Abstract
Colonization of plant roots by symbiotic arbuscular mycorrhizal fungi frequently leads to the accumulation of several apocarotenoids. The corresponding carotenoid precursors originate from the plastidial 2-C-methyl-d-erythritol 4-phosphate pathway. We have cloned and characterized 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR), catalyzing the first committed step of the pathway, from maize (Zea mays). Functional identification was accomplished by heterologous expression of sequences coding for the mature protein in Escherichia coli. DXR is up-regulated in maize roots during mycorrhization as shown at transcript and protein levels, but is also abundant in leaves and young seedlings. Inspection of sequenced genomes and expressed sequence tag (EST) databases argue for a single-copy DXR gene. Immunolocalization studies in mycorrhizal roots using affinity-purified antibodies revealed a DXR localization in plastids around the main symbiotic structures, the arbuscules. DXR protein accumulation is tightly correlated with arbuscule development. The highest level of DXR protein is reached around maturity and initial senescence of these structures. We further demonstrate the formation of a DXR-containing plastidial network around arbuscules, which is highly interconnected in the mature, functional state of the arbuscules. Our findings imply a functional role of a still unknown nature for the apocarotenoids or their respective carotenoid precursors in the arbuscular life cycle.
Arbuscular mycorrhiza (AM) is a widespread mutual symbiosis between the roots of more than 80% of terrestrial plants and about 150 fungal species. The fungi belong mainly to the recently described order Glomerales in the new phylum Glomeromycota (formerly Glomales within the Zygomycota; Schüssler et al., 2001). The main benefit for both partners is the exchange of nutrients: the fungal microsymbiont supplies mineral nutrients and receives photosynthetic carbohydrates in return (Smith and Read, 1997). Other benefits for the plant partner include an increased resistance to root pathogens (Gianinazzi-Pearson et al., 1996; Cordier et al., 1998) or to drought stress (Augé, 2001). The main symbiotic organ of the interaction is the so-called arbuscule, a highly branched tree-like fungal structure that develops within root cortex cells. The plant host cell undergoes major changes in morphology and metabolism, including the formation of a periarbuscular membrane around the arbuscule to separate it from the host protoplast. With its large surface, the arbuscule is thought to serve as the main symbiotic interface, at least for the exchange of mineral nutrients from the fungus to the plant (Harrison, 1999). Arbuscules are highly dynamic structures that develop and degenerate in a single cell in a period of about 7 to 10 d as estimated for maize (Zea mays) and other cereals (Alexander et al., 1988).
Despite the widespread distribution and ecological significance of the symbiosis, knowledge about signaling compounds or the role of secondary metabolites in the symbiosis is still poor (for review, see Strack et al., 2003). A long-known phenomenon is the accumulation of yellow compounds in the roots of many plants upon mycorrhization (Jones, 1924; Fester et al., 2002a). The respective chromophore component of this so-called “yellow pigment” was isolated and identified as a linear C14 polyenic acid of putative carotenoid origin called mycorradicin (Klingner et al., 1995). Simultaneously, a number of C13 cyclohexenone isoprenoid derivatives were shown to accumulate in mycorrhizal roots of cereals (Maier et al., 1995, 1997), tobacco (Nicotiana tabacum), and tomato (Lycopersicon esculentum; Maier et al., 1999, 2000). Retrobiosynthetic studies suggested that these cyclohexenone compounds are also derived from carotenoids (Maier et al., 1998). These and other results led to the hypothesis that cyclohexenones and mycorradicin are apocarotenoids formed from a common carotenoid precursor by oxidative cleavage (Walter et al., 2000; Fester et al., 2002b). They also indicated that mycorrhizal colonization of roots may be a suitable system to study the regulation of isoprenoid biosynthesis.
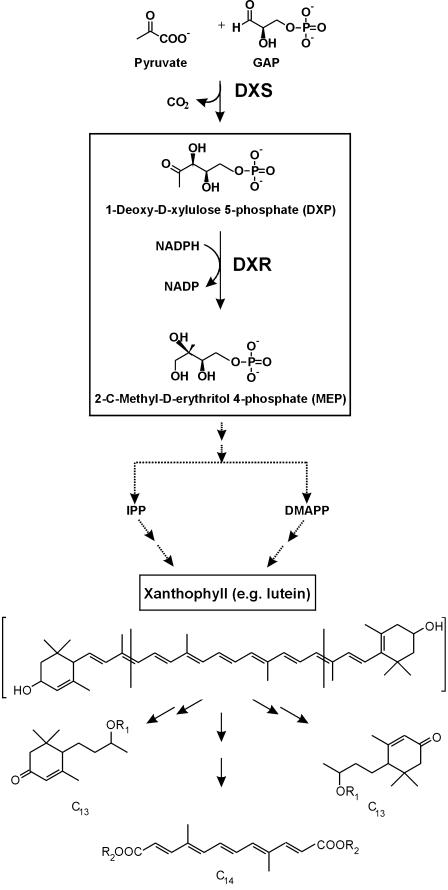
All isoprenoids, including carotenoids, are derived from the ubiquitous C5 building blocks isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). These precursors can be synthesized by two different routes: the classical mevalonate pathway in the cytoplasm or the alternative nonmevalonate pathway in plastids (Rohmer et al., 1993; Schwarz, 1994; Arigoni et al., 1997). Metabolic crosstalk between the pathways can be observed under certain conditions (Hemmerlin et al., 2003). The plastidial pathway, now known as 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway, has been fully elucidated by a combination of biochemical and genomic approaches (Rodríguez-Concepcíon and Boronat, 2002). It provides the precursors for monoterpenes, diterpenes, carotenoids, tocopherols, and the prenyl moiety of chlorophyll (Eisenreich et al., 2001). In the first reaction, catalyzed by 1-deoxy-d-xylulose 5-phosphate synthase (DXS), 1-deoxy-d-xylulose 5-phosphate (DXP) is synthesized from glyceraldehyde 3-P and pyruvate in a condensation/decarboxylation reaction. The DXS reaction is also the first step for thiamine and pyridoxol biosynthesis. Subsequently, DXP is converted into MEP, the first committed metabolite of the pathway, by the NADPH-dependent enzyme DXP reductoisomerase (DXR; see Fig. 1, top part). Genes coding for this enzyme were first identified in bacteria (Kuzuyama et al., 1998b; Takahashi et al., 1998). By now, cDNAs or genes encoding plant DXRs have been cloned from Arabidopsis (Schwender et al., 1999; Carretero-Paulet et al., 2002), peppermint (Mentha x piperita; Lange and Croteau, 1999), Catharanthus roseus (Veau et al., 2000), and tomato (Rodríguez-Concepcíon et al., 2001).
Figure 1.
Proposed pathway to the apocarotenoids C13 cyclohexenones and C14 mycorradicin. The early steps of the MEP pathway to IPP and DMAPP, including the reaction catalyzed by DXR, are highlighted. Subsequently, (oxo) carotenoids (xanthophylls) are synthesized and cleaved to lead to C13 cyclohexenone derivatives (R1 = glycosyl residues) and C14 mycorradicin (R2 = H). Mycorradicin is presumably esterified to cyclohexenone glycosides, forming an insoluble complex (Fester et al., 2002a).
Although DXR catalyzes the first committed step toward IPP and DMAPP formation, its role in the control of metabolic flux through the pathway remains unclear. Results obtained so far show that DXR catalyzes a rate-limiting step in some systems (Veau et al., 2000; Mahmoud and Croteau, 2001), whereas in others, e.g. in ripening fruits of tomato, DXR transcript and protein levels remained unaffected during carotenoid accumulation (Rodríguez-Concepcíon et al., 2001). Our earlier work on apocarotenoid biosynthesis in roots of maize, barley (Hordeum vulgare), and rice (Oryza sativa) indicated a concomitant increase in whole-root transcript levels of DXS and DXR upon mycorrhization preceding the massive accumulation of apocarotenoids (Walter et al., 2000). In the present paper, we report on the cloning and characterization of a maize DXR cDNA. Generation of an affinity-purified DXR antibody by way of recombinant protein and its use in immunolocalization studies allowed a high-resolution tracing of DXR protein accumulation and its localization to plastids during the development and degradation of arbuscules in mycorrhizal maize roots.
RESULTS
Isolation and Characterization of a Maize DXR Clone
To clone a mycorrhiza-induced DXR cDNA, about 360,000 clones of a mycorrhizal maize root cDNA library were screened with a rice expressed sequence tag (EST) sequence (S11168) that showed high homology to known plant DXR sequences. After the first round of purification, 45 clones were subjected to further analysis, which resulted in 16 clones. Restriction mapping of the clones allowed the classification into three different groups, which was also confirmed by partial sequencing. The longest representatives of each group were fully sequenced on both strands. Two groups of clones (ZmDXR64 and ZmDXR88) showed frame-shifting insertions and therefore were not further analyzed. The third group is represented by ZmDXR33 (deposited in GenBank accession no. AJ297566), a 1,678-bp cDNA clone with an open reading frame of 1,419 bp, a 5′-untranslated region of 113 bp, and a 3′-untranslated region of 126 bp. It encodes a 473-amino acid protein with a calculated molecular mass of 51.2 kD and a theoretical pI of 6.7. BLAST analysis of the clone against GenBank revealed highest similarities to other published DXR sequences from plants, e.g. 91% identity to the DXR from rice (GenBank accession no. AF367205), 88% identity to the barley DXR (GenBank accession no. AJ583446), recently identified in our laboratory, or 74% identity to the peppermint DXR (GenBank accession no. AF116825). As the MEP pathway is plastid-located in plants, the sequence was subjected to software analysis for target signal prediction. The deduced ZmDXR protein is predicted to contain a signal sequence for plastid import, with the most probable cleavage site between residues C48 and C49. Interestingly, C49 belongs to a highly conserved CS-X motif (X: A, V, or M; Carretero-Paulet et al., 2002) that is present in all plant DXR sequences at the predicted cleavage site. The plastid targeting signals show only a very low degree of sequence conservation. The predicted mature protein of ZmDXR consists of 423 amino acids with a calculated mass of 45.8 kD and a pI of 5.6.
Clone ZmDXR33 Encodes a Functional DXR
The assignment of the cloned cDNA as DXR was originally based solely on the high degree of sequence identity to published plant DXR sequences. Therefore, a DNA fragment of ZmDXR33 containing only the predicted mature part of the protein was cloned in frame behind a 6×-His tag into pQE30. The recombinant protein was partially purified by metal affinity chromatography. After elution from the Talon column, fractions were checked by SDS-PAGE and western-blot analysis using a monoclonal anti-His-Tag antibody for the presence of recombinant protein. Those fractions containing recombinant 6×-His-ZmDXR were used for enzyme assays with NADPH and enzymatically generated DXP as substrate. The conversion of NADPH to NADP was measured by an absorption decrease at 340 nm. Initial rates were used for calculation of specific enzyme activities (Table I). The oxidation of NADPH was specific for the DXR reaction, as shown by assays omitting the substrate or adding 1 μm fosmidomycin, a specific inhibitor of DXR activity (Kuzuyama et al., 1998a). These results indicate that ZmDXR33 encodes a functional DXR protein.
Table 1.
Enzyme activity of recombinant DXR from maize and Synechococcus leopoliensis
Activity assays were performed with 5 μg of protein of each partially purified fraction of 6×His-ZmDXR33 and S. leopoliensis pQEdxr by monitoring NADPH consumption at 340 nm. Specificity for the DXR reaction is shown by various controls as indicated. n.d., Not detectable.
| Sample | Activity |
|---|---|
| nkat mg-1 | |
| ZmDXR | |
| Standard | 28.02 |
| Denatured protein | n.d. |
| -DXP | n.d. |
| +Fosmidomycin | n.d. |
| SIDXR | |
| Standard | 34.94 |
| +Fosmidomycin | n.d. |
For antibody production, the recombinant 6×-His-ZmDXR was purified from inclusion bodies after overnight induction with isopropyl β-d-thiogalactoside (IPTG). Approximately 1 mg of the protein was used for the production of a polyclonal antiserum in rabbits. Dot-blot assays with serial dilutions of the antigen on a polyvinylidene difluoride (PVDF) membrane showed a sensitivity of the crude antiserum of less than 7 pg of antigen (not shown). However, the antiserum exhibited a slight cross reactivity to several proteins in total protein extracts from maize. For that reason, the antibody was affinity purified on immobilized antigen, resulting in the recognition of a single band with the correct Mr in western-blot analysis.
DXR Is Differentially Expressed in Maize Tissues
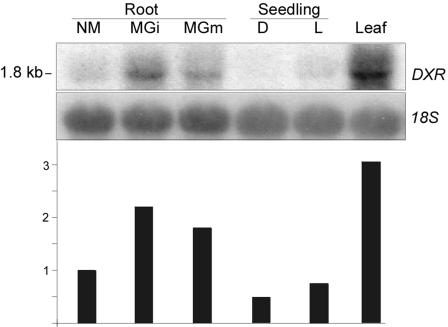
The full-length sequence of ZmDXR33 was used as a probe in a northern-blot analysis with RNA from different maize tissues. The probe recognized a single band at about 1.8 kb. DXR exhibits differential transcript accumulation upon mycorrhization, with an approximately 2-fold increase of DXR mRNA for two different AM fungi compared with nonmycorrhizal roots (Fig. 2). The rate of root colonization was slightly higher for Glomus intraradices (approximately 80%) as compared with Glomus mosseae (approximately 60%), in accordance with a slightly stronger signal in the blot analysis in the first sample. A similar correlation of colonization intensity, including arbuscule frequency as well as metabolite accumulation and transcript abundance of DXR, has been observed in other experiments (not shown) and in earlier investigations with a heterologous DXR probe on wheat (Triticum aestivum) samples (Walter et al., 2000). The highest accumulation of DXR transcripts can be found in leaves (Fig. 2). Lower levels are detectable in dark- and light-grown seedlings. Transcripts from ZmDXR64 and ZmDXR88 (calculated sizes 1.92 and 1.98 kb, respectively) were not detected.
Figure 2.
Northern-blot analysis of maize RNA samples using the DXR probe. Five micrograms total RNA was loaded per lane and was hybridized with a 1.7-kb EcoRI/XhoI fragment of ZmDXR33. Quantification was done by PhosphorImager (Molecular Dynamics, Sunnyvale, CA) after normalization to 18S rRNA as loading control. Quantitative data (bars) are normalized to nonmycorrhizal roots. NM, Nonmycorrhizal; M Gi, mycorrhizal (G. intraradices); M Gm, mycorrhizal (G. mosseae); D, dark grown; L, light grown.
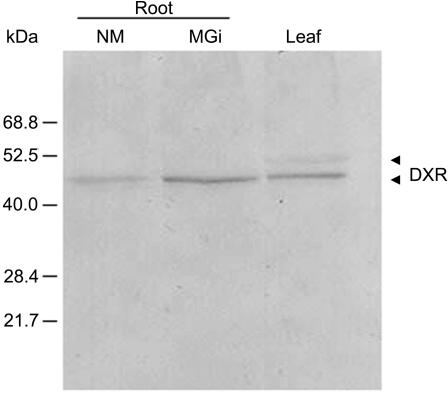
For roots and leaves of maize cv Garant, the results obtained by RNA-blot analysis could be verified on the protein level. Due to its faster and more consistent root colonization, only G. intraradices was used in this and the subsequent experiments. Mycorrhizal roots contain significantly higher amounts of DXR protein than nonmycorrhizal samples (Fig. 3). The level of DXR protein in leaves is similar to that in mycorrhizal roots. However, the antibody detects a faint band with a slightly higher molecular mass matching the calculated value for the preprotein (51.2 kD), which may represent the DXR before plastid import and signal peptidase processing.
Figure 3.
Western-blot analysis of total protein extracts from maize tissues for DXR. Twenty-five micrograms of total protein was loaded per lane, blotted onto PVDF membranes, and immunodecorated with affinity-purified anti-6×-His-ZmDXR antibodies (1:2,000) followed by an anti-rabbit immunoglobulin G (IgG) conjugated with alkaline phosphatase conjugate (1:5,000). NM, Nonmycorrhizal; M Gi, mycorrhizal (G. intraradices).
DXR Is Localized around Arbuscules in Mycorrhizal Maize Roots
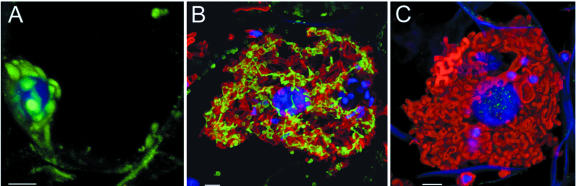
To study the accumulation of DXR on a cell-specific and subcellular level, we used affinity-purified anti-6×-His-ZmDXR antibodies in immunolocalization studies with maize root sections. Figure 4 shows the localization of DXR in maize root cortex cells. In nonmycorrhizal roots, signals for DXR can be found in plastid aggregations in the area around the nucleus and in single distinct plastids located in the cytoplasm close to the cell wall (Fig. 4A). If a cell is colonized by an arbuscule, the abundance and intensity of signals for the DXR protein is markedly increased compared with noncolonized cells. DXR-containing plastids are tightly associated with fungal structures (Fig. 4B). Plastid morphology is also altered: The formation of so-called stromules (stromafilled tubules; Köhler et al., 1997; Tobin, 1997) can be observed (Figs. 4B and 5C). These stromules interconnect plastids, leading to the formation of a plastid network that tightly surrounds the arbuscule. No signal for DXR can be observed when sections are incubated with the preimmune serum, demonstrating the specificity of primary and secondary antibodies (Fig. 4C). The fungal structure is clearly visible after WGA-TRITC staining. In mycorrhizal roots, there is a clear difference between arbusculated and nonarbusculated cells. The latter cells exhibit the same pattern of DXR localization as root cortex cells in nonmycorrhizal plants (not shown).
Figure 4.
Immunolocalization of DXR in maize roots visualized by confocal laser scanning micrographs of root sections. A, Cortex cell of a nonmycorrhizal root after incubation of root section with affinity-purified antiserum against 6×-His-ZmDXR (1:1,000); B, Arbusculated cell (anti-6×-His-ZmDXR, 1:1,000); C, Arbusculated cell after incubation with preimmune serum (1:1,000). The green fluorescence obtained with the anti-DXR/Alexa488 stain is indicative of the presence of DXR, whereas fungal structures are shown in red after staining with wheat germ agglutinin coupled to tetramethyl rhodamine isothiocyanate (WGA-TRITC). DNA is represented by blue color resulting from 4′,6′-diamidine 2-phenyl indole (DAPI) staining. For each micrograph, projections of 25 optical sections are superimposed. Bars = 5 μm.
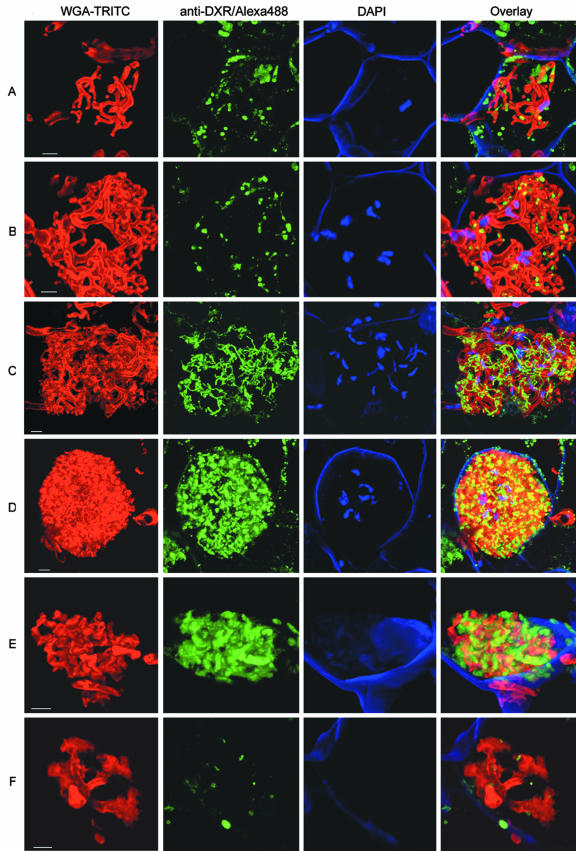
Figure 5.
DXR accumulation in plastid networks during arbuscule development. The confocal laser scanning micrographs of mycorrhizal maize root sections show stages from the life cycle of arbuscules ranging from very young (A) through fully mature (D) to collapsed structures (F). Three types of stainings are shown separately and as an overlay: i, fungal structures, visualized by WGA-TRITC staining (red channel); ii, DXR, detected by anti-6×-His-ZmDXR/anti-rabbit IgG-Alexa488 (green channel); and iii, DNA using DAPI counterstain (blue channel). Projections of 25 optical sections are shown. Bars = 5 μm.
Arbuscular development is characterized by extensive ramification of fungal hyphae to the tree-like functional structure and the eventual collapse of the fine branches during the decay. To visualize possible alterations in DXR localization during arbuscule development, changes in abundance and morphology of DXR-containing plastids were analyzed in dependence of different developmental stages of arbuscules. For easier interpretation, individual stainings by WGA-TRITC (fungal cell wall), anti-6×-His-ZmDXR/Alexa488 (DXR protein), and DAPI (plant and fungal DNA) are shown separately and as an overlay (Fig. 5). The early stages of the arbuscular cycle exhibit few, relatively thick, unbranched hyphae (Fig. 5, A and B). Here, DXR-containing plastids are not yet interconnected and are mostly distributed in the distal areas of the growing arbuscule. In the following developmental stage, arbuscules exhibit a more intense branching of the distal hyphae (Fig. 5C). This is accompanied by the formation of the plastidial network by interconnecting stromules around these arbuscules. The highest density of this network is reached at stages of mature and presumably highly active arbuscules that fill almost completely the cellular space (Fig. 5D). The beginning degeneration of the fungal structure is characterized by collapsing distal hyphae, concomitant with a condensation of the signals for DXR around the arbuscules (Fig. 5E). In this stage, the highest labeling intensity for DXR can be observed. In the latest stage of the arbuscule life cycle, the distal branches have nearly completely collapsed and no signal for DXR-containing plastids can be observed (Fig. 5F). Overall, arbuscule development in a root cortex cell is accompanied by a strong increase in plastid and DXR protein abundance. Furthermore, the formation and degeneration of the plastidial network is tightly associated with the arbuscular life cycle.
DISCUSSION
The Genes Encoding the Early Steps of the MEP Pathway Are Organized and Regulated Differently
As part of our ongoing work on the regulation of MEP pathway enzymes and genes during mycorrhization, we have now analyzed the DXR as the second reaction but the first committed step of the pathway. Excluding two cDNAs with incomplete open reading frames probably originating from pseudogenes or splicing errors, only a single type of DXR cDNA (ZmDXR33) was found in maize. A search in the available EST collections of maize from various tissues (about 360,000 sequences in ZmDB as of October 2003, Dong et al., 2003) afforded only sequences that matched exactly ZmDXR33 but not ZmDXR64 or ZmDXR88 sequences. Inspection of the published rice genome also argues for the single-copy nature of DXR. Earlier analyses of the Arabidopsis genome have indicated a single DXR gene as well (Rodríguez-Concepcíon and Boronat, 2002; Lange and Ghassemian, 2003). Furthermore, the gene-specific probe used in the RNA-blot analysis detected only a single band (Fig. 2), demonstrating high transcript levels in mycorrhizal roots and photosynthetic leaves. These results lead to the conclusion that a single DXR copy is active in all physiological situations that were investigated and that there is no regulation via different isogenes.
In contrast, our recent work on DXS regulation has identified two distantly related genes and transcripts for this transketolase-like enzyme displaying differential and largely complementary expression patterns (Walter et al., 2002). DXS1 expression is correlated with synthesis of primary isoprenoids during housekeeping functions like photosynthesis, whereas DXS2 transcript levels are elevated in a number of situations of secondary isoprenoid production, including apocarotenoid biosynthesis in mycorrhizal roots and monoterpene volatile formation in peppermint species. The conservation of amino acid sequences for each of the mature proteins of DXR, DXS1, and DXS2 between plant species is extremely high (up to 95%), but is rather low between DXS1 and DXS2 (about 70%), arguing for an early separation of DXS1 and DXS2 and a possible coevolution of the three genes (Carretero-Paulet et al., 2002; Walter et al., 2002).
As it catalyzes the first committed reaction, DXR is a potential regulatory step in the MEP pathway. However, the possible role of DXR as a flux-controlling step has been addressed in only a few experimental systems. In mycorrhizal roots, a concomitant induction of DXS and DXR by AM fungal colonization has been shown previously at the transcript level for several cereals using heterologous probes (Walter et al., 2000). We can confirm DXR induction now with a homologous probe for the maize DXR at the transcript level (Fig. 2), and were also able to demonstrate an increase in extractable DXR protein level in roots upon mycorrhization (Fig. 3). The DXR cDNA probe also detected high levels of DXR transcripts in leaves in contrast to a maize DXS2 probe (Walter et al., 2002). This corroborates again the notion that a single DXR gene is activated in photosynthetic leaves and mycorrhizal roots. High levels of DXR transcripts in leaves are not reflected in similarly high amounts of DXR protein. This, together with the detection of the putative preprotein form of DXR, could be seen as a hint for a possible higher protein turnover in leaves compared with roots.
In Arabidopsis, there is a direct light regulation and a developmental modulation of DXR, as shown by RNA-blot analysis and promoter::β-glucuronidase fusion experiments (Carretero-Paulet et al., 2002). DXR is strongly up-regulated upon induction of monoterpenoid indole alkaloid production in cell cultures of C. roseus (Veau et al., 2000). Expression of a homologous DXR cDNA in sense orientation in transgenic peppermint led to variable phenotypes caused by overexpression or cosuppression outcomes (Mahmoud and Croteau, 2001). Transgenic plants with constitutively elevated DXR transcript levels and enzyme activity accumulated substantially more essential oil (about 50% yield increase), with no change in monoterpene composition compared with wild type. The only system of carotenoid synthesis reported to not exhibit regulation at the level of DXR is the ripening tomato fruit. Constitutive levels of DXR transcripts and protein were found at the mature green, orange, and red ripe stages (Rodríguez-Concepcíon et al., 2001). Even in the phase of strong carotenoid accumulation in the fruits, no up-regulation of DXR transcript or protein was detectable. In contrast, DXS transcript levels were markedly elevated at the orange and red ripe stages (Lois et al., 2000).
DXR Accumulation and Plastid Proliferation Correlates with Arbuscule Development
Total organ analyses such as RNA blots or immunoblots underestimate the extent of transcript or protein accumulation in physiological situations that are limited to few single cells like colonization by arbuscular mycorrhizal fungi. Therefore, histological methods are required to obtain more detailed results about abundance and distribution of transcripts and/or proteins on a cellular or subcellular level. For this reason, we used DXR antibodies to localize the protein in sections of maize roots. In nonmycorrhizal roots and in noncolonized cells of mycorrhizal roots, only a few DXR-containing plastids could be found that were anisotropically distributed or were located adjacent to the cell nucleus (Fig. 4). These results are in agreement with the findings of Fester et al. (2001) in transgenic tobacco roots showing plastid-targeted green fluorescent protein (GFP) in a ring-shaped area around the nucleus of noncolonized cells. However, the abundance of GFP-labeled plastids in nonmycorrhizal tobacco root cells seems to be higher than the number of DXR-labeled plastids in the respective maize root cells. This might represent a difference between the two plant species under investigation. It is tempting to speculate that not all plastids in root cells are producing IPP and DMAPP, so that only the plastid population with an active MEP pathway can be detected using DXR as a marker.
Upon mycorrhization, abundance and distribution of DXR-labeled plastids change significantly in maize root cortex cells. With the beginning colonization of the cell, plastids start to multiply and to aggregate until, finally, a dense network around the arbuscule is formed (Fig. 5). Similar arbuscule-located networks were observed by Fester et al. (2001) in transgenic tobacco using GFP. Our results demonstrate the occurrence of such plastidial network formation now also for a monocot, and they show a close correlation of MEP pathway activity and plastid proliferation in mycorrhizal roots. It would be interesting to investigate mycorrhizal hosts from lower phylogenetic groups such as ferns or mosses for the presence of such networks or to investigate other forms of mycorrhizal symbioses like ericoid mycorrhizas. As plastids are sites of biosynthesis for a number of essential compounds (e.g. fatty acids, purines, and pyrimidines) and of important processes like nitrogen assimilation, it can be assumed that the close proximity of these plastids with arbuscules is necessary for a functioning symbiosis. The plastid multiplication is known to be accompanied by many other changes in cell morphology upon colonization of the host cell (Gianinazzi-Pearson, 1996).
Arbuscule development reportedly occurs in a time span of 7 to 10 d in cereals (Alexander et al., 1988). As there is no molecular marker available yet for a functioning symbiosis in maize, the different developmental stages of the fungal structure are assigned using morphological features like degree of branching of hyphae, and size and diameter of the hyphal branches (Kinden and Brown, 1975, 1976). Here, we show a tight correlation between arbuscule development and the formation of the DXR-containing plastid network, pointing to a highly regulated exchange of compounds and/or signals between the two partners. The peak of DXR accumulation around arbuscules is reached at a stage where the fungal structure is just beyond the apparent peak of maturity and functionality, characterized by a maximum in hyphal branching. However, there are already strong signals at the actual peak of arbuscule maturity. Moreover, in the later stage, the antibody might also detect, in part, residual inactive DXR protein, which has not yet undergone turnover. The timing of DXR maximal occurrence around maturity and initial degeneration of arbuscules appears to be slightly shifted to the later stages as compared with the appearance and disappearance of the mycorrhiza-specific phosphate transporter MtPT4 in roots of Medicago truncatula colonized by Glomus versiforme as detected by immunolocalization (Harrison et al., 2002), and to the presence of a H+-ATPase as shown by activity staining (Gianinazzi-Pearson et al., 1991). However, additional experiments are required to substantiate a true difference. The fate of DXR-containing plastids after the collapse of arbuscular structures remains unclear. In mycorrhizal transgenic tobacco roots, GFP-containing plastid networks are degraded during arbuscule degeneration (Fester et al., 2001). In maize, we could observe a diminishing staining for DXR in the late stages of the arbuscular cycle arguing for a similar disappearance of these structures.
The Role of DXR Induction and Apocarotenoid Accumulation in Mycorrhizal Roots Is Still Enigmatic
In the apocarotenoid biosynthetic pathway, we have now characterized the MEP pathway genes DXS2 (Walter et al., 2002) and DXR (this paper) as being affected by mycorrhization. Further on in the pathway, the phytoene desaturase (PDS) step within the carotenoid-specific route also appears to be upregulated, as indicated by PDS promoter activity specifically in colonized roots of tobacco (Fester et al., 2002b). Therefore, the whole route to apocarotenoids may be up-regulated in arbuscule-containing cells.
The temporal and spatial correlation of DXR accumulation with proteins having a defined role in arbuscule function (MtPT4 and H+-ATPase) is in agreement with a postulated but as yet unknown role of DXR and (apo) carotenoids in arbuscule physiology. It is still an open question whether carotenoids or their apocarotenoid cleavage products are the functional molecules. A functional significance for the accumulating end products seems rather unlikely because they are found in osmiophilic droplets in the cytoplasm or in the vacuole of cells harboring degenerating or collapsed arbuscules (Fester et al., 2002a). During attempts to detect and identify carotenoids in mycorrhizal roots, only very low levels of ζ-carotene could be found (Fester et al., 2002a, 2002b), suggesting a high flux rate in the carotenoid pathway. Only upon blocking the PDS reaction with norflurazone, could measurable levels of phytoene be identified in several plant species (Fester et al., 2002b). The view of a function for carotenoids in mycorrhizal structures is supported by results obtained with two maize mutants deficient in carotenoid biosynthesis that showed reduced numbers of fungal structures in their roots (Fester et al., 2002b). However, because these mutants are also compromised in photosynthesis, a secondary effect on mycorrhization cannot be excluded.
Carotenoids are potent general antioxidants and they might play a role in the detoxification of reactive oxygen species generated during the life cycle of arbuscules (Salzer et al., 1999). They can also directly protect membranes from oxidative damage or profoundly affect the rigidity of membranes (Hara et al., 1999; Sujak et al., 1999). This feature might be important for a correct function of the periarbuscular membrane. Perhaps carotenoids or their cleavage products are simply essential for the biogenesis of mycorrhiza-related plastids to properly carry out the various reactions located exclusively within these unique organelles. One example mentioned already is plant fatty acid biosynthesis, which is still unexplored in the context of mycorrhization. Gene suppression experiments of carotenoid biosynthetic genes using RNAi approaches are expected to provide initial clues toward the function of these compounds, whose end products often accumulate in such massive amounts.
MATERIALS AND METHODS
Plant Cultivation and Fungal Inoculation
Maize (Zea mays cv Garant) was grown in expanded clay (Lecaton, 2–5 μm particle size; Fibo Exclay, Pinneberg, Germany) in 250-mL plastic pots under a 16-h light/8-h dark regime in a phytochamber. Fertilization was done once per week using Long Ashton solution with 10% of the original phosphate content. Fungal inocula of Glomus intraradices Schenk and Smith (isolate 49; provided by Henning von Alten (Institut für Pflanzenkrankheiten und Pflanzenschutz, Universität Hannover, Hannover, Germany) or of Glomus mosseae (Nicolson and Gerdemann) Gerd. and Trappe (obtained from Biorize, Dijon, France) consisted of propagules in expanded clay that were enriched by previous cocultivation with leek (Allium porrum) plants. Mycorrhization was achieved by cocultivation of plants with the inoculum mixed with sterile expanded clay (1:4, v/v) for at least 4 weeks. Rates of colonization were estimated after staining roots with trypan blue according to standard procedures as described by Maier et al. (1995).
cDNA Isolation and Sequence Analysis
A cDNA library prepared from mycorrhizal maize roots (cv dwarf1 colonized by G. intraradices) in the λ vector Uni-ZAP XR (Stratagene, La Jolla, CA) was screened with a 1.8-kb NotI/SalI fragment of rice (Oryza sativa) EST S11168 (GenBank accession no. D46469, courtesy of Takuji Sasaki, MAAF DNA Bank, Tsukuba, Ibaraki, Japan) as probe. After two rounds of purification, positive clones were subjected to in vivo excision according to the manufacturer's instructions. The clones were further characterized by restriction mapping and partial sequencing (Medigenomix, Martinsried, Germany). The longest clones were fully sequenced on both strands.
Sequence analysis was performed using the DNAstar LaserGene (DNAstar, Madison, WI) and BioEdit (Hall, 1999) software packages. Plastid-targeting sequences and putative cleavage sites were predicted using ChloroP V1.1 (http://www.cbs.dtu.dk/services/ChloroP/; Emanuelsson et al., 1999).
Heterologous Expression of ZmDXR33
The coding part of the ZmDXR33 cDNA excluding the predicted plastidtransit peptide was amplified using the primers KpnDXR (5′-CAGGTACCCAACAGGCTCCACC-3′) and PstDXR (5′-CACTGCAGTTATGCAGGGACAGG-3′) to create a KpnI-site at the 5′ end and a PstI-site at the 3′ end. The resulting amplicon was subcloned into pGEM T Easy (Promega, Madison, WI) and was subsequently cloned in-frame behind an N-terminal 6×-His-tag into the KpnI/PstI-sites of pQE30 (Qiagen, Hilden, Germany), thus creating pQEdxr. Heterologous expression was carried out in the Escherichia coli M15 pREP4 strain (Qiagen). For overexpression, 800 mL of M15 pREP4 pQEdxr were induced with 1 mm IPTG at 37°C overnight. After harvest, bacterial cell pellets were resuspended in 100 mm Tris HCl, pH 7.5, 300 mm NaCl, and 1 mm β-mercaptoethanol. The suspension was treated with ultrasound (two 30-s bursts, 60% duty cycle). The clarified supernatant was discarded and the remaining pellet was resuspended in 8 m urea, 10 mm Tris HCl, and 100 mm NaH2PO4, pH 8.0. After another round of ultrasound treatment and centrifugation, the supernatant was loaded onto a 2-mL Ni-NTA agarose column (Qiagen) and was washed with 30 mm imidazole. The recombinant protein was eluted with 250 mm imidazole. Elution fractions were pooled, concentrated, and approximately 1 mg of recombinant protein was used for the production of a polyclonal antiserum in rabbits (Eurogentec, Seraing, Belgium). The obtained antiserum was affinity-purified by absorption to immobilized antigen and elution of the specific antibody with an acidic Gly buffer (0.1 m Gly, pH 2.5).
For functional heterologous expression of ZmDXR33, 2.4 L of culture of E. coli M15 pREP4 pQEdxr cells were induced with 0.2 mm IPTG for 6 h at 30°C. After harvest, cells were resuspended on ice in 100 mm Tris HCl, pH 7.5, 300 mm NaCl, 2.5 mm imidazol, 1 mm β-mercaptoethanol, and 10% (w/v) glycerol. Cells were disrupted by ultrasound (3 × 45 s, 60% duty cycle, on ice) and insoluble material was pelleted by centrifugation (20,000g at 4°C for 15 min). The supernatant was incubated with 2 mL of 50% (v/v) Talon matrix (Clontech, Palo Alto, CA) at 4°C overnight with slight agitation. Column chromatography was performed according to the manufacturer's instructions, with the recombinant protein eluting in 150 mm imidazole.
RNA and Immunoblot Analysis
RNA from maize tissues was isolated using a guanidinium thiocyanate-based method. Briefly, ground tissue was suspended in extraction buffer (4 m guanidinium thiocyanate, 20 mm EDTA, 20 mm MES, 0.5% [w/v] N-lauryl sarcosine, 5% [w/v] polyvinylpolypyrrolidone, and 1% [v/v] β-mercaptoethanol, pH 7) and was subsequently subjected to a phenol/chloroform treatment. After phase separation, nucleic acids were ethanol-precipitated, redissolved, and RNA was selectively precipitated using 2 m LiCl. Residual polysaccharides were removed by heating the redissolved RNA for 5 min to 65°C, followed by 5 min incubation on ice; polysaccharides were then pelleted by centrifugation at 20,000g. Total RNA was separated on a denaturing agarose gel according to Sambrook et al. (1989). After separation, the RNA was blotted onto a Hybond N membrane (Amersham Biosciences, Piscataway, NJ) by a reverse capillary blot method using an alkaline transfer buffer (10 mm NaOH and 5× SSC). The blots were hybridized with an [α32P]dATP-labeled 1.7-kb EcoRI/XhoI fragment of ZmDXR33 in 7% (w/v) SDS, 250 mm NaPi, pH 7.0, 250 mm NaCl, and 1 mm EDTA at 60°C overnight. Final washes were done in 0.5× SSC and 0.1% (w/v) SDS at 65°C. Blots were exposed to films or PhosphorImager (Molecular Dynamics) screens. Radioactivity was quantified using a PhosphorImager (Storm 860; Molecular Dynamics) and the ImageQuant software (Molecular Dynamics).
Total protein was extracted from maize tissues by a phenol-based method (Meyer et al., 1988). Protein pellets were redissolved in sample buffer (68 mm Tris HCl, pH 6.8, 2% [w/v] SDS, 0.7 m β-mercaptoethanol, and 10% [w/v] glycerol) and protein content was determined according to Esen (1978). Equal amounts of protein were loaded onto SDS-PAGE gels and were electrophoretically separated. Protein was subsequently blotted onto a PVDF membrane (SequiBlot; Bio-Rad, Hercules, CA) using Towbin's buffer (25 mm Tris and 192 mm Gly) with 20% (w/v) methanol. After blocking with 5% (w/v) fat-free dry milk (Bio-Rad) in Tris-buffered saline (TBS; 100 mm Tris HCl, pH 7.5, and 150 mm NaCl), the membrane was incubated overnight with the primary antibody (anti-6×-His-ZmDXR or anti-His-tag [Novagen, Madison, WI]; 1:2,000 in 5% [w/v] dry milk/TBS). Detection was performed using anti-rabbit IgG alkaline phosphatase conjugate (Chemicon, Temecula, CA; 1:5,000 in TBS) or anti-mouse IgG alkaline phosphatase conjugate (Chemicon; 1:5,000 in TBS) according to the manufacturer's instructions with p-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate as substrates.
DXR Activity Assays
Substrate for DXR reactions (DXP) was synthesized according to Miller et al. (2000) in an overnight production assay with the purified recombinant DXS from Synechococcus leopoliensis (clone E. coli TG1 pQEdxs, a kind gift of Wolfgang Zimmer, Garmisch-Partenkirchen, Germany). Assay mixtures contained 50 mm Tris HCl, pH 7.5, 1 mm MnCl2, 5 mm β-mercaptoethanol, 20- to 50-μL aliquots of DXP, and aliquots of recombinant DXR protein fractions. Reactions were started by adding 0.125 mm NADPH and they were monitored on a photometer at 340 nm for 5 min. Positive control reactions were performed with purified recombinant DXR from S. leopoliensis (TG1 pQEdxr, a kind gift of Wolfgang Zimmer, Garmisch-Partenkirchen). Reaction rates were determined by applying linear fits to the data.
Immunolocalization of ZmDXR33
Freshly harvested roots of 4- to 6-week-old maize plants were immediately fixed in 3% (w/v) paraformaldehyde/0.1% (v/v) Triton X-100 in phosphate-buffered saline (PBS; 135 mm NaCl, 3 mm KCl, 1.5 mm KH2PO4, and 8 mm Na2HPO4, pH 7.1). After vacuum infiltration, fixation was performed for 2 h at room temperature. After dehydration of the samples in a graded series of ethanol, the material was embedded in polyethylene glycol (PEG; mixture of PEG 1500 and PEG 4000, 2:1, v/v) and was cut as described by Hause et al. (2002). Sections of 20 μm were put into tissue culture inserts with an 8-μm mesh (Nunc, Wiesbaden, Germany) for facilitated transfer between incubation solutions. Plant cell walls were digested with 1% (w/v) cellulase and 0.1% (w/v) pectolyase in PBS for 15 min at room temperature. After blocking the sections with 5% (w/v) bovine serum albumin in PBS for 30 min, primary antibody incubation (anti-6×-His-ZmDXR33; 1:1,000 in 5% [w/v] bovine serum albumin/PBS) was performed overnight at 4°C. An anti-rabbit IgG-Alexa488 (Molecular Probes, Eugene, OR) conjugate was used as secondary antibody (1:500 in PBS). Fungal structures were visualized by staining with 0.05 mg mL–1 WGA-TRITC (Molecular Probes) in PBS for 30 min at room temperature. After counterstaining with DAPI (10 μg mL–1 in PBS for 15 min at room temperature), the sections were transferred onto poly-l-lysine-coated slides and were covered with antifading agent (Slow Fade Antifade kit; Molecular Probes). Analysis was performed on a confocal laser scanning microscope (LSM 510 Meta; Zeiss, Jena, Germany) in multitrack mode using the 351-nm (DAPI), 488-nm (Alexa488), and 543-nm (TRITC) laser lines, respectively, for excitation. Series of optical sections (z series) were acquired by scanning 25 sections with a distance of 0.4 to 0.7 μm on the z axis. Image analysis and z series projections were done with the LSM software and the LSM Viewer program (Zeiss).
Acknowledgments
We thank Takuji Sasaki (MAAF DNA Bank) for the rice EST clone S11168 and Wolfgang Zimmer (Garmisch-Partenkirchen) for E. coli TG1 pQEdxs and TG1 pQEdxr. Fosmidomycin was a kind gift of Wilhelm Boland (Jena, Germany). We also thank Kerstin Manke for excellent technical assistance.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.032342.
This work was supported by the Deutsche Forschungsgemeinschaft within the priority program (Schwerpunktprogramm) 1084 Molecular Basics of Mycorrhizal Symbioses (grant nos. Wa536/3–1 and Wa536/3–2 to M.H.W.).
References
- Alexander T, Meier R, Toth R, Weber HC (1988) Dynamics of arbuscule development and degeneration in mycorrhizas of Triticum aestivum L. and Avena sativa L. with reference to Zea mays L. New Phytol 110: 363–370 [Google Scholar]
- Arigoni D, Sagner S, Latzel C, Eisenreich W, Bacher A, Zenk MH (1997) Terpenoid biosynthesis from 1-deoxy-d-xylulose in higher plants by intramolecular skeletal rearrangement. Proc Natl Acad Sci USA 94: 10600–10605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augé RM (2001) Water relations, drought and VA mycorrhizal symbiosis. Mycorrhiza 11: 3–42 [Google Scholar]
- Carretero-Paulet L, Ahumada I, Cunillera N, Rodríguez-Concepcíon M, Ferrer A, Boronat A, Campos N (2002) Expression and molecular analysis of the Arabidopsis DXR gene encoding 1-deoxy-d-xylulose 5-phosphate reductoisomerase, the first committed enzyme of the 2-C-methyl-d-erythritol 4-phosphate pathway. Plant Physiol 129: 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordier C, Pozo MJ, Barea JM, Gianinazzi S, Gianinazzi-Pearson V (1998) Cell defense responses associated with localized and systemic resistance to Phytophtora parasitica induced in tomato by an arbuscular mycorrhizal fungus. Mol Plant-Microbe Interact 11: 1017–1028 [Google Scholar]
- Dong Q, Roy L, Freeling M, Walbot V, Brendel V (2003) ZmDB, an integrated database for maize genome research. Nucleic Acids Res 31: 244–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich W, Rohdich F, Bacher A (2001) Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci 6: 78–84 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen A (1978) A simple method for quantitative, semiquantitative and qualitative assay of protein. Anal Biochem 89: 264–273 [DOI] [PubMed] [Google Scholar]
- Fester T, Hause B, Schmidt D, Halfmann K, Schmidt J, Wray V, Hause G, Strack D (2002a) Occurrence and localization of apocarotenoids in arbuscular mycorrhizal plant roots. Plant Cell Physiol 43: 256–265 [DOI] [PubMed] [Google Scholar]
- Fester T, Schmidt D, Lohse S, Walter MH, Giuliano G, Bramley PM, Fraser PD, Hause B, Strack D (2002b) Stimulation of carotenoid metabolism in arbuscular mycorrhizal roots. Planta 216: 148–154 [DOI] [PubMed] [Google Scholar]
- Fester T, Strack D, Hause B (2001) Reorganization of tobacco root plastids during arbuscule development. Planta 213: 864–868 [DOI] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V (1996) Plant cell responses to arbuscular mycorrhizal fungi: getting to the roots of the symbiosis. Plant Cell 8: 1871–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V, Dumas-Gaudot E, Gollotte A, Tahiri-Alaoui A, Gianinazzi S (1996) Cellular and molecular defence-related responses to invasion by arbuscular mycorrhizal fungi. New Phytol 133: 45–57 [Google Scholar]
- Gianinazzi-Pearson V, Smith SE, Gianinazzi S, Smith FA (1991) Enzymatic studies on the metabolism of vesicular-arbuscular mycorrhizas. New Phytol 117: 61–74 [Google Scholar]
- Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Hara M, Yuan HQ, Yang Q, Hoshino T, Yokoyama A, Miyake J (1999) Stabilization of liposomal membranes by thermozeaxanthins: carotenoidglucose esters. Biochim Biophys Acta 1461: 147–154 [DOI] [PubMed] [Google Scholar]
- Harrison MJ (1999) Biotrophic interfaces and nutrient transport in plant/fungal symbioses. J Exp Bot 50: 1013–1022 [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Maier W, Miersch O, Kramell R, Strack D (2002) Induction of jasmonate biosynthesis in arbuscular mycorrhizal barley roots. Plant Physiol 130: 1213–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerlin A, Hoeffler JF, Meyer O, Tritsch D, Kagan IA, Grosdemange-Billiard C, Rohmer M, Bach TJ (2003) Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J Biol Chem 278: 26666–26676 [DOI] [PubMed] [Google Scholar]
- Jones FR (1924) A mycorrhizal fungus in the roots of legumes and some other plants. J Agric Res 29: 459–470 [Google Scholar]
- Kinden DA, Brown MF (1975) Electron microscopy of vesicular-arbuscular mycorrhizae of yellow poplar: host-endophyte interactions during arbuscule development Can J Microbiol 21: 1930–1939 [DOI] [PubMed] [Google Scholar]
- Kinden DA, Brown MF (1976) Electron microscopy of vesicular-arbuscular mycorrhizae of yellow poplar: host-endophyte interactions during arbuscule deterioration. Can J Microbiol 22: 64–75 [DOI] [PubMed] [Google Scholar]
- Klingner A, Bothe H, Wray V, Marner FJ (1995) Identification of a yellow pigment formed in maize roots upon mycorrhizal colonization. Phytochemistry 38: 53–55 [Google Scholar]
- Köhler RH, Cao J, Zipfel WR, Web WW, Hanson MR (1997) Exchange of protein molecules through connections between higher plant plastids. Science 276: 2039–2042 [DOI] [PubMed] [Google Scholar]
- Kuzuyama T, Shimizu T, Takahashi S, Seto H (1998a) Fosmidomycin, a specific inhibitor of 1-deoxy-d-xylulose 5-phosphate reductoisomerase in the nonmevalonate pathway for terpenoid biosynthesis. Tetrahedron Lett 39: 7913–7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuyama T, Takahashi S, Watanabe H, Seto H (1998b) Direct formation of 2-C-methyl-d-erythritol 4-phosphate from 1-deoxy-d-xylulose 5-phosphate by 1-deoxy-d-xylulose 5-phosphate reductoisomerase, a new enzyme in the nonmevalonate pathway to isopentenyl diphosphate. Tetrahedron Lett 39: 4509–4512 [Google Scholar]
- Lange BM, Croteau R (1999) Isoprenoid biosynthesis via a mevalonate-independent pathway in plants: cloning and heterologous expression of 1-deoxy-d-xylulose 5-phosphate reductoisomerase from peppermint. Arch Biochem Biophys 365: 170–174 [DOI] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51: 925–948 [DOI] [PubMed] [Google Scholar]
- Lois LM, Rodriguez-Concepcion M, Gallego F, Campos N, Boronat A (2000) Carotenoid biosynthesis during tomato fruit development: regulatory role of 1-deoxy-d-xylulose 5-phosphate synthase. Plant J 22: 503–513 [DOI] [PubMed] [Google Scholar]
- Mahmoud SS, Croteau R (2001) Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc Natl Acad Sci USA 98: 8915–8920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier W, Hammer K, Dammann U, Schulz B, Strack D (1997) Accumulation of sesquiterpenoid cyclohexenone derivatives induced by an arbuscular mycorrhizal fungus in members of the Poaceae. Planta 202: 36–42 [Google Scholar]
- Maier W, Peipp H, Schmidt J, Wray V, Strack D (1995) Levels of a terpenoid glycoside (blumenin) and cell wall-bound phenolics in some cereal mycorrhizas. Plant Physiol 109: 465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier W, Schneider B, Strack D (1998) Biosynthesis of sesquiterpenoid cyclohexenone derivatives in mycorrhizal barley roots proceeds via the glyceraldehyde 3-phosphate/pyruvate pathway. Tetrahedron Lett 39: 521–524 [Google Scholar]
- Maier W, Schmidt J, Nimtz M, Wray V, Strack D (2000) Secondary products in mycorrhizal roots of tobacco and tomato. Phytochemistry 54: 473–479 [DOI] [PubMed] [Google Scholar]
- Maier W, Schmidt J, Wray V, Walter MH, Strack D (1999) The mycorrhizal fungus, Glomus intraradices, induces the accumulation of cyclohexenone derivatives in tobacco roots. Planta 207: 620–623 [Google Scholar]
- Meyer I, Grosset J, Chartier Y, Cleyet-Maral JC (1988) Preparation by two-dimensional electrophoresis of proteins for antibody production: Antibody against proteins whose synthesis is reduced by auxin in tobacco mesophyll protoplasts. Electrophoresis 9: 704–712 [DOI] [PubMed] [Google Scholar]
- Miller B, Heuser T, Zimmer W (2000) Functional involvement of a deoxy-d-xylulose 5-phosphate reductoisomerase gene harboring locus of Synechococcus leopoliensis in isoprenoid biosynthesis. FEBS Lett 481: 221–226 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepcíon M, Ahumada I, Diez-Juez E, Sauret-Güeto S, Lois LM, Gallego F, Carretero-Paulet L, Campos N, Boronat A (2001) 1-deoxy-d-xylulose 5-phosphate reductoisomerase and plastid isoprenoid biosynthesis during tomato fruit ripening. Plant J 27: 213–222 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepcíon M, Boronat A (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids: a metabolic milestone achieved through genomics. Plant Physiol 130: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M, Knani M, Simonin P, Sutter B, Sahm H (1993) Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J 295: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer P, Corbière H, Boller T (1999) Hydrogen peroxide accumulation in Medicago truncatula roots colonized by the arbuscular mycorrhizal fungus Glomus intraradices. Planta 208: 319–325 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schüssler A, Schwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105: 1413–1421 [Google Scholar]
- Schwarz MK (1994) Terpenbiosynthese in Gingko biloba. PhD thesis. Eidgenössische Technische Hochschule, Zürich, Switzerland
- Schwender J, Müller C, Zeidler J, Lichtenthaler HK (1999) Cloning and heterologous expression of a cDNA encoding 1-deoxy-d-xylulose 5-phosphate reductoisomerase from Arabidopsis thaliana. FEBS Lett 455: 140–144 [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ (1997) Mycorrhizal Symbiosis, Ed 2. Academic Press, New York
- Strack D, Fester T, Hause B, Schliemann W, Walter MH (2003) Arbuscular mycorrhiza: biological, chemical and molecular aspects. J Chem Ecol 29: 1955–1979 [DOI] [PubMed] [Google Scholar]
- Sujak A, Gabrielska J, Grudzinski W, Borc R, Mazurek P, Gruszecki WI (1999) Lutein and zeaxanthin as protectors of lipid membranes against oxidative damage: the structural aspects. Arch Biochem Biophys 371: 301–307 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Kuzuyama T, Watanabe H, Seto H (1998) A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc Natl Acad Sci USA 95: 9879–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin EM (1997) Renewing an old view of chloroplasts. Trends Plant Sci 2: 405–406 [Google Scholar]
- Veau B, Courtois M, Oudin A, Chénieux JC, Rideau M, Clastre M (2000) Cloning and expression of cDNAs encoding two enzymes of the MEP pathway in Catharanthus roseus. Biochim Biophys Acta 1517: 159–163 [DOI] [PubMed] [Google Scholar]
- Walter MH, Fester T, Strack D (2000) Arbuscular mycorrhizal fungi induce the non-mevalonate methylerythritol phosphate pathway of isoprenoid biosynthesis correlated with the accumulation of the “yellow pigment” and other apocarotenoids. Plant J 21: 571–578 [DOI] [PubMed] [Google Scholar]
- Walter MH, Hans J, Strack D (2002) Two distantly related genes encoding 1-deoxy-d-xylulose 5-phosphate synthases: differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J 31: 243–254 [DOI] [PubMed] [Google Scholar]