Abstract
The highly-conserved protein, IscU, serves as the scaffold for iron-sulfur cluster assembly in the ISC system common to bacteria and eukaryotic mitochondria. The apo-form of IscU from Escherichia coli has been shown to populate two slowly interconverting conformational states: one structured (S) and one dynamically disordered (D). Furthermore, single-site amino acid substitutions have been shown to shift the equilibrium between the metamorphic states. Here, we report three-dimensional structural models derived from NMR spectroscopy for the S-state of wild-type (WT) apo-IscU, determined under conditions where the protein was 80% in the S-state and 20% in the D-state, and for the S-state of apo-IscU(D39A), determined under conditions where the protein was ~ 95% in the S-state. We have used these structures in interpreting the effects of single site amino acid substitutions that alter %S = (100×[S])/([S]+[D]). These include different residues at the same site, %S: D39V > D39L > D39A > D39G ≈ WT, and alanine substitutions at different sites, %S: N90A > S107A≈E111A > WT. Hydrophobic residues at residue 39 appear to stabilize the S-state by decreasing the flexibility of the loops that contain the conserved cysteine residues. The alanine substitutions at positions 90, 107, and 111, on the other hand stabilize the protein without affecting the loop dynamics. In general, the stability of the S-state correlates with the compactness and thermal stability of the variant.
Keywords: conformational equilibrium, effects of single amino acid substitutions, solution structure determination, dynamics, NMR
Iron-sulfur (Fe-S) clusters are ancient and ubiquitous prosthetic groups involved in a wide variety of biological processes.1 In prokaryotes, the ISC (iron-sulfur cluster) biogenesis system, encoded by the isc operon (IscR, IscS, IscU, IscA, HscB, HscA, and ferredoxin) is mainly responsible for the ‘house-keeping’ generation of iron-sulfur clusters.2 The complex interactions of these proteins are essential for efficient Fe-S cluster biogenesis.3 A very similar ISC system is found in the mitochondria of eukaryotes.4 IscU is the scaffold protein for cluster assembly and transfer. The amino acid sequence of IscU is highly conserved over diverse organisms.2 More recently, it was found that a single intronic mutation in the human ISCU gene is associated with a myopathy resulting from the deficiencies in Fe-S proteins succinate dehydrogenase and aconitase.5,6 Complete removal of the iscu gene in mice resulted in an early embryonic death.7
Early NMR investigations of Thermotoga maritima IscU suggested that the apo-form of this protein has a ‘molten globule-like’ structure.8 IscU from Haemophilus influenzae showed a much less structured conformation in its apo-state than in its Zn2+-bound state.9 Thus, initial structural investigations of IscU focused on the Zn2+-bound form of the protein.9,10 The X-ray crystal structure of Aquifex aeolicus holo-IscU revealed a homotrimer with only one of the three subunits containing a [2Fe-2S] cluster. The two apo-IscU subunits adopted different conformations in the crystal.11 It has been reported that the IscU(D39A) substitution increases the stability of the Fe-S cluster in holo-IscU.12–14
We found that E. coli apo-IscU exists in solution as two interconverting conformations, one structured (S) and one dynamically disordered and lacking secondary structure (D).15,16 The two forms interconvert on time scale of about one second, and at pH 8.0 and 25 °C, the free energy between the two states is ~ 0.5 kcal/mol.16 A separate study from our group has demonstrated that the S⇆D transition involves the conversion of two peptidyl-prolyl peptide bonds from trans in the S-state to cis in the D-state (Dai Z, Tonelli, M, and Markley JL, submitted for publication). Thus, although the D-state appears disordered from the lack of dispersion of its backbone NMR chemical shifts,16 it does contain a fold that stabilizes two high-energy cis peptide bonds. The highly cooperative transition between the S- and D-states, which involves two concerted peptide bond isomerizations and a change in secondary structure, explain the high activation barrier and the consequent slow rates of interchange between the two states. Thus, IscU can be categorized as a metamorphic protein.17
Both states have found to be physiologically important. The D-state of IscU is the substrate for IscS, the PLP-dependent cysteine desulfurase that catalyzes the conversion of cysteine to alanine and transfers the sulfane sulfur generated to IscU.16 Once a cluster has been assembled, IscU converts to the S-state.11 The S-state of IscU binds preferentially to HscB (the DnaJ-like co-chaperone),15 and HscB targets the complex to HscA(ATP) (the DnaK-like chaperone in its ATP-bound state). Finally, upon ATP hydrolysis HscA(ADP) binds to the D-state of IscU ensuring cluster release to an acceptor protein (Kim JH et al., submitted for publication). IscU appears to have evolved to undergo the S⇆D interconversion and to have an optimal set point for this equilibrium. We found that certain single amino acid substitutions (D39A, N90A, S107A, E111A) stabilize the S-state of IscU, whereas others (K89A, N90D) stabilize the D-state.16 In vitro Fe-S cluster assembly studies showed that IscU variants with %S = (100×[S])/([S]+[D]) higher than wild-type (WT) assemble clusters slowly with an initial lag and that variants with %S lower than WT assemble less stable clusters at a slower rate without a lag.16 Thus it is of considerable interest to determine the structures of the S- and D-states along with the features of the IscU sequence that are responsible for its metamorphic properties and for setting the position of the equilibrium.
We report here the solution structures of the S-states of apo-IscU(WT) and apo-IscU(D39A) determined by NMR spectroscopy. The structure of apo-IscU(WT) was determined under conditions in which %S ≈ 80; thus we had to contend with signals from the D-state of the protein. By contrast, apo-IscU(D39A) had %S ≈ 95. Comparison of structures revealed that the wild-type protein is more dynamic than the D39A variant, particularly in the cysteine-containing loops. This result suggests that the higher %S can be explained by stabilization of the S-state by the D39A substitution. The two structures have provided a framework for our analysis of data from several other single-site amino acid substitutions that alter %S.
EXPERIMENTAL PROCEDURES
Production and purification of proteins
We used a QuikChange II Site-Directed Mutagenesis Kit (Stratagene) to mutate the pTrcIscU(WT) expression vector18 for the production of IscU variants. Unlabeled, [U-15N]- and [U-13C,U-15N]-labeled apo-IscU samples were produced and purified as described previously.15,18 The elution profile of apo-IscU variants in the size exclusion chromatography using a HiLoad 16/60 Superdex 75 column (GE-Healthcare) did not show any evidence for heterogeneous oligomeric species16 (and data not shown).
NMR spectroscopy
The solvent used for NMR samples contained 20 mM Tris·HCl pH 8.0, 0.5 mM EDTA, 5 mM dithiothreitol, 150 mM NaCl, 7% D2O, 0.7 mM 2,2-dimethyl-2-silapentane-5-sulfonate, and 0.02% sodium azide. A sample of 0.6–0.8 mM [U-15N]-apo-IscU was employed to obtain 2D NMR spectra, while a sample of 1.5–2.0 mM [U-13C,U-15N]-apo-IscU was used to collect 3D NMR spectra. For measuring residual dipolar couplings (RDCs), 12–13 mg/mL Pf1 phage (Cedarlane Labs, Burlington, Ontario, Canada) was added to the [U-15N]-apo-IscU sample.19
Unless specified otherwise, all NMR spectra were acquired at 25 °C with 600 MHz or 800 MHz Varian VNMRS spectrometers equipped with a z-gradient cryogenic probe. We used NMRPipe20 to process raw NMR data and SPARKY21 for data analysis. The following data sets were collected to assign the backbone and side-chain resonances from the samples of [U-13C,U-15N]-apo-IscU(WT) and [U-13C,U-15N]-IscU(D39A): 2D 15N-HSQC, 2D 13C-CT-HSQC, 3D HNCO, 3D CBCA(CO)NH, 3D HNCACB, 3D C(CO)NH, 3D HBHA(CO)NH, 3D HC(CO)NH, 3D HCCH-TOCSY, 3D 15N-NOESY-HSQC, and 3D 13C-NOESY-HSQC. For assigning backbone signals of IscU(D39V), IscU(N90A), IscU(S107A), and IscU(E111A), the following data sets were collected: 2D 15N-HSQC, 3D HNCO, 3D CBCA(CO)NH, and 3D HNCACB. ‘%S’ of IscU variants were calculated from the relative S- and D-state 1H-15N peak intensities from the K128 backbone or the W76 side chain by using the equation: %S = (100×[S])/([S]+[D]). In cases where both measurements could be made, they resulted in similar %S values. 1DNH RDC measurements of IscU(WT) and IscU(D39A) were achieved by collecting 2D IPAP 15N-HSQC spectra22 at 25 °C on a 750 MHz Bruker DMX spectrometer equipped with a cryogenic probe.
15N R1 and R2 relaxation rates and 1H-15N heteronuclear NOEs of IscU variants were measured with a 600 MHz Varian VNMRS spectrometer at 25 °C. The water flip-back scheme using water-selective pulses was employed to prevent saturation of the water resonance as well as to suppress water signals.23 Relaxation delays of 0.1, 0.2, 0.3, 0.4, 0.6, 0.8, and 1.2 s were used to measure R1 relaxation rates, and relaxation delays of 0.01, 0.03, 0.05, 0.07, 0.09, 0.11, and 0.13 s were used to measure R2. For 1H-15N NOE experiments, 1H excitation time was set to 3 s, and peak intensities of the 2D 15N-HSQC spectra collected with and without 1H excitation were compared to obtain 1H-15N NOE data. Two identical experiments were repeated to estimate standard deviations of R1, R2, and 1H-15N NOEs. The rotational correlation time (τc) of each IscU variant was calculated from the average R2/R1 value of the residues 27–33, 43–59, 73–97, and 110–125, which are not located in the flexible loops, with the following equation:24,25 τc≈[6(R2/R1)−7]1/2÷4πυN), where υN is the resonance frequency of 15N (60.775 MHz in our case). The relaxation rate measurements were carried out with the S-state peaks, and because the relaxation times were shorter than the lifetime of the S-state (>1.3 s), contributions from S⇆D exchange are expected to be small.
Structure Determination
To determine the solution structures of apo-IscU(WT) and apo-IscU(D39A), backbone and side-chain signals of IscU(D39A) were manually assigned with SPARKY.21 The resonance assignments of IscU(D39A) were used to discriminating signals from the S-state of IscU(WT) from those of the D state. Subsequently, the peaks of 3D 15N-NOESY-HSQC and 3D 13C-NOESY-HSQC spectra of apo-IscU(WT) and apo-IscU(D39A) were manually picked, and CYANA 3.026 was used to assign them, generate distance constraints, and calculate initial structural models. TALOS+ software27 was used to obtain backbone dihedral angle (φ and ψ) constraints. After an initial structure calculation, all assignments of the two NOESY spectra were manually inspected to re-pick peaks and modify assignments when necessary. 1DNH RDC constraints were included at the structure refinement step. The quality of the final structure was checked with the Protein Structure Validation Software Suite.28 The statistics summarizing quality of the structure, CYANA target functions and violations are summarized in Table 1.
Table 1.
Statistics for the structure determinations of the structured states of E. coli apo-IscU(WT) and apo-IscU(D39A).
| IscU Variants [PDB] | WT [2L4X] | D39A [2KQK] |
|---|---|---|
| Distance constratins | ||
| Intraresidue (i = j) | 218 | 305 |
| Sequential (i–j = 1) | 446 | 559 |
| Medium range (1 < (i–j) ≤ 5) | 416 | 525 |
| Long range (i–j > 5) | 621 | 713 |
| Total | 1701 | 2102 |
| Dihedral angle constraints | ||
| φ | 82 | 77 |
| ψ | 83 | 77 |
| Number of 1DNH RDC constraints | 22 | 16 |
| Number of constraints per residue | 14.9 | 17.9 |
| Number of long range constraints per residue | 4.9 | 5.6 |
| CYANA target function (Å) | 1.00 (± 0.10) | 0.98 (± 0.18) |
| Average number of distance constraints violations per conformer (> 0.2 Å) | 0 | 0 |
| Average number of dihedral angle constraints violations per conformer (°) | 0 | 0 |
| Average r.m.s.d for ordered residuesa to the mean structure (Å) | ||
| Backbone atoms (N, Cα, C’) | 0.5 | 0.4 |
| All atoms | 1.0 | 0.9 |
| Ramachandran plot summary for ordered residuesa from PROCHECK 32 | ||
| Most favoured regions | 91.4 % | 91.1 % |
| Additionally allowed regions | 7.8 % | 8.8 % |
| Generously allowed regions | 0.8 % | 0.1 % |
| Disallowed regions | 0.0 % | 0.0 % |
| PROCHECK raw scorea (φ and ψ/all dihedral angle) | −0.50/−0.75 | −0.47/−0.73 |
| PROCHECK z-scorea (φ and ψ/all dihedral angle) | −1.65/−4.44 | −1.53/−4.32 |
Residues 24–34, 41–58, and 72–127 of IscU(WT) and residues 26–38, 40–62, and 68–27 of IscU(D39A) were considered ordered by the Protein Structure Validation Software Suite.28
Data Deposition
The 3D structural models of apo-IscU(WT) and apo-IscU(D39A) have been deposited in the Protein Data Bank (PDB ID: 2L4X and PDB: 2KQK, respectively), and the related NMR data sets have been deposited at the BioMagResBank (BMRB accession numbers 17282 and 16603, respectively). The chemical shifts and R1, R2, and 1H-15N NOE data for apo-IscU(D39V), apo-IscU(N90A), apo-IscU(S107A), and apo-IscU(E111A) have been deposited in BioMagResBank (BMRB accession numbers, 18359, 18361, 18362, 18360, respectively ).
CD Spectroscopy
The buffer used for circular dichroism (CD) spectroscopy was 20 mM Tris·H2SO4 pH 8.0 containing 0.5 mM TCEP. We used 1-mm pathlength cuvettes. The concentration of apo-IscU was 10–20 µM. The far-UV CD spectra of IscU variants were recorded at 25 °C and 69 °C. Thermal denaturation curves were obtained by following CD signals at 222 nm with increasing the temperature by 3 °C steps from 9 °C to 69 °C. The protein samples were incubated for 5 min at each temperature. After the spectrum at 69 °C was taken, the temperature was lowered to 25 °C, and a spectrum was taken to determine the reversibility of the reaction. The reversibility was found to be on the order of 90 %. Because of the variable secondary structure content of the IscU variants, the final thermal denaturation curves were normalized so that ‘0’ represented the maximal S-state while ‘1’ represented the minimal S- state (Figure 2S of the Supporting Information).
RESULTS
Solution structures of the S-state conformations of apo-IscU(WT) and apo-IscU(D39A)
We overcame the difficulty posed by the heterogeneous conformation of apo-IscU9,15 by taking advantage of the S-state stabilization of apo-IscU(D39A).15 We first assigned the signals of apo-IscU(D39A), whose NMR spectra were much simpler to analyze than those of apo-IscU(WT). We then assigned many of the NMR signals of apo-IscU(WT) on the basis of their similarity to those of apo-IscU(D39A). These assignments gave us starting points for extending the assignments to other signals of apo-IscU(WT) by reference to data from conventional 3D NMR experiments.
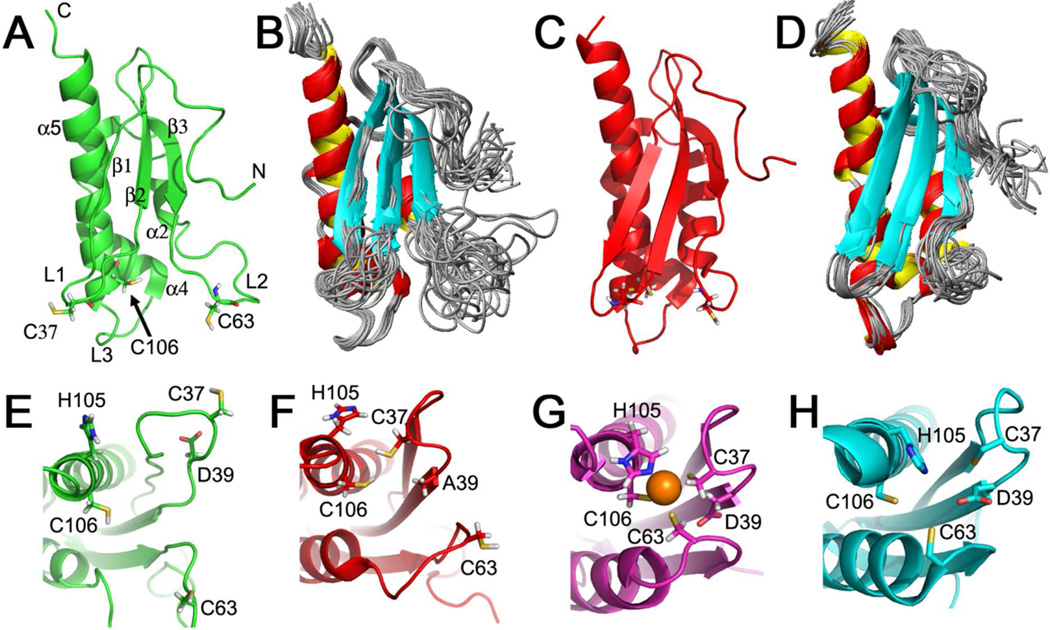
In the end, we obtained structural models for the S-states of both apo-IscU(WT) (Figure 1A,B) and apo-IscU(D39A) (Figure 1C,D). The overall fold of apo-IscU resembles those previously reported for H. influenzae Zn2+-IscU (NMR),9 A. aeolicus holo-IscU (X-ray),11 and E. coli IscU in complex with IscS (X-ray).29 Our results showed that apo-IscU has a more flexible structure than these complexes. The backbone 1H-15N cross-peaks of several residues (C37–K42, T60–S71, E98, L99, and H105) located in loops L1–L3 of apo-IscU(WT) (as denoted in Figure 1A) were not observed in 2D 15N-HSQC spectra. We interpret this result to indicate that the loops adopt multiple conformations that interconvert on a time scale of ms and that exchange broadening renders the peaks unobservable. The flexibility of these loops is reduced somewhat in apo-IscU(D39A) (Figure 1D) as evidenced by the fewer number of unobserved backbone 1H-15N cross-peaks (A36, T60, Y61, G64–A68, and H105). The residues that have been proposed to coordinate an iron-sulfur cluster9,10 (C37, C63, C106 and D39 or H105) are poorly packed in apo-IscU(WT) (Figure 1E), are closer together in apo-IscU(D39A) (Figure 1F), and are tightly packed in the structure of H. influenzae Zn2+-IscU 9 (Figure 1G).
Figure 1.
NMR-based models for the structured forms of apo-IscU variants. (A) Lowest energy conformer of apo-IscU(WT). (B) Overlaid 20 conformers that represent the structure of apo-IscU(WT). (C) Lowest energy conformer of apo-IscU(D39A). (D) Overlaid 20 conformers representing the structure of apo-IscU(D39A). Note that residues 1–14 are disordered, and not shown in this figure. The N- and C-terminals are denoted in panel A, along with the secondary structural elements as assigned previously.9 The loops that include highly-conserved cysteine residues are also assigned as follows: the loop containing C37 is L1(35–38); the loop containing C63 is L2(60–66); the loop containing the L99PPVK103 motif33 and flanking C106 is L3(99–103). The side chains of the three cysteine residues are shown in stick form in panels A and C. Enlargements are shown of the putative iron-sulfur cluster binding residues of (E) apo-IscU(WT) from this study; (F) apo-IscU(D39A) from this study; (G) NMR structure of Zn2+-bound IscU9 [PDB 1R9P]; and (H) X-ray structure of apo-IscU complexed with IscS29 [PDB 3LVL] (only IscU is shown). The side chains of the residues proposed to be involved in cluster coordination9,10 are labeled and shown in stick form. The orange sphere in panel G represents a Zn2+ ion.
Ordering effects of different amino acids at position 39 of IscU
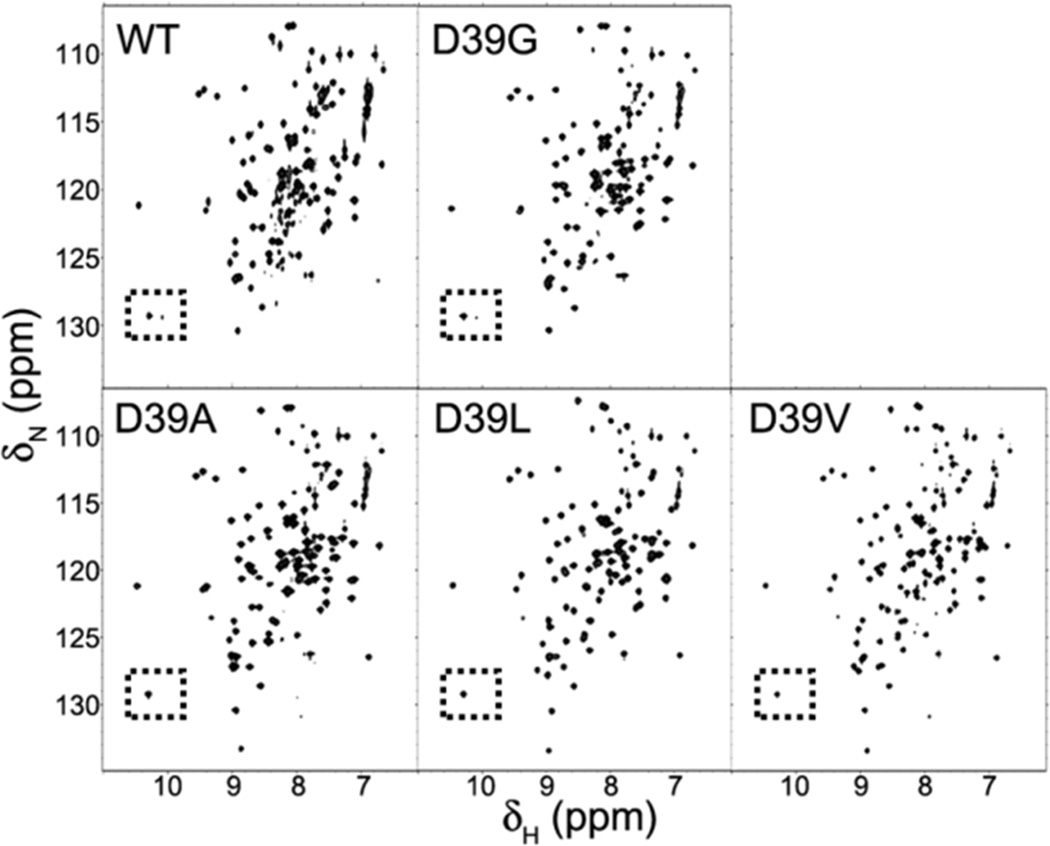
Because the substitution of a charged residue with a hydrophobic residue at position 39 resulted in a more highly ordered S-state, we decided to investigate the effects of inserting amino acids of different hydrophobicity (G, A, L, V) at that site. 2D 15N-HSQC spectra of these variants labeled uniformly with 15N (Figure 2) showed that, whereas the %S of apo-IscU(D39G) was similar to that of apo-IscU(WT), all the hydrophobic substitutions increased %S. The effect is easily seen by comparing the intensities of the 1Hε1-15Nε1 signals from the side chain of the single tryptophan residue (W76) in IscU (the signal on the left is from the S-state, and that on the right is from the D-state)15,16 (boxed areas in Figure 2). We used circular dichroism (CD) spectroscopy as an independent measure of the conformational states of these variants. The far-UV CD spectra of the variants (Figure S1 of the Supporting Information) showed that apo-IscU(WT) and apo-IscU(D39G) are less structured than apo-IscU(D39A), apo-IscU(D39L), or apo-IscU(D39V). We used CD spectroscopy to determine the relative thermal stabilities of S-states of the variants (Figure S2 of the Supporting Information): D39V > D39L > D39A > D39G ≈ WT.
Figure 2.
2D 15N-HSQC NMR spectra of [U-15N]-apo-IscU variants at 25 °C, pH 8.0 and 150 mM NaCl. The relative populations of the S-state and D-state forms of these variants can be gauged from the W76 side-chain signals (boxed; left peak from the S state and right peak from the D state) and by the level of poorly-dispersed peaks at the center of the spectrum from the D state. The spectra of the more structured IscU variants (D39A, D39L, and D39V) lack the D-state peak of the W76 side-chain and several poorly-dispersed D-state peaks at the center of the spectrum.
NMR signal assignments of IscU variants
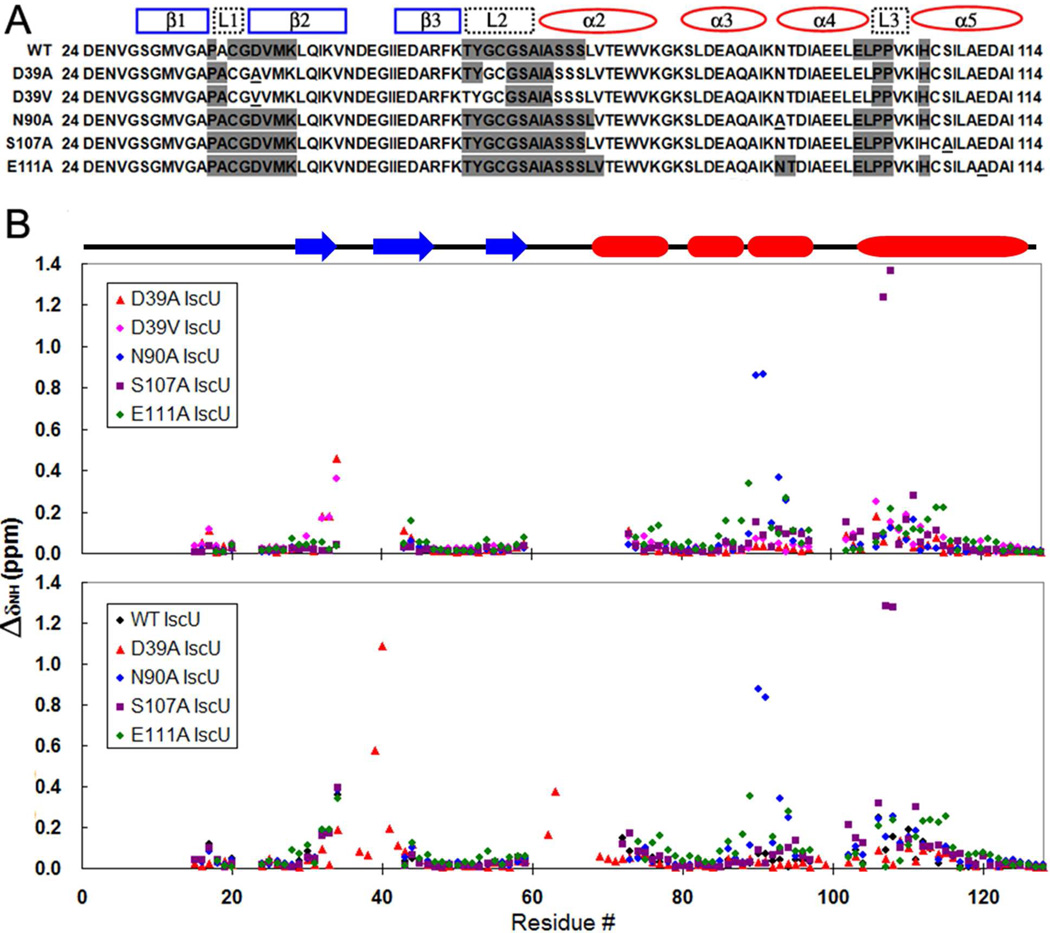
To address the question of how single amino acid substitutions perturb the S⇆D conformational equilibrium of apo-IscU, we assigned the backbone 1H-15N NMR signals for variants with different %S values. This enabled us to determine which residues are involved in ms time-scale dynamics leading to disappearance of their signals (Figure 3A). For variants at position 39, we found a correlation between the number of observable cross peaks from loop residues and the %S (or thermal stability). For example, cross peaks C37–K42, G62, C63, S69–S71, E98, and L99, which were not observed in the spectrum of WT but were found in the spectrum of D39A, were also observable in the spectrum of D39V. However, D39V exhibited additional peaks from T60 and Y61 (Figure 3A).
Figure 3.
Analysis of assigned 1H-15N backbone signals of IscU(WT), IscU(D39A), IscU(D39V), IscU(N90A), IscU(S107A), and IscU(E111A). The secondary structural elements as observed in the structure of IscU(D39A) are marked in both panels (α-helix, red; β-strand, blue). (A) Results for residues D24–I114, the stretch showing the largest chemical shift differences between variants. Residues in this stretch whose NMR signals were not observed in 2D 15N-HSQC spectra are shaded gray. The residue substituted in each IscU variant is underlined. (B) The chemical shift perturbations of IscU(WT) (black), IscU(D39A) (red), IscU(D39V) (pink), IscU(N90A) (blue), IscU(S107A) (purple), and IscU(E111A) (green) referenced to the chemical shift of IscU(WT) (upper panel) or referenced to the chemical shift of IscU(D39V), the most stable variant studied (lower panel). Aggregate 1H and 15N chemical shift perturbations were calculated from the equation: ΔδNH = [(ΔδN/6)2+(ΔδH)2]1/2.
This correlation did not hold, however, for variants at other locations. Although apo-IscU variants N90A, S107A, and E111A exhibited %S greater than WT, their 15N-HSQC spectra lacked the same 1H-15N cross peaks as WT. In some cases, these variants exhibited more dynamic loops than WT. Of the peaks observable in WT, N90A lacked the peak from L72, and E111A lacked signals from L72, V73, N90, and T91. It is clear that the S-states of these variants are stabilized by mechanisms different from those of the residue 39 variants.
The chemical shifts of several IscU variants are shown referenced to the corresponding chemical shifts of apo-IscU(WT) (Figure 3B, upper panel) and to those of apo-IscU(D39V), the most thermally stable variant (Figure 3B, lower panel). As expected, the largest differences are around the sites of the substitutions. In general, the chemical shift differences in other regions are small, suggesting that the overall S-state conformations of IscU variants are similar. Still, it is noteworthy that the chemical shifts of residues located in loops L1 (C37, G38) and L2 (G62, C63), in β-strands β1 (V32–A35) and β2 (M41–Q44), and in helix α5 (C106–A110, D112–I114) deviate noticeably between IscU variants (Figure 3B). For example, substitutions at residue 39 not only affect the functionally-important loops (L1–L3) but also are transmitted to more remote secondary structural elements.
Relaxation parameters and NOEs
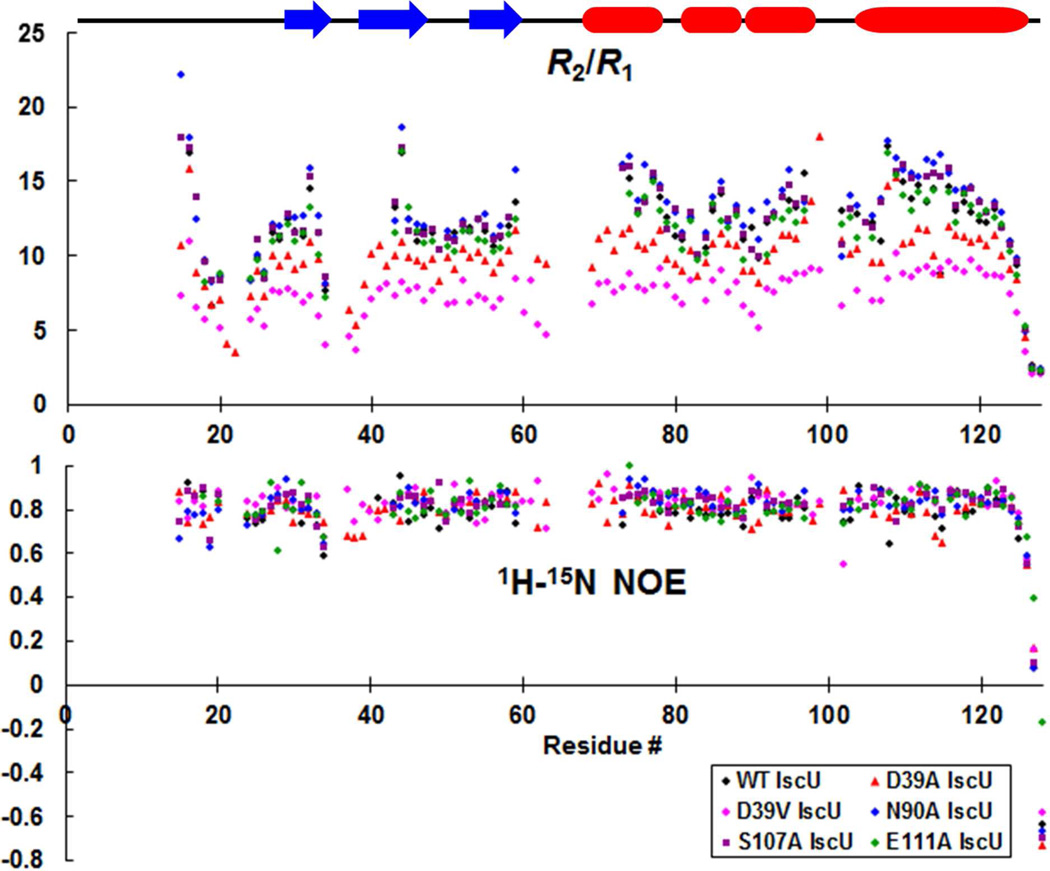
We measured 15N R1, R2 values and 1H-15N heteronuclear NOEs to investigate the dynamic features of several apo-IscU variants (WT, D39A, D39V, N90A, S107A, E111A) (Figure 4). D39V and, to a lesser extent, D39A exhibited overall R2/R1 ratios significantly lower than WT. The R2/R1 ratio of a macromolecule in solution correlates with its rotational correlation time (τc).24,25 From the R2/R1 ratios of residues not in the flexible loop regions (see Experimental Procedures for details), we determined estimates of τc for each apo-IscU variant: N90A, 11.4 ns; S107A, 11.1 ns; WT, 10.9 ns; E111A, 10.6 ns; D39A, 9.8 ns, D39V, 8.3 ns. Notably, the variants with the smallest molecular sizes in solution (D39A and D39V) are the ones with the most stabilized loops. By contrast, the 1H-15N NOEs of the apo-IscU variants were highly similar; this result indicates that the amino acid substitutions that alter %S (or thermal stability) do not affect dynamics on the ps–ns time scale.
Figure 4.
15N R2/R1 relaxation ratios (upper panel) and 1H-15N heteronuclear NOE values (lower panel) of IscU(WT) (black), IscU(D39A) (red), IscU(D39V) (pink), IscU(N90A) (blue), IscU(S107A) (purple), and IscU(E111A) (green). Data were collected at 25 °C with a 600 MHz NMR spectrometer. The errors, which were estimated from the standard deviation of two replicate measurements, are not shown to simplify the figure, but have been deposited along with the chemical shift data in the BioMagResBank. The secondary structural elements (α-helix, red; β-strand, blue) as determined in the structure of IscU(D39A) are marked.
DISSCUSSION
We previously documented that E. coli apo-IscU populates two conformational states that interconvert on a sub-second time scale, one structured (S) state and one dynamically disordered (D).15,16 We also observed that single substitutions of a few highly-conserved residues (D39A, N90A, S107A, and E111A) stabilize the S-state of apo-IscU.16 In the present study, we took advantage of the S-state stabilization of variant D39A as an aid to assigning the S-state signals of WT under conditions where the protein exists as 80% S and 20% D. The 1H, 13C, and 15N NMR spectral assignments of these two proteins led to determination of their solution structures.
Comparison of the two structures shows that the major structural differences center on the three loops (L1–L3), which are more ordered in apo-IscU(D39A) than in apo-IscU(WT). This result prompted us to investigate a series of variants with residues of differing hydrophobicities at position 39. These variants exhibited differences in the dynamics of loop residues, and led us to correlate the degree of rigidity of the loops with the %S (or thermal stability) of the variant. The relative thermal stabilities of the S-states of the apo-IscU variants (D39V > D39L > D39A > D39G ≈ WT) (Figure 2S of the Supporting Information) suggest that a hydrophobic residue at this position favors the S state. The smaller rotational correlation times of D39A and D39V indicate that these apo-IscU variants are more compact than WT. Our structural model of IscU(D39A) indicates that the side chain of the substituted residue (A39) takes part in a hydrophobic cluster (Figure 1F). Apparently the hydrophobic packing energy and the entropic gain from desolvation of residues participating in the cluster overcomes the entropic loss from reduced loop flexibility. The negative charge on Asp39 appears to prevent this region from forming a stable and well-packed structure. Asp39 is a potential cluster ligand30 and has been found to ligate the metal in the X-ray structure of a Zn2+-IscU complex (PDB 2QQ4). On the other hand, H105 was found to be the cluster ligand in the NMR structures of another Zn2+-IscU complex9 and in the X-ray structure of [2Fe-2S]-IscU(D39A).11 If H105 is the fourth ligand, the negative charge on D39 will be countered by the positive charge on the assembled cluster.
The L1–L3 loops of IscU are well-packed in the NMR structure of the Zn2+-IscU complex9 (Figure 1G) and in the X-ray crystal structure of apo-IscU complexed with IscS29 (Figure 1H). We have shown by NMR spectroscopy that the D-state of apo-IscU binds to and is stabilized by the cysteine desulfurase (IscS).16 In the X-ray structure of apo-IscU:IscS,29 crystal packing forces appear to have trapped apo-IscU in the S-state, which in solution becomes populated only during or after Fe-S cluster formation. The recent X-ray structure of the [2Fe-2S]-IscS-IscU(D35A) complex from Archaeoglobus fulgidus30 demonstrated that IscU is in the S-state in this complex. The two conformational states of apo-IscU (S and D) are selected by different proteins involved in Fe-S cluster assembly and delivery, and their existence appears not to be an artifact resulting from the absence of a cofactor or partner protein as has been proposed.31
Our results show that the flexibility of the L1–L3 loops of the S-state of apo-IscU(WT) is dictated by the protein sequence. The highly-conserved and solvent-exposed hydrophobic residues Met31 and Val40, which have been shown to interact with HscB,15 are better packed in IscU(D39A) than in the wild-type protein. HscB interacts preferentially with the S-state of apo-IscU,15 and its physiological role is to bind to holo-IscU, which is stabilized in the S-state. It appears that the interaction between HscB and IscU is stronger when the region containing Met31, Asp39, and Val40 is stably packed.
Our studies of apo-IscU variants (N90A, S107A, E111A) show that the S-state can be stabilized by mechanisms that do not involve the loops. Differences between the chemical shifts of these apo-IscU variants and WT suggest that interactions between helices α3-α4 and helix α5 may be responsible for stabilizing the S-state. Because D39A and D39V also showed perturbed chemical shifts in these regions, L1–L3 loop stabilization may be transmitted to the helices as part of the mechanism for increasing %S.
Supplementary Material
Abbreviations used
- EDTA
2,2',2",2"'-(ethane-1,2-diyldinitrilo)tetraacetic acid
- HSQC
heteronuclear single-quantum correlation
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser enhancement
- TCEP
3,3',3"-phosphanetriyltripropanoic acid
- PLP
pyridoxal-5’-phosphate
- WT
wild type
Footnotes
This work was supported by U.S. National Institutes of Health (NIH) grants R01 GM58667 and U01 GM94622 and was a collaboration with the National Magnetic Resonance Facility at Madison which is supported by NIH grants from the National Center for Research Resources (5P41RR002301-27) and the National Institute of General Medical Sciences (8 P41 GM103399-27).
SUPPORTING INFORMATION AVAILABLE
Far-UV CD spectra of apo-IscU variants (Figure S1), and their thermal stabilities measured with CD spectroscopy (Figure S2). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Beinert H, Holm RH, Munck E. Iron-sulfur clusters: Nature's modular, multipurpose structures. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 4.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 5.Mochel F, Knight MA, Tong WH, Hernandez D, Ayyad K, Taivassalo T, Andersen PM, Singleton A, Rouault TA, Fischbeck KH, Haller RG. Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am. J. Hum. Genet. 2008;82:652–660. doi: 10.1016/j.ajhg.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olsson A, Lind L, Thornell LE, Holmberg M. Myopathy with lactic acidosis is linked to chromosome 12q23.3-24.11 and caused by an intron mutation in the ISCU gene resulting in a splicing defect. Hum. Mol. Genet. 2008;17:1666–1672. doi: 10.1093/hmg/ddn057. [DOI] [PubMed] [Google Scholar]
- 7.Nordin A, Larsson E, Thornell LE, Holmberg M. Tissue-specific splicing of ISCU results in a skeletal muscle phenotype in myopathy with lactic acidosis, while complete loss of ISCU results in early embryonic death in mice. Hum. Genet. 2011;129:371–378. doi: 10.1007/s00439-010-0931-3. [DOI] [PubMed] [Google Scholar]
- 8.Bertini I, Cowan JA, Del Bianco C, Luchinat C, Mansy SS. Thermotoga maritima IscU. Structural characterization and dynamics of a new class of metallochaperone. J. Mol. Biol. 2003;331:907–924. doi: 10.1016/s0022-2836(03)00768-x. [DOI] [PubMed] [Google Scholar]
- 9.Ramelot TA, Cort JR, Goldsmith-Fischman S, Kornhaber GJ, Xiao R, Shastry R, Acton TB, Honig B, Montelione GT, Kennedy MA. Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J. Mol. Biol. 2004;344:567–583. doi: 10.1016/j.jmb.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Oganesyan N, Shin DH, Jancarik J, Yokota H, Kim R, Kim SH. Structural characterization of an iron-sulfur cluster assembly protein IscU in a zinc-bound form. Proteins. 2005;59:875–881. doi: 10.1002/prot.20421. [DOI] [PubMed] [Google Scholar]
- 11.Shimomura Y, Wada K, Fukuyama K, Takahashi Y. The asymmetric trimeric architecture of [2Fe-2S] IscU: implications for its scaffolding during iron-sulfur cluster biosynthesis. J. Mol. Biol. 2008;383:133–143. doi: 10.1016/j.jmb.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Unciuleac MC, Chandramouli K, Naik S, Mayer S, Huynh BH, Johnson MK, Dean DR. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry. 2007;46:6812–6821. doi: 10.1021/bi6026665. [DOI] [PubMed] [Google Scholar]
- 13.Wu G, Mansy SS, Wu Sp SP, Surerus KK, Foster MW, Cowan JA. Characterization of an iron-sulfur cluster assembly protein (ISU1) from. Schizosaccharomyces pombe, Biochemistry. 2002;41:5024–5032. doi: 10.1021/bi016073s. [DOI] [PubMed] [Google Scholar]
- 14.Foster MW, Mansy SS, Hwang J, Penner-Hahn JE, Surerus KK, Cowan JA. A mutant human IscU protein contains a stable [2Fe-2S](2+) center of possible functional significance. J. Am. Chem. Soc. 2000;122:6805–6806. [Google Scholar]
- 15.Kim JH, Füzéry AK, Tonelli M, Ta DT, Westler WM, Vickery LE, Markley JL. Structure and dynamics of the iron-sulfur cluster assembly scaffold protein IscU and its interaction with the cochaperone HscB. Biochemistry. 2009;48:6062–6071. doi: 10.1021/bi9002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JH, Tonelli M, Markley JL. Disordered form of the scaffold protein IscU is the substrate for iron-sulfur cluster assembly on cysteine desulfurase. Proc. Natl. Acad. SciU.SA. 2012;109:454–459. doi: 10.1073/pnas.1114372109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murzin AG. Biochemistry - Metamorphic proteins. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 18.Hoff KG, Silberg JJ, Vickery LE. Interaction of the iron-sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2000;97:7790–7795. doi: 10.1073/pnas.130201997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen MR, Mueller L, Pardi A. Tunable Alignment of Macromolecules by Filamentous Phage Yields Dipolar Coupling Interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 20.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPIPE - A Multidimensional Spectral Processing System Based on UNIX Pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 21.Goddard TD, Kneller DG. Sparky 3. San Francisco: University of California, San Francisco; 2008. [Google Scholar]
- 22.Ottiger M, Delaglio F, Bax A. Measurement of J and Dipolar Couplings from Simplified Two-Dimensional NMR Spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 23.Grzesiek S, Bax A. The Importance of Not Saturating H2O in Protein NMR. Application to Sensitivity Enhancement and NOE measurements. J. Am. Chem. Soc. 1993;115:12593–12594. [Google Scholar]
- 24.Aramini JM, Tubbs JL, Kanugula S, Rossi P, Ertekin A, Maglaqui M, Hamilton K, Ciccosanti CT, Jiang M, Xiao R, Soong TT, Rost B, Acton TB, Everett JK, Pegg AE, Tainer JA, Montelione GT. Structural basis of O6-alkylguanine recognition by a bacterial alkyltransferase-like DNA repair protein. J. Biol. Chem. 2010;285:13736–13741. doi: 10.1074/jbc.M109.093591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kay LE, Torchia DA, Bax A. Backbone Dynamics of Proteins as Studied by 15N Inverse Detected Heteronuclear NMR Spectroscopy: Application to Staphylococcal Nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 26.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 27.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 29.Shi R, Proteau A, Villarroya M, Moukadiri I, Zhang L, Trempe JF, Matte A, Armengod ME, Cygler M. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 2010;8:e1000354. doi: 10.1371/journal.pbio.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marinoni EN, de Oliveira JS, Nicolet Y, Raulfs EC, Amara P, Dean DR, Fontecilla-Camps JC. (IscS-IscU)2 Complex Structures Provide Insights into Fe2S2 Biogenesis and Transfer. Angew. Chem. Int. Ed. 2012;51:1–5. doi: 10.1002/anie.201201708. [DOI] [PubMed] [Google Scholar]
- 31.Prischi F, Pastore C, Carroni M, Iannuzzi C, Adinolfi S, Temussi P, Pastore A. Of the vulnerability of orphan complex proteins: the case study of the E. coli IscU and IscS proteins. Protein Expr. Purif. 2010;73:161–166. doi: 10.1016/j.pep.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK - a program to check the stereochemical quality of protein structures. J. App. Cryst. 1993;26:283–291. [Google Scholar]
- 33.Hoff KG, Cupp-Vickery JR, Vickery LE. Contributions of the LPPVK motif of the iron-sulfur template protein IscU to interactions with the Hsc66-Hsc20 chaperone system. J. Biol. Chem. 2003;278:37582–37589. doi: 10.1074/jbc.M305292200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.