. invadens were associated with different ecological and biogeographical conditions (i.e. different climate, altitude, vegetational community, etc.) (S.A. Mohamed et al., unpublished data), they showed similar morphology to the African samples, distinct from the other Bactrocera species but clustering with the B. dorsalis s.s. and supports the fact that the two species are closely related. In another study, Tan et al. [22] using chemoecological analysis of phenylpropanoid volatiles in male rectal pheromone gland, showed that males of laboratory-raised B. invadens accumulated two metabolites, 2-allyl-4,5-dimethoxyphenol (DMP) and (E)-coniferyl alcohol (E-CF) similar to B. dorsalis in almost equal quantities, in the rectal sac. On the basis of this finding, the authors concluded that the two pest species are biologically identical. Ongoing mating compatibility studies between B. invadens and B. dorsalis should further shed more light as to whether the two species belong to the same clade.
In our studies, we also observed that the Mahalanobis distance between B. invadens and B. kandiensis was short (8.1) indicating that the two species are closely related. Drew et al. [4] in the description of B. invadens stated that this exotic species in Africa was morphologically similar to B. kandiensis based on wing and abdominal characters. The only differentiating characters reported by Drew et al. [4] was femora that was entirely fulvous in B. invadens and the variation in microtrichia pattern in the basal area of cell br, above cell bm. In a more recent study Drew et al. [5] did not find any difference in the thorax length between B. kandiensis (3.10 mm) and B. invadens (3.10 mm); wing length, B. kandiensis (6.18 mm) and B. invadens (6.21 mm); wing vein, B. kandiensis (2.25 mm) and B. invadens (2.25 mm); and hind tibia length, B. kandiensis (1.86 mm) and B. invadens (1.85 mm). The results of our findings therefore, concur with those of Drew et al. [5].
Although morphological characters are primarily used to define species, the genetic and behavioural boundaries of species need to be understood and elucidated. This is particularly true for groups of economically important species such as those in the B. dorsalis complex. In this regard, DNA barcoding that involves retrieval of a standard region of mitochondrial gene, Cytochrome c oxidase 1 (CO1) at its 5′ end containing ≈650 base pairs gene (acting as barcode) for identification and delineation of all animal life [39] has shown to be potentially a useful tool to separate members of Tephritid groups of fruit flies [16], [46], [47]. The use of COI sequences together with quantitative support in terms of bootstraps and divergence values provides a better resolution for fruit flies identification than was possible with other methods like the PCR-RFLP [46], [48].
The use of DNA barcodes utilizing the CO1 gene has enabled the interpretation of the relationship between B. invadens and the other Bactrocera species. In this study, the smallest genetic distance (0.025) was detected between populations of B. invadens and B. dorsalis, B. invadens and B. kandiensis (0.057) and B. dorsalis and B. kandiensis (0.06). This is a typical scenario of divergences between congeneric species which are normally higher than within species [39], [40], [47], [49]. Similar studies by Tan et al. [22] using COI gene clustered B. invadens and B. dorsalis in the same branch. The molecular findings in the current study further substantiate our morphometric data obtained above, in which the Canonical variate plots separated B. invadens populations from all the other Bactrocera species but matched B. invadens populations with B. dorsalis s.s. and B. kandiensis. This is also evident in the PCA plots using the genetic distances, where B. invadens populations clustered with the B. dorsalis s.s. and B. kandiensis.
Although we recorded some levels of concordance between the molecular and the morphometric results, large divergence at micro-geographic scale was observed among populations of B. invadens. For instance, some individuals belonging to B. invadens populations of Sri Lanka clustered together with B. kandiensis in the NJ trees generated in the study. Therefore, the NJ tree did not fully discriminate the populations belonging to the B. dorsalis complex. This is also true in the study by Armstrong and Ball [46] where the COI gene could not confidently separate some of the species within the B. dorsalis complex, and hence, have suggested the use of additional gene regions to overcome this limitation.
Some representatives of the B. dorsalis complex are extremely polyphagous and highly invasive pests, thus it is one of the most important pest species complexes in world agriculture [1]. Because of their economic and quarantine importance, species-level taxonomic work and diagnostics in the B. dorsalis complex is relatively advanced [50], [20], [21], but much effort in this regard is still required. Since the primary goal of DNA barcoding is to develop an accurate, rapid, cost-effective, and universally accessible DNA-based system for species identifications [39], [40], this method could be adopted as a standard method of identification of invasive alien species that pose a high risk on the global economy [46].
The use of DNA barcoding in conjunction with other molecular diagnostic tools, for biosecurity will bridge the limitations of previous molecular tools such as inconsistency in technology use, and finite taxas in some of the invasive species [46]. Several studies have demonstrated the effectiveness of DNA barcoding in different insect groups [39], [40], [51], [52], [46]. These projects have shown that >95% of species possess unique COI barcode sequences; thus species-level identifications are regularly attained [53]. Although DNA barcoding is a current molecular tool of choice and to a certain extent can provide answers to molecular identification (e.g. DNA from incomplete libraries [47]), conclusive phylogeny should include an array of molecular diagnostic tools. Our study has contributed to unraveling the identity of B. invadens, a finding which could facilitate its placement in the right phylogeny, potentially easing quarantine restrictions and improvement in the management of this invasive pest. Nevertheless, divergent views still exist. Hence additional studies on the pest chemoecology, behaviour, morphometry and genetics are warranted.
Materials and Methods
Ethics Statement
No specific permits were required for the described field studies.
No specific permissions were required for these locations/activities.
The locations are not privately-owned or protected in any way.
The field studies did not involve endangered or protected species.
Sample collection
Male samples of B. invadens were collected (using Methyl eugenol baited traps) from different countries in Africa and South East Asia. The collected insects were preserved in ethanol (70% for morphometric analysis and 95% for DNA analysis). In Africa, the sampling area included regions from the East to West Africa (Data not included). Some representative samples included in the study are from East Africa: from Kenya (Nguruman) and Uganda (Kawanda) and a sample from West Africa, Zaria, Nigeria (Table 5). The South East Asian sample is represented by Sri Lanka (the presumed aboriginal home of B. invadens). Also included in this study, were other Bactrocera species belonging to the B. dorsalis species complex namely B. dorsalis sensu stricto (Hendel) (Hawaii) and B. kandiensis (Drew & Hancock) (Sri Lanka). Other Bactrocera species considered in the study that do not belong to B. dorsalis complex were: B. correcta (Bezzi) (India), B. cucurbitae (Coquillett) (Kenya), B. oleae Gmelin (Kenya) and B. zonata (Saunders) (Mauritius). The specimens were identified by M.K. Billah (Department of Zoology, University of Ghana, Legon).
Table 5. Key of the veins used in morphometrics analysis.
| Representation | Description | |
| vein 1 | a_m | Wing length |
| vein 2 | a_b | Humeral break – Subcostal break |
| vein 3 | b_c | Subcostal break – vein R1 |
| vein 4 | d_e | r – m |
| vein 5 | e_f | Upper length of dm-cell |
| vein 6 | f_g | Basal height of dm-cell |
| vein 7 | g_h | Lower length of dm-cell |
| vein 8 | h_i | Apical height of dm-cell |
| vein 9 | i_e | Upper length of dm-cell |
| vein 10 | c_j | Vein R1 – Vein R2+3 |
| vein 11 | d_k | R4+5 |
| vein 12 | i_l | M |
| vein 13 | J_k | C |
| vein 14 | n_o | Wing width |
Morphometry
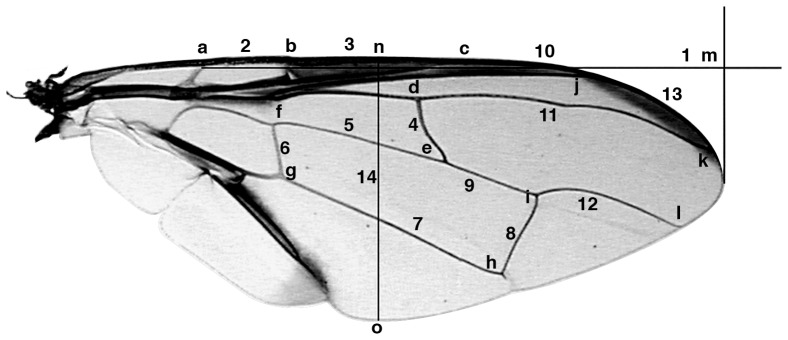
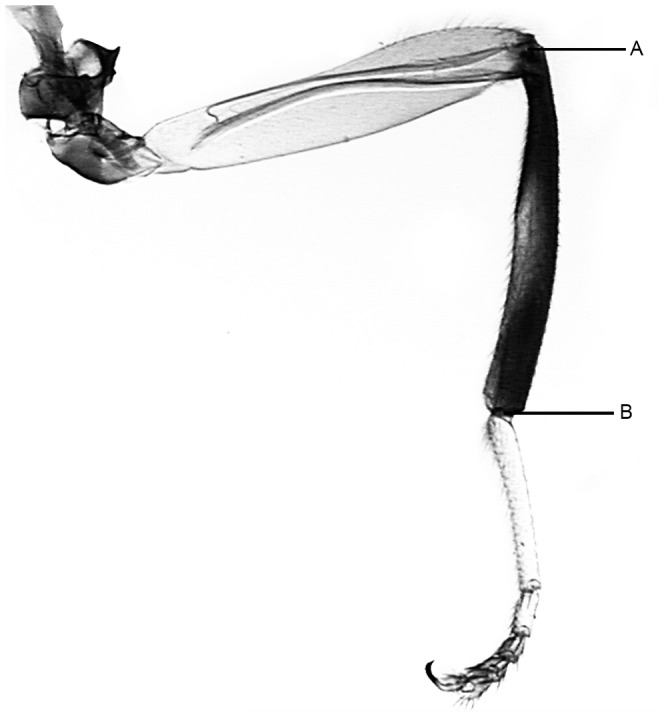
Specimens of the different Bactrocera species were prepared following the general procedure for slide preparation [54] with modifications according to the needs or state of the specimen. Images of the right wing and right hind tibia from the slide mounted specimens were captured using video microscopy – Leica MZ 125 Microscope, fitted with Toshiba 3CCD camera using the Auto Montage software (Syncroscopy, Synoptics group, 2004) at magnification ×25 for total length and width of the wing, ×50 for the wing veins and ×63 for the tibia. Measurements were taken by the program Image-Pro® Plus version 4.1 for Windows™ (Media Cybernetics, 1999) and the data exported directly to an Excel data sheet. For all parts, measurements were taken in triplicate (to an accuracy of 0.001 mm). Fourteen wing distances between 15 selected landmarks on the wing were computed to characterize the shape and size of the wings for differentiation. These distances are: the Humeral break – Subcostal break, Subcostal break – vein R1, r – m, Upper length of dm-cell, Basal height of dm-cell, Lower length of dm-cell, Apical height of dm-cell, Vein R1 – Vein R2+3, R4+5, M, C, the wing length and width, and tibia length (Figure 6 & 7; Table 5). Voucher specimens of all insects and slides are deposited at the Biosystematics Unit of the International Centre of Insect physiology and Ecology (icipe), Nairobi, Kenya.
Figure 6. Wing showing points of reading taken for morphometric analysis.
Figure 7. Tibia points of measurement (measurement taken from point A to point B).
DNA extraction and PCR
The insects used for DNA barcoding were photographed laterally, dorsally and ventrally and appropriately labeled prior to DNA extraction. DNA from whole insects was extracted using the Qiagen DNeasy® Blood and Tissue Kit (Qiagen, GmbH-Hilden, Germany) as per manufacturer's instructions. The extracted DNA was stored at −20°C until required for amplification. PCR was carried out using universal primers, Forward primer (LCO1490) 5′-GGTCAACAAATCATAAAGATATTGG-3′ and reverse primer (HCO2198) 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′ [55] to amplify a 658 bp fragment of the COI gene. The PCR amplification was carried out in a 20 µl volume containing 1× reaction Buffer, 200 µm of dNTP mix, 0.4 pmol/µl of each primer, 2.5 mM, MgCl2, 1 unit Taq DNA polymerase (Genescript) and 1 ng DNA template. Standard cycling conditions of 5 min at 94°C, then 35 cycles of 30 s at 94°C, 1 min at annealing temperature of 45°C and 1 min at 72°C, followed by a final elongation step of 5 min at 72°C were used. The products were purified using QIAquick PCR purification kit (Qiagen, GmbH-Hilden, Germany) according to the manufacturer's instructions and subsequently sequenced in both directions using ABI 3700 genetic analyzers. The COI sequences were submitted to the Barcode of Life database (BOLD) and deposited in GenBank (Accession numbers are found in Table 6). The DNA voucher specimens are kept in icipe Molecular biology and biotechnology department.
Table 6. Collection data of B. invadens populations and other Bactrocera species used in this study.
| Region/Country | Sample name | Sample site | Collection | Coordinates | GenBank Accession numbers |
| Bactrocera invadens | |||||
| Africa | |||||
| Kenya | Ke | Nguruman | ME | 01°48′32S, 036°03′35E | JQ692820, JQ692701, JQ692801, JQ692845, JQ692688, JQ692664, JQ692811, JQ692781, JQ692805, JQ692780 |
| Uganda | Ug | Kawanda | ME | 00°49′52S, 031°55′05″E | JQ692633,JQ692709, JQ692824, JQ692854, JQ692650, JQ692794, JQ692844, JQ692752, JQ692681, JQ692841 |
| Nigeria | Nig | Zaria | ME | 11°06′N, 07°42′E | JQ692727, JQ692723, JQ692698, JQ692742, JQ692867, JQ692816, JQ692825, JQ692719, JQ692731, JQ692636, JQ692684, JQ692812 |
| Asia | |||||
| Sri Lanka | SL | Ranbukpitiya | Tropical almond | JQ692669, JQ692818, JQ692757, JQ692661, JQ692741, JQ692737, JQ692708, JQ692838, JQ692722, JQ692639, JQ692764, JQ692835 | |
| Bactocera correcta | Bcor | Sri Lanka-Anuradhapura | ME | 08°21′0″N, 080°23′1″E | JQ692856,JQ692753, JQ692641, JQ692784, JQ692787 |
| Bactrocera cucurbitae | Bcu | Kenya-Nairobi | LT & Cu Lure | 01°13′952S, 036°51′314E | JQ692734, JQ692803, JQ692772, JQ692685, JQ692740 |
| Bactrocera dorsalis s.s | Bdo | Hawaii | Laboratory reared | - | JQ692775, JQ692829, JQ692694, JQ692790, JQ692747, JQ692706, JQ692864, JQ692758, JQ692678 |
| Bactrocera kandiensis | Bka | Sri Lanka-Kandy | ME | 07°16′753N, 80°35′731E | JQ692767, JQ692836, JQ692837, JQ692692, JQ692806, JQ692673, JQ692813, JQ692674, JQ692686 |
| Bactrocera oleae | Bole | Kenya-Burguret forest | Ex-fruits (olives) | 00°06′720S, 37°02′342E | JQ692833, JQ692687, JQ692778, JQ692762, JQ692808 |
| Bactrocera zonata | Bzo | Mauritius | Laboratory reared | - | JQ692749, JQ692662, JQ692819, JQ692799, JQ692705 |
Data analysis
Morphometry
Principal component analysis (PCA), a multivariate statistical procedure commonly used to reveal patterns in measured correlated variables, was used to determine if there was any clustering in the fruit fly species populations based on the wing veins measurements. The data were transformed (log10) prior to PCA to stabilize the variance of the measured variables and thus give the variables approximately equal weights in the PCA [56], [57]. Since PCA is inherently a single-group procedure and is not guaranteed to find group differences even if they exist, the log-transformed morphometric data were also subjected to canonical discriminant analysis, a method for analyzing group structure in multivariate data. Bartlett's χ2 was used to test for significance of principal components and canonical variates [58], [59]. Mahalanobis squared distances between fruit fly species were obtained as a measure of distance between species based on means, variances and covariances [60]. The analyses were performed using R 2.13.1 [61].
COI sequence data analysis
Sequences were assembled and edited using Chromas version 2.13 (Technelysium Pty ltd, Queensland, Australia), and aligned using ClustalX version 1.81 [62]. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 5 [43] and a Neighbour-joining tree constructed [63] with bootstrapping and using the Kimura 2 distance matrix [42]. A table of between species distances was also constructed using MEGA version 5 [43]. This table of distances was used to generate the principal component plots using GenAlEx 6.41 [44].
Acknowledgments
We thank J. Gitau for slides preparation. We would also like to acknowledge Nathalie Erbout for her assistance in the morphometric work. We are also grateful to numerous partners of the African Fruit Fly Program for providing insects from Africa and other parts of the world.
Funding Statement
The authors are grateful to the International Atomic Energy Agency (IAEA) Vienna, Austria (Contract number 16072), and the German Ministry of Economic Cooperation and Development (BMZ) through Deutsche Gesellschaft für Internationale Zusammenarbeit (GIZ) (Contract number 81132021) for grant support for this investigation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Clarke AR, Armstrong KF, Carmichael AE, Milne JR, Raghu S, et al. (2005) Invasive phytophagous pests arising through a recent evolutionary radiation: the Bactrocera dorsalis complex of fruit flies. Annual Review of Entomology 50: 293–319. [DOI] [PubMed] [Google Scholar]
- 2.McPheron BA, Steck GJ (1996) Fruit Fly Pests: A world assessment of their biology and management. St Lucie Press, Delray Beach, FL.
- 3. Lux SA, Copeland RS, White IM, Manrakhan A, Billah MK (2003) A New Invasive Fruit Fly Species from the Bactrocera dorsalis (Hendel) Group Detected in East Africa. Insect Science and Its Application 23 4:355–361. [Google Scholar]
- 4. Drew RAI, Tsuruta K, White IM (2005) A new species of pest fruit fly (Diptera: Tephritidae: Dacinae) from Sri Lanka and Africa. African Entomology 13: 149–154. [Google Scholar]
- 5. Drew RAI, Raghu S, Halcoop P (2008) Bridging the morphological and biological species concepts: studies on the Bactrocera dorsalis (Hendel) complex (Diptera: Tephritidae: Dacinae) in South-east Asia. Biological Journal of the Linnean Society 93: 217–226. [Google Scholar]
- 6.French C (2005) The new invasive Bactrocera species pp. 19–20 in Insect Pest Control Newsletter, No. 65. International Atomic Energy Agency, Vienna, Austria.
- 7. Vayssières JF, Goergen G, Lokossou O, Dossa P, Akponon C (2005) A new Bactrocera species in Benin among mango fruit fly (Diptera: Tephritidae) species. Fruits 60: 371–377. [Google Scholar]
- 8. Ekesi S, Nderitu PW, Rwomushana I (2006) Field infestation, life history and demographic parameters of the fruit fly Bactrocera invadens (Diptera: Tephritidae) in Africa. Bulletin of Entomological Research 96: 379–386. [PubMed] [Google Scholar]
- 9. Mwatawala MW, De Meyer M, Makundi RH, Maerere AP (2006a) Biodiversity of fruit flies (Diptera, Tephritidae) at orchards in different agro-ecological zones of the Morogoro region, Tanzania. Fruits 61: 321–332. [Google Scholar]
- 10.Ekesi S, Billah MK (2007) A field guide to the management of economically important tephritid fruit flies in Africa. ICIPE Science Press, Nairobi, Kenya.
- 11. Abanda F-XN, Quilici S, Vayssiéres JF, Kouodiekong L, Woin N (2008) Inventory of fruit flies species on guava in the area of Yaounde, Cameroon. Fruits 63: 19–26. [Google Scholar]
- 12. Rwomushana I, Ekesi S, Gordon I, Ogol CKPO (2008) Host plant and host plant preference studies for Bactrocera invadens (Diptera: Tephritidae) in Kenya, a new invasive fruit fly species in Africa. Annals of the Entomological Society of America 101: 331–340. [Google Scholar]
- 13. Mwatawala MW, De Meyer M, Makundi RH, Maerere AP (2009) Host range and distribution of fruit-infesting pestiferous fruit flies (Diptera, Tephritidae) in selected areas of Central Tanzania. Bulletin of Entomological Research 1–13. [DOI] [PubMed] [Google Scholar]
- 14. Goergen G, Vayssières J-F, Gnanvossou D, Tindo M (2011) Bactrocera invadens (Diptera: Tephritidae), a new invasive fruit fly pest for the Afrotropical region: Host plant range and distribution in west and central Africa. Entomological Society of America 40 4:844–854 2011. DOI: 10.1603/EN11017. [DOI] [PubMed] [Google Scholar]
- 15. Vayssières JF, Korie S, Ayegnon D (2009) Correlation of fruit fly (Diptera Tephritidae) infestation of major mango cultivars in Borgou (Benin) with abiotic and biotic factors and assessment of damage. Crop protection 28: 477–488. [Google Scholar]
- 16. Van Houdt JKJ, Breman FC, Virgilio M, De Meyer M (2010) Recovering full DNA barcodes from natural history collections of Tephritid fruitflies (Tephritidae, Diptera) using mini barcodes. Molecular Ecology Resources 10: 459–465. [DOI] [PubMed] [Google Scholar]
- 17. Drew RAI, Hancock DL (1994) The Bactrocera dorsalis complex of fruit flies (Diptera: Tephritidae: Dacinae) in Asia. Bulletin of Entomological Research, Supplement Series 2: 1–68. [Google Scholar]
- 18.Lawson AE, McGuire DJ, Yeates DK, Drew RAI, Clarke AR (2003) Dorsalis: an interactive identification tool to fruit flies of the Bactrocera dorsalis complex. CD-ROM Publication, Griffith University, Brisbane, Australia.
- 19.Armstrong KF, Cameron CM (2000) Species identification of tephritids across a broad taxonomic range. In: Tan KH, ed. Area-wide control of fruit flies and other insect pests. London: CABI, 703–710.
- 20. Muraji M, Nakahara S (2002) Discrimination among pest species of Bactrocera (Diptera: Tephritidae) based on PCR-RFLP of the mitochrondrial DNA. Applied Entomology and Zoology 37: 437–446. [Google Scholar]
- 21. Nakahara S, Kato H, Kaneda M, Sugimoto T, Muraji M (2001) Identification of Bactrocera dorsalis complex species (Diptara: Tephritidae) by PCR-RFLP analysis. II.Astudy of genetic variation in B. dorsalis complex (Philippines population) and B. dorsalis (Taiwan population). Res Bull Plant Prot Serv Jpn 37: 69–73. [Google Scholar]
- 22.Tan KH, Tokushima I, Ono H, Nishida R (2010) Comparison of phenylpropanoid volatiles in male rectal pheromone gland after methyl eugenol consumption, and molecular phylogenetic relationship of four global pest fruit fly species: Bactrocera invadens, B. dorsalis, B. correcta and B. zonata. Chemoecology, Springer. DOI 10.1007/s00049-010-0063-1.
- 23. Schutze MK, Jessup A, Clarke AR (2011) Wing shape as a potential discriminator of morphologically similar pest taxa within the Bactrocera dorsalis species complex (Diptera: Tephritidae). Bulletin of Entomological Research 1–9 Doi:10.1017/S0007485311000423. [DOI] [PubMed] [Google Scholar]
- 24.Reyment RA, Blackith RE, Campbell NA (1984) Multivariate morphometrics (second edition). Academic Press, London.
- 25.Perero JG, Nasca AJ, Stilinovic D (1984) Introducción a un estudio morfológico-taxonómico de especímenes de Anastrepha fraterculus Wiedemann colectados sobre distintos hospederos en la Provincia de Tucumán. pp. 421–455 in Anales II Congreso Internacional de Biomatemáticas, Facultad de Ciencias Exactas y Naturales, Buenos Aires, Argentina.
- 26. Willig RM, Owen RD, Colbert RL (1986) Assessment of morphometric variation in natural populations: the inadequacy of the univariate approach. Systematic Zoology 35: 195–203. [Google Scholar]
- 27. McNamee S, Dytham C (1993) Morphometric discrimination of the species Drosophila melanogaster (Meigen) and D. simulans (Sturtevant) (Diptera: Drosophilidae). Systematic Entomology 18: 231–236. [Google Scholar]
- 28.Selivon D (1996) Estudo sobre a diferenciaçâo populacional em Anastrepha fraterculus (Wiedemann) (Diptera: Tephritidae). 137 pp. PhD thesis (unpublished), Instituto de Biociencias, Universidade de Sâo Paulo, Brazil.
- 29. Adsavakulchai A, Baimai V, Prachyabrued W, Grote PJ, Lertlum S (1999) Morphometric study for identification of the Bactrocera dorsalis complex (Diptera: Tephritidae) using wing image analysis. Biotropica 13: 37–48. [Google Scholar]
- 30. De Meyer M (2005) Phylogenetic relationships within the fruit fly genus Ceratitis MacLeay (Diptera: Tephritidae), derived from morphological and host plant evidence. Insect Systematics and Evolution 36: 459–480. [Google Scholar]
- 31. Drew RAI, Dorji C, Romig MC, Loday P (2006) Attractiveness of various combinations of colors and shapes to females and males of Bactrocera minax (Diptera: Tephritidae) in a commercial mandarin grove in Bhutan. Journal of Economic Entomology 99: 1651–1656. [DOI] [PubMed] [Google Scholar]
- 32. Armstrong KF, Cameron CM, Frampton ER (1997) Fruit fly (Diptera: Tephritidae) species identification: a rapid molecular diagnostic technique for quarantine application. Bulletin of Entomological Research 87: 111–118. [Google Scholar]
- 33. De Meyer M (2000) Systematic revision of the subgenus Ceratitis MacLeay s. s (Diptera: Tephritidae). Zoological Journal of the Linnean Society 128: 439–467. [Google Scholar]
- 34.McPheron BA (2000) Population genetics and cryptic species. In Tan, K.-H., (Ed). Area wide control of fruit flies and other insect pests. Pernerbit Universiti sains Malaysia, Peneng, p. 483–490, 782p.
- 35. Sonvinco A, Manso F, Quesada-Allue LA (1996) Discrimination between the immature stages of Ceratitis capitata and Anastrepha fraterculus (Diptera: Tephritidae) populations by random amplified polymorphic DNA polymerase chain reactiopn. Journal of Economic Entomology 89: 1208–1212. [DOI] [PubMed] [Google Scholar]
- 36. Morrow J, Scott L, Congdon B, Yeates D, Frommer M, et al. (2000) Close genetic similarity between two sympatric species of tephritid fruit fly reproductively isolated by mating time. Evolution 54: 899–910. [DOI] [PubMed] [Google Scholar]
- 37. Barr NB, Copeland RS, De Meyer M, Masiga D, Kibogo HG, et al. (2006) Molecular diagnostics of economically important Ceratitis fruit fly species (Diptera: Tephritidae) in Africa using PCR and RFLP analyses. Bulletin of Entomological Research 96: 505–521 DOI: 10.1079/BER2006452. [PubMed] [Google Scholar]
- 38. Ratnasingham S, Hebert PDN (2007) BOLD: The Barcode of Life Data System (www.barcodinglife.org). Molecular Ecology Notes 7: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hebert PD, Cywinska A, Ball SL, de Waard JR (2003a) Biological identifications through DNA barcodes. Proceedings of the Royal Society B 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hebert PDN, Ratnasingham S, de Waard JR (2003b) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B Biol Sci 270 Suppl.:S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 42. Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 43. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA 5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hernandez-ortiz V, Bartolucci AF, Morales-Valles P, Frias D, Selivon D (2012) Cryptic species of the Anastrepha fraterculus complex (Diptera: Tephritidae): A multivariate approach for the recognition of the South American morphotypes. Entomological society of America 105: 305–318. [Google Scholar]
- 46. Armstrong KF, Ball SL (2011) DNA barcodes for biosecurity: invasive species identification. Phil Trans R Soc B 2005 360: 1813–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Virgilio M, Jordaens K, Breman FC, Backeljau T, De Meyer M (2012) Identifying Insects with Incomplete DNA Barcode Libraries, African Fruit Flies (Diptera: Tephritidae) as a Test Case. PLoS ONE 7 2:e31581 doi:10.1371/journal.pone.0031581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barr NB (2009) Pathway analysis of Ceratitis capitata (Diptera: Tephritidae) using mitochondrial DNA. Journal of Economical Entomolology 102: 401–411. [DOI] [PubMed] [Google Scholar]
- 49. Barr NB, Islam MS, De Meyer M, McPheron BA (2012) Molecular identification of Ceratitis capitata (Diptera: Tephritidae) using DNA sequences of the COI barcode region. Annals of the Entomological Socociety of America 105 2:339–350 DOI: http://dx.doi.org/10.1603/AN11100. [Google Scholar]
- 50. Muraji M, Nakahara S (2001) Phylogenetic relationships among fruit flies, Bactrocera (Diptera, Tephritidae), based on the mitochondrial rDNA sequences. Insect Mol Biol 10: 549–59. [DOI] [PubMed] [Google Scholar]
- 51. Hebert PD, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004a) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences USA 101: 14 812–14 817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PDN (2006) DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proc Natl Acad Sci U S A 103: 3657–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hajibabaei M, Singer GA, Hebert PDN, Hickey DAC (2007) DNA barcoding: how it complements taxonomy, molecular Phylogenetics and population genetics. Science direct 10: 1–6. [DOI] [PubMed] [Google Scholar]
- 54. Billah MK, Kimani-Njogu S, Overholt WA, Wharton RA, Wilson DD, et al. (2005) The effect of host larvae on three Psyttalia species (Hymenoptera: Braconidae), parasitoids of fruit nesting flies (Diptera: Tephritidae). Int J Trop Insect Sci 25: 168–175. [Google Scholar]
- 55. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biological Biotechnology 3: 294–299. [PubMed] [Google Scholar]
- 56. Baxter MJ (1995) Standardization and Transformation in Principal Component Analysis, with Applications to Archaeometry. Applied Statististics V 44 4:513–527. [Google Scholar]
- 57.Sokal RR, Rohlf FJ (1995) Biometry: The principles and Practice of Statistics in Biological Research. 3rd Edition. W. H. Freeman & Company, USA. 887 pp.
- 58. Bartlett MS (1950) Tests of significance in factor analysis. British Journal of Psychology 3: 77–85. [Google Scholar]
- 59. Bartlett MS (1951) A further note on tests of significance. British Journal of Psychology 4: 1–2. [Google Scholar]
- 60.Zar JH (1999) Biostatistical analysis. 4th edition. Prentice Hall, Upper Saddle River, NJ.
- 61.R Development Core Team (2011) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/.
- 62. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]