Abstract
Asthma is a common disease with a complex risk architecture including both genetic and environmental factors. We performed a meta-analysis of North American genome-wide association studies (GWAS) of asthma in 5,416 asthma cases representing European Americans, African Americans/African Caribbeans, and Latinos, and replicated five regions among the most significant signals in 12,649 individuals from the same ethnic groups. Four were at previously reported loci on 17q21, and near the IL1RL1, TSLP, and IL33, genes, but we report for the first time that these loci are associated with asthma risk in three ethnic groups. In addition, we identified a novel association with asthma in the PYHIN1, gene that was specific to individuals of African descent (p=3.9×10−9). These results suggest that some asthma susceptibility loci are robust to differences in ancestry when sufficiently large samples sizes are investigated, and that ancestry-specific associations also contribute to the complex genetic architecture of asthma.
Asthma is a common, complex disease that affects over 300 million people worldwide1. In the United States, the prevalence of asthma in 2001–2003 varied between ethnic groups, ranging from 7.7% in European Americans, 12.5% in African Americans, and 3.9–14.5% in Latino Americans2. Estimates of heritability indicate that 35–80% of the variation in risk is attributable to genetic variation3,4, yet attempts to identify asthma susceptibility alleles by candidate gene and positional cloning studies had relatively few successes5,6. Most genome-wide association studies (GWAS) of asthma were conducted in relatively small samples of Europeans7, European Americans 8–10, Mexicans11, Puerto Ricans12, and African Americans and African Caribbeans13 and did not reveal significant associations or associations that replicated across studies. In contrast, the association at the IKZF3/ZPBP2/GSDMB/ORMDL3 locus on chromosome 17q21 (henceforth referred to as the 17q21 locus) has been consistently replicated as an asthma susceptibility locus in ethnically diverse subjects from Europe, North America, and Asia14–22. Recently, a large meta-analysis of GWAS in European populations, called the GABRIEL Study23, reported significant associations with single nucleotide polymorphisms (SNPs) at six loci, including the 17q21 locus and five other regions not identified in previous GWASs of asthma. However, it is currently unknown whether the additional associations, implicating HLA-DQ, IL1RL1/IL18R1, IL33, SMAD3, and IL2RB1, play a significant role in asthma risk in European populations only or whether the earlier studies were just underpowered to detect associations with these loci.
The combined results of the previous GWAS for asthma highlight the challenges in the search for genetic risk factors for asthma. These include the large number of statistical tests performed (with millions of polymorphisms), missing genotype information (especially for rare variants, and due to inadequately designed genotyping platforms for non-European populations), modest effect sizes for associated alleles, heterogeneity in the clinical definition of disease, and the effects of a large number of potentially important environmental exposures, all of which will reduce power to detect associations. The simplest solution for increasing power is to pool data from many studies in a meta-analysis of genome-wide data sets for asthma, as in the GABRIEL Study in Europeans23. The goal of the EVE Consortium, which includes GWAS datasets from nine research groups in the United States (Table 1), is not only to increase the power to identify SNPs, genes and pathways associated with asthma risk by combining studies, but also to provide a better understanding of the patterns of variation in asthma risk genes or variants in the three major ethnic groups in the United States and to provide a more comprehensive understanding of the differences in genetic risk patterns between European American, African American/African Caribbean, and Latino individuals. Only studies of diverse populations will allow for the discovery of both robust associations that replicate across ethnic groups and unique associations that contribute to the heterogeneity in disease prevalence across different ethnic groups. Moreover, studying populations with diverse ancestries can increase the resolution of associated regions as a result of different patterns of linkage disequilibrium (LD) between racial/ethnic groups. The results reported here are based on analyses of >2 million SNPs in 3246 asthma cases, 3385 non-asthmatic controls, 1702 asthma case-parent trios, and 355 family-based cases and 468 family-based controls, comprising three ethnic groups: European American, African American/African Caribbean, and Latino (Table 1).
Table 1.
Baseline characteristics of subjects in meta-analysis.
| Sample^ | Genotyping Platform+ | N Asthma Cases | Sex Ratio (M:F) | AGE OF ONSET (YRS) (ASTHMA) | AGE AT ENROLLMENT (YRS) | % Atopic* | ||
|---|---|---|---|---|---|---|---|---|
| Mean (SD) | Median (range) | Mean (SD) | Median (range) | |||||
| European Americans | ||||||||
| CAG/CSGA/SARP | 1Mv1 | 843 | 321:522 | 13.1 (13.7) | 8.0 (3–66) | 32.0 (15.6) | 31 (6–74) | 79.3 |
| CARE@ | 6.0 | 217 | 148:69 | 2.1 (2.4) | 1.1 (0–12) | 6.0 (4.1) | 3.8 (1–17) | 68.1 |
| CAMP@ (ref.8) | 550K | 403 | 242:143 | 3.1 (2.5) | 2.5 (0–11) | 8.8 (2.1) | 8.6 (5–13) | 87.5 |
| CHS | 550K, 610K | 643 | 348:295 | 7.0 (5.0) | 7.1 (0–18) | 8.4 (2.1) | 9.0 (5–14) | NA |
| African American/African Caribbeans | ||||||||
| Barbados% (ref.13) | 650K | 355 | 176:179 | 8.2 (10.6) | 5.0 (0–63) | 20.8 (12.9) | 16 (3–73) | 71.0 |
| CAG/CSGA/SARP | 1Mv1 | 644 | 266:378 | 9.8 (12.4) | 5.0 (0–63) | 26.4 (15.8) | 24 (6–18) | 84.2 |
| GRAAD (ref.13) | 650K | 464 | 211:253 | 11.9 (13.2) | 7.0 (0–65) | 24.4 (17.6) | 18 (4–76) | 85.2 |
| SAPPHIRE | 6.0 | 149 | 60:89 | 10.5 (11.6) | 7.0 (0–50) | 32.3 (13.3) | 31 (12–56) | 91.3 |
| Latino | ||||||||
| CARE@ | 6.0 | 52 | 41:11 | 1.4 (1.6) | 1.0 (0–8) | 4.8 (3.2) | 3.7 (1–13) | 58.8 |
| CHS | 550K, 610K | 606 | 339:267 | 6.8 (4.8) | 7.6 (0–18) | 7.4 (1.7) | 6.9 (5–14) | NA |
| GALA@ | 6.0 | 538 | 302:236 | 5.7 (6.8) | 3.0 (0–39) | 14.8 (7.7) | 12.0 (7–40) | 79.1 |
| MCCAS@(ref.11) | 550K | 492 | 289:283 | # | # | 9.0 (2.4) | 9 (5–17) | 92.0 |
There were a total of 3246 cases, 3385 controls, 1702 case-parent trios, and 355 family-based cases and 468 family-based controls included in the meta-analysis. Unless otherwise indicated samples are unrelated asthma cases and non-asthma controls. Descriptions of the individual study samples are in the Supplementary Methods online. References for published GWAS in the individual samples are shown in the first column. NA, information not available.
CARE, Clinical Asthma Research Network; CAMP, Childhood Asthma Management Program; CHS, Children's Health Study; CAG, Chicago Asthma Genetics Study; CSGA, Collaborative Studies on the Genetics of Asthma; SARP, Severe Asthma Research Program; GRAAD, Genetic Research on Asthma in the African Diaspora; SAPPHIRE, Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-Ethnicity; GALA, Genetics of Asthma in Latino Americans; MCCAS, Mexico City Childhood Asthma Study
Illumina arrays: 1Mv1, 550k, 610k, 650k; Affymetrix arrays: 500k, 6.0
Asthma case-parent trios
Family-based cases and controls
1 or more positive allergen skin test or specific IgE, or high total IgE
Age of onset was queried as a categorical varaiable. Percentages in each category were: <1 yr=7.1%, 2–3 yrs=25.6%, 4–5 yrs=29.1%, 6–7 yrs=17.7%, 8–9 yrs=11.6%, >9=8.9%.
We performed four genome-wide investigations: one meta-analysis in each of three ethnic groups and one in the combined sample. The quantile-quantile (QQ) plots of the results (see Supplementary Figs. 1–4 online) indicate that none of the studies showed inflation in test statistics, but also revealed an abundance of small p-values, especially in the combined sample. In each of the four meta-analyses, we expected (on average) approximately two p-values to be smaller than 10−6 by chance alone. We observed 34 SNPs with p-values smaller than 10−6 in the European American, four in the African American/African Caribbean, 32 in the Latino, and 75 in the combined meta-analyses (see Supplementary Fig. 5 online). In the European Americans, 33 of the SNPs are at the 17q21 asthma locus; one additional SNP on chromosome 17 (rs9891949) is 27 Mb from the 17q21 locus. In the African Americans/African Caribbeans, two SNPs are in the PYHIN1 gene on chromosome 1q23 and two are in the intergenic region between the NNMT and C11orf71 loci on chromosome 11q23. In the Latinos, 13 SNPs are on chromosome 3q27 around the RTP2 gene, one is on chromosome 5q33 in the GALNT10 gene, 12 are at the chromosome 17q21 locus, and two are on chromosome 19q12 between the CCNE1 and C19orf2 loci. One SNP in an intron of RTP2, rs2017908, reached genome-wide significance in the Latino samples (p=4.4×10−9). The 75 SNPs with p<10−6 in the combined meta-analysis occur in 15 chromosomal regions; the most significant SNP in each region with at least one p<10−6 in the combined sample or in one of the ethnic groups is shown in Table 2, with risk allele frequencies shown in Supplementary Table 1 online. Among the 75 SNPs, those at the 17q21, IL1RLI, and TSLP loci reached genome-wide significance in the combined sample (p<2×10−8). The results for all SNPs with p<10−6 can be found as Supplementary Tables 2 and 3 online.
Table 2.
Results of meta-analyses.
| P-value | OR (95% CI) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Position | SNP | #SNPs | Nearest Gene | Eur Am | Afr Am/Afr Carib | Latino | Meta | Eur Am | Afr Am/Afr Carib | Latino | Overall |
| rs4845783 | 1 | 150,759,183 | G/A | 3 | CRCT1 | 0.0023 | 2.3×10−4 | 0.048 | 5.6×10−7 | 1.18 (1.04, 1.33) | 1.35 (1.02, 2.04) | 1.09 (0.94, 1.30) | 1.16 (1.06, 1.28) |
| rs1102000 | 1 | 155,745,980 | T/C | 2 | PYHIN1 | NA | 3.6×10−7 | 0.12 | 3.6×10−6 | NA | 1.34 (1.19, 1.49) | 1.04 (0.47, 1.62) | 1.32 (1.17, 1.46) |
| rs4653433 | 1 | 224,041,154 | A/G | 1 | SRP9 | 0.0026 | .027 | 6.2×10−4 | 5.4×10−7 | 1.13 (1.03, 1.23) | 1.12 (0.99, 1.24) | 1.16 (1.04, 1.29) | 1.14 (1.07, 1.2) |
| rs10173081* | 2 | 102,323,780 | C/T | 13 | IL1RL1 | 1.3×10−4 | 0.07 | 4.5×10−5 | 1.4×10−8 | 1.28 (1.13, 1.42) | 1.1 (0.96, 1.23) | 1.28 (1.08, 1.48) | 1.2 (1.11, 1.29) |
| rs2017908+ | 3 | 188,900,662 | A/C | 13 | RTP2 | 0.24 | 0.67 | 4.4×10−9 | 0.0049 | 0.93 (0.79, 1.08) | 1.03 (0.9, 1.17) | 1.63 (1.43, 1.82) | 1.09 (1, 1.18) |
| rs11735820 | 4 | 66,181,611 | G/T | 2 | EPHA5 | 1.5×10−4 | 0.003 | 0.033 | 2.3×10−7 | 1.18 (1.08, 1.28) | 1.15 (1.02, 1.27) | 1.14 (1, 1.28) | 1.16 (1.09, 1.23) |
| rs1837253* | 5 | 110,429,771 | C/T | 1 | TSLP | 1.1×10−5 | 0.0023 | 0.0018 | 7.3×10−10 | 1.23 (1.12, 1.35) | 1.2 (1.06, 1.35) | 1.14 (1.01, 1.27) | 1.19 (1.12, 1.27) |
| rs10064618 | 5 | 153,752,482 | G/A | 1 | GALNT10 | 0.76 | 0.36 | 5.7×10−7 | 4.4×10−4 | 1.01 (0.93, 1.12) | 1.08 (0.93, 1.28) | 1.25 (1.09, 1.47) | 1.10 (1.02, 1.19) |
| rs2453626 | 8 | 101,207,073 | C/T | 1 | FBXO43 | 0.0044 | 2.1×10−4 | 0.016 | 2.8×10−7 | 1.15 (1.05, 1.24) | 1.24 (1.11, 1.37) | 1.13 (1, 1.26) | 1.17 (1.1, 1.23) |
| rs2381416 | 9 | 6,183,455 | C/A | 2 | IL33 | 0.011 | 0.0022 | 6.6×10−4 | 2.5×10−7 | 1.18 (1.04, 1.35) | 1.14 (0.99, 1.32) | 1.22 (1.04, 1.47) | 1.18 (1.08, 1.28) |
| rs11214966 | 11 | 113,736,465 | C/T | 2 | C11orf71 | 0.76 | 3.5×10−7 | 0.59 | 0.0012 | 1.02 (0.83, 1.21) | 1.35 (1.2, 1.51) | 1.04 (0.86, 1.22) | 1.16 (1.06, 1.26) |
| rs16929496 | 12 | 25,885,071 | T/C | 3 | RASSF8 | 3.2×10−4 | 0.025 | 0.0027 | 2.6×10−7 | 1.23 (1.06, 1.45) | 1.10 (0.94, 1.33) | 1.25 (1.02, 1.61) | 1.19 (1.08, 1.33) |
| rs9891949 | 17 | 8,039,908 | A/G | 1 | AURKB | 7.5×10−7 | 0.68 | 0.73 | 4.5×10−4 | 1.25 (1.15, 1.36) | 1.06 (0.94, 1.18) | 1.02 (0.89, 1.14) | 1.12 (1.05, 1.19) |
| rs11078927* | 17 | 35,317,931 | C/T | 49 | GSDMB | 8.6×10−7 | 0.0012 | 3.1×10−7 | 1.2×10−14 | 1.25 (1.15, 1.35) | 1.31 (1.13, 1.5) | 1.29 (1.16, 1.42) | 1.27 (1.2, 1.34) |
| rs335016 | 19 | 35,065,232 | A/C | 2 | C19orf2 | 0.16 | 0.68 | 2.6×10−7 | 5.8×10−5 | 1.05 (0.94, 1.16) | 1 (0.86, 1.14) | 1.25 (1.12, 1.38) | 1.09 (1.02, 1.17) |
The most significant SNP in each region with at least one p-value <1×10−6 in the combined sample or in at least one ancestry group, sorted by chromosome. Results for all SNPs with p-values <1×10−6 are shown in Supplementary Table 4 online (p-values) and Supplementary Table 5 online (minor allele frequencies and imputation quality scores). #SNPS, number of SNPs in a 1 Mb region with p-values <1×10−6. NA, not available (SNP not polymorphic);
genome-wide significant in the combined sample
genome-wide significant in Latinos only.
We selected one SNP from each of the 15 regions with at least one p-value <10−6 for replication studies (as described in Table 2). The samples used for replication are described in the Supplementary Note and Supplementary Table 4 online, and the results of those studies are shown in Table 3. Two SNPs (rs4653433 near the SRP9 gene on chromosome 1q and rs9891949 near the AURK gene on chromosome 17p) could not be assayed in the replication samples. Using a Bonferroni-corrected (for 13 tests) p-value of p<0.0038 as the threshold for significance in the replication studies, SNPs in five regions were significantly associated with asthma in the replication samples (Table 3).
Table 3.
Replication study results.
| SNP | Chr | Position | Nearest Gene | Replication Meta-analysis P-values | Meta-analysis + Replication P-values | |||
|---|---|---|---|---|---|---|---|---|
| Eur Am | Afr Am/Afr Carib | Latino | Overall | |||||
| Replicated Associations | ||||||||
| rs1101999 | 1 | 157,199,179 | PYHIN1 | NA | 0.0028 | 0.28 | 0.64 | 3.9×10−9^ |
| rs3771180 | 2 | 102,323,780 | IL1RL1 | 0.17 | 0.0018 | 2.5×10−5 | 5.3×10−7 | 1.5×10−15 |
| rs1837253 | 5 | 110,429,771 | TSLP | 0.0076 | 0.012 | 0.0010 | 1.59×10−6 | 1.0×10−14 |
| rs2381416 | 9 | 6,183,455 | IL33 | 0.0026 | 0.010 | 0.0034 | 1.3×10−6 | 1.7×10−12 |
| rs11078927 | 17 | 35,317,931 | GSDMB | 7.4×10−7 | 0.86 | 4.1×10−7 | 1.5×10−8 | 2.2×10−16 |
| Non-replicated Associations | ||||||||
| rs4845783 | 1 | 150,759,183 | CRCT1 | 0.25 | 0.74 | 0.24 | 0.15 | 6.2×10−6 |
| rs2017908 | 3 | 188,900,662 | RTP2 | 0.25 | 0.56 | 0.24 | 0.88 | 0.0030+ |
| rs11735820 | 4 | 66,181,611 | EPHA5 | 0.29 | 0.37 | 0.52 | 0.15 | 0.010 |
| rs10064618 | 5 | 153,752,482 | GALNT10 | 0.30 | 0.69 | 0.52 | 0.32 | 1.7×10−4+ |
| rs2453626 | 8 | 101,207,073 | FBXO43 | 0.71 | 0.66 | 0.06 | 0.28 | 0.0049 |
| rs11214966 | 11 | 113,736,465 | C11orf71 | 0.72 | 0.18 | 0.15 | 0.48 | 6.3×10−7^ |
| rs16929496 | 12 | 25,885,071 | RASSF8 | 0.44 | 0.59 | 0.28 | 0.97 | 3.7×10−4 |
| rs335016 | 19 | 35,065,232 | C19orf2 | 0.83 | 0.88 | 0.75 | 1.00 | 0.0018+ |
SNP rs1101999 in PYHIN1 is in perfect LD with rs1102000 (Table 2) in African Americans (r2=1.0), and was used as a surrogate SNP in the replication studies (neither SNP is polymorphic in European Americans); rs3771180 used as a surrogate SNP for rs10173081 near IL1RL1. Two associated SNPs could not be assayed in the replication samples: rs4653433 near SRP9 and rs9891949 near AURKB. NA, not available.
P-value is combined for only the African American and African Caribbean populations
P-value is combined for only the Latino populations
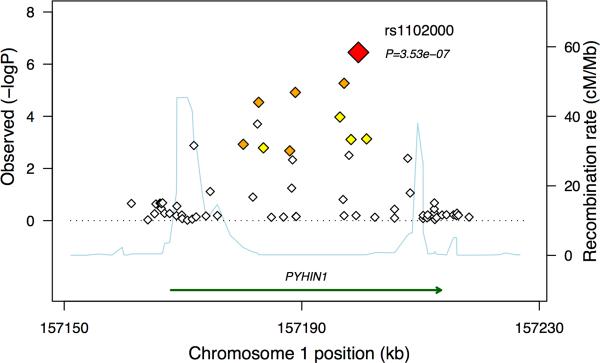
SNPs near the 17q21 locus and the IL1RL1, TSLP, and IL33 genes were associated with asthma in all three ethnic groups in the replication studies, whereas the PYHIN1 association was specific to the African American replication samples. Interestingly, the associated SNPs in PYHIN1 occur with minor allele frequencies of 0.26–0.29 in the African American/African Caribbean controls, but they are not polymorphic in European Americans and occur at low frequencies (<0.05) in the Latino populations. However, multiple other SNPs in the PYHIN1 region showed evidence for association only in the African American/African Caribbean sample (Figure 1), whereas none showed evidence of association with asthma in Latinos or European Americans (see Supplementary Fig. 6 online), suggesting that this association may be specific to populations of African descent. In fact, rs1102000 in PYHIN1 (also known as IFIX) has a relatively large effect size (OR=1.34 in the GWAS meta-analysis, OR=1.23 in the replication samples), suggesting an important role for this gene in risk for asthma in African American and African Caribbean populations.
Figure 1. Results of Meta-Analysis on the Chromosome 1 Region of Association in African Americans/African Caribbeans.
The relative location of the PYHIN1 gene and direction of transcription are shown in the lower portion of the figure, and the chromosomal position on the x-axis. The light blue line shows the recombination rate across the region (right y-axis), and the left y-axis shows the significance of the associations. The large red diamond shows the p-value for rs1102000. The small diamonds in show the p-values for all other SNPs, color-coded according to the level of LD with rs1102000 in the HapMap YRI (red, r2 > 0.9; orange, r2 = 0.7–0.9; yellow, r2 = 0.5–0.7). Results for the European American, Latino, and combined meta-analysis are shown in Supplementary Fig. 6 online.
The associations with SNPs in PYHIN1 (pyrin and HIN domain family member 1; IFIX, interferon inducible nuclear protein X) with asthma are the first genome-wide significant associations reported in African Americans or African Caribbeans and may be the first asthma susceptibility gene specific to populations of African descent. The associated SNPs are in an LD bin in HapMap YRI samples, spanning ~30kb within the seventh intron of the gene, suggesting that the causal variation is contained within this intron. Imputation of additional SNPs in PYHIN1 using pilot data from the 1000 Genomes Project yielded no additional signals of association (see Supplementary Fig. 7 online). However, the incomplete coverage of PYHIN1 makes any conclusions on causal variation inaccurate (see Supplementary Fig. 8 online). Interestingly, the most strongly associated variants discovered in this study are not present in populations of European descent. Although, it is possible that rare variants in PHYIN1 that are not tagged by the SNPs included in this study are risk alleles for asthma in European American and Latino populations, none of the multiple SNPs in this gene that were associated with asthma in the African American/African Caribbean populations showed evidence for association in the European Americans or Latinos. This further suggests that the African-specific variants may be causal or in LD with other African-specific causal variants in this gene. Moreover, estimates of local ancestry at PYHIN1 in the African American/African Caribbean samples did not differ between cases and controls for any of the studies (combined p=0.77; see Supplementary Table 6 online for more details), indicating that the African-American specific association is present in the absence of an admixture signal and that the observed association is not due to uncorrected local ancestry. At present, little is known about the function of PYHIN1, although it is expressed in both adult leukocytes and lung tissues and the pyrin domain is a protein-protein interaction domain that is present in many interferon-inducible proteins that functions in both apoptotic and inflammatory pathways (see URL). This family of genes has been previously associated with autoimmunity24, but to date PYHIN1 has not been implicated in asthma pathogenesis.
Lastly, we examined the evidence for association with SNPs that were associated with asthma in previous GWAS other than those included in the EVE meta-analysis (see Supplementary Table 5 online). These included SNPs in or near HLA-DQ9,23, the 17q12 asthma locus23, IL3323, IL1RL123, SMAD323, IL2RB23, RORA23, SLC22A523, IL139,23, RAD509, and DENND1B10. We were able to replicate at p<0.05 associations with SNPs at the HLA-DQ locus in all three ethnic groups and in the combined sample, although no single SNP was associated in all three groups. The two SNPs associated at the 17q21 asthma locus in the GABRIEL Study were also associated with asthma in our study, although one (rs2894194) showed little evidence for association in the African American/African Caribbean samples. Among the remaining SNPs associated in the GABRIEL study, we replicated associations with the same SNPs in or near the IL18RL1, IL33, SLC22A5, SMAD3, and RORA genes. SNPs at the latter three loci were associated only in the European American sample. Lastly, we replicated associations with two SNPs in RAD50, with the signals coming largely from the Latino and African American/African Caribbean samples. A SNP at the IL2RB locus that was associated with asthma in the GABRIEL Study was nearly significant in the European American EVE sample (p=0.06). We did not replicate associations with SNPs at the IL139,23 and DENNDB110 loci.
The results reported here highlight the importance of studying large datasets of diverse populations in several ways. First, the large sample size allowed us to first discover and then replicate loci with modest effects. For example, SNPs at the IL1RL1, TSLP, and IL33 loci did not reach genome-wide significance in any of the ethnic-specific meta-analyses, but SNPs in IL1RL1 and TSLP reached this threshold of significance, and IL33 approached genome-wide significance, in the overall analysis (Table 2; see Supplementary Figs. 10–12 online). The modest, but real, effect of variants on asthma risk likely explains why this locus was not identified as an asthma susceptibility locus in previous genome wide association studies in individual samples7–11,13, but was among the most significant associations in the EVE and GABRIEL meta-analyses.
Second, the diverse ancestries of the EVE Consortium samples provided a more accurate and more complete picture of asthma risk by identifying common variants at four loci (17q21 locus, IL1RL1, TSLP, and IL33) that increase risk in all ethnic groups and at least one locus (PYHIN1) that may contribute to risk only in populations of African descent. Replication of previously associated variants in the EVE samples may also provide additional insights into those genes. For example, the SNPs in or near SMAD3 and RORA reported in the GABRIEL Study meta-analysis were modestly associated with asthma in the European American EVE samples but showed no association in the African American/African Caribbean samples, suggesting that these may be risk variants only in populations of European descent. We anticipate that ongoing studies in the EVE Consortium data sets will identify other loci contributing to asthma risk and, ultimately provide a better understanding of the molecular pathways and networks that are common to the risk architecture of asthma in diverse populations, and those that are specific to certain groups.
Methods
GWAS subjects, genotyping and statistical analyses
Asthma cases, unaffected controls, asthmacase parent trios, and extended families were recruited in clinics in the U.S, Mexico, and Barbados. Twelve samples with GWAS data were included in this study (Table 1). Detailed descriptions of the individual studies, ascertainment schemes, genotyping platforms, quality control (QC) protocols, and statistical analyses for the primary association testing are described in the Supplementary Note online.
Meta-analysis: QC and testing for association
Summary files on a common set of SNPs were shared among the EVE investigators. The common set of SNPs consisted of all Phase 2, Release 21 consensus HapMap variants. Prior to genotype imputation, each center oriented their SNPs to the plus strand, and filtered for call rates (> 95%) and consistency with Hardy-Weinberg expectations (p > 10−5 for case/control studies, p > 10−6 for trio studies). Genotype imputation using HapMap reference panels were performed separately in each sample with the program MACH25, and associations were tested using the genotype dosages that are part of the imputation algorithms output, with adjustments for admixture in the African American and Latino case-control samples (see Supplementary Note online). The shared summary files contained the SNP identifiers (rs number, chromosome, position, alleles), SNP QC metrics (call rate, Hardy-Weinberg equilibrium p-value, imputation quality metrics), and information related to the test for association (allele frequencies in cases and controls or in the transmitted and untransmitted alleles in trios, association p-value, odds ratios with standard errors). Reference alleles were assigned as the allele coded 0 in the HapMap release 21 phased consensus haplotypes during genotype imputation in MACH. The QC checks performed on summary file data included consistency in reference allele and strand orientation (see Supplementary Figs. 13−15 online), and imputation quality (see Supplementary Fig. 16 online). SNPs with imputation quality scores below a threshold were removed from the analysis (Rsq < 0.3 or < 0.5 for the Barbados study). QQ plots were visually inspected to compare the distribution of association p-values for genotyped and imputed markers separately for each cohort.
The meta-analysis searched for asthma susceptibility variants for which the same allele was associated with asthma in the different studies. For each study, we constructed a test statistic that has a standard normal distribution under the null hypothesis of no association and captures the direction of the effect (i.e., the statistic was positive if the reference allele was associated with an increased risk of asthma). The meta-analysis test statistic was calculated as a linear combination of the individual study scores with weights proportional to the square root of the number of cases (or trios). P-values were obtained using normal approximations. Odds ratios were calculated by combining linearly log odds ratios with weights reflecting the standard errors from the genome-wide association studies.
Replication samples and genotyping
The replication cohorts and sample sizes are shown in Supplementary Table 4 online. Detailed descriptions of these samples, genotyping technologies, QC protocols, and statistical analyses are described in the Supplementary Note online.
Association testing in replication samples
The combined analysis of the replication samples was performed in a similar manner to the meta-analysis. For each study, we constructed a test statistic that had standard normal distribution under the null hypothesis of no association, and that also captured the direction of the effect. A combined test statistic was calculated as a linear combination of the individual study scores with weights proportional to the square root of the number of cases, and p-values were obtained using normal approximations.
Imputation of pilot data from the 1000 Genomes Project
Pilot data from the 1000 Genomes Project (August 2010 haplotypes) was used to impute a 2Mb region surrounding the PYHIN1 gene in all studies using impute226. The EUR haplotypes (European) were used as a reference for the European American studies (assuming an effective population size [Ne] of 11418), the EUR and AFR (African) haplotypes were used as a reference for the African American studies (assuming Ne=15000), and the EUR, AFR, and ASN (Asian haplotypes) were used as a reference for the Latino studies (assuming Ne=15000). Individual genotypes were filtered for probabilities > 90%, and only SNPs with call rates > 90% were included in allelic tests of association. For family-based studies, SNPs with Mendelian errors were removed.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Heart, Lung, and Blood Institute (HL101651 to C.O. and D.L.N.; HL087665 to D.L.N.; HL70831, HL087665, HL072414, and HL49596 to C.O.; HL064307 and HL064313 to F.D.M.; HL075419, HL65899, HL083069, HL066289, HL087680, HL101543, and HL101651 to S.T.W.; HL079055 to L.K.W.; HL087699, HL49612, HL075417, HL04266, and HL072433 to K.C.B.; HL061768, HL076647, to F.D.G.; HL087680 to W.J.G.; HL078885 and HL088133 to E.G.B.; HL87665 to D.A.M.; and HL69116, HL69130, HL69149, HL69155, HL69167, HL69170, HL69174, HL69349 to D.A.M., E.R.B., W.W.B., W.J.C., M.C., K.F.C., S.C.E., E.I., and S.E.W.), the National Institutes of Allergy and Infectious Disease (AI070503 to C.O.; AI079139 and AI061774 to L.K.W.; AI50024, AI44840, and AI41040 to K.C.B.; and AI077439 to E.G.B.), the National Institute of Diabetes and Digestive and Kidney Diseases to L.K.W. (DK064695); the National Institutes of Environmental Health Sciences (ES09606, ES018176, and ES015903 to K.C.B.; ES007048, ES009581, R826708, RD831861, and ES011627 to F.D.G.; ES015794 to E.G.B.; and the Division of Intramural Research, Z01 ES049019, to S.J.L.); the National Center for Research Resources (RR03048 to K.C.B.), the Environmental Protection Agency (83213901 and R-826724 to K.C.B.), the American Asthma Foundation and the Fund for Henry Ford Hospital (to L.K.W.), Mary Beryl Patch Turnbull Scholar Program (to K.C.B.), the National Council of Science and Technology (Mexico) (26206-M to I.R.), the Centers for Disease Control, U.S. (to I.R.), the Eudowood Foundation (to N.N.H.); and the Flight Attendant Medical Research Institute (FAMRI), RWJF Amos Medical Faculty Development Award, the American Asthma Foundation, and the Sandler Foundation (to E.G.B.).
The authors gratefully acknowledge the support from Dr. James Kiley, Dr. Susan Banks-Schlegel, and Dr. Weiniu Gan at the National Heart, Lung, and Blood Institute, and all the patients and families that participated in these studies.
References
- 1.Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59:469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 2.Moorman JE, et al. National surveillance for asthma--United States, 1980–2004. MMWR Surveill Summ. 2007;56:1–54. [PubMed] [Google Scholar]
- 3.Duffy DL, Martin NG, Battistutta D, Hopper JL, Mathews JD. Genetics of asthma and hay fever in Australian twins. Am Rev Respir Dis. 1990;142:1351–1358. doi: 10.1164/ajrccm/142.6_Pt_1.1351. [DOI] [PubMed] [Google Scholar]
- 4.Nieminen MM, Kaprio J, Koskenvuo M. A population-based study of bronchial asthma in adult twin pairs. Chest. 1991;100:70–75. doi: 10.1378/chest.100.1.70. [DOI] [PubMed] [Google Scholar]
- 5.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006;7:95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 6.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 7.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 8.Himes BE, et al. Genome-wide association analysis identifies PDE4D as an asthma susceptibility gene. Am J Hum Genet. 2009;84:581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLADR/DQ regions. J Allergy Clin Immunol. 2010;125:328–335.e11. doi: 10.1016/j.jaci.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sleiman PM, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 11.Hancock DB, et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in mexican children. PLoS Genet. 2009;5:e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhry S, et al. Genome-wide screen for asthma in Puerto Ricans: evidence for association with 5q23 region. Hum Genet. 2008;123:455–468. doi: 10.1007/s00439-008-0495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathias RA, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125:336–346.e4. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouzigon E, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–1994. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 15.Galanter J, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halapi E, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18:902–908. doi: 10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung TF, et al. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy. 2009;64:621–628. doi: 10.1111/j.1398-9995.2008.01873.x. [DOI] [PubMed] [Google Scholar]
- 18.Madore AM, Tremblay K, Hudson TJ, Laprise C. Replication of an association between 17q21 SNPs and asthma in a French-Canadian familial collection. Hum Genet. 2008;123:93–95. doi: 10.1007/s00439-007-0444-x. [DOI] [PubMed] [Google Scholar]
- 19.Sleiman PM, et al. ORMDL3 variants associated with asthma susceptibility in North Americans of European ancestry. J Allergy Clin Immunol. 2008;122:1225–1227. doi: 10.1016/j.jaci.2008.06.041. [DOI] [PubMed] [Google Scholar]
- 20.Tavendale R, Macgregor DF, Mukhopadhyay S, Palmer CN. A polymorphism controlling ORMDL3 expression is associated with asthma that is poorly controlled by current medications. J Allergy Clin Immunol. 2008;121:860–863. doi: 10.1016/j.jaci.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Wu H, et al. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64:629–635. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bisgaard H, et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med. 2009;179:179–185. doi: 10.1164/rccm.200809-1436OC. [DOI] [PubMed] [Google Scholar]
- 23.Moffatt MF, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choubey D, et al. Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: role in inflammation and autoimmunity. J Interferon Cytokine Res. 2010;30:371–380. doi: 10.1089/jir.2009.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, GR A. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. American Journal of Human Genetics. 2006;S79:2290. [Google Scholar]
- 26.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.