Abstract
My previous theoretical research shows that the rotating circular genetic code is a viable tool to make easier to distinguish the rules of variation applied to the amino acid exchange; it presents a precise and positional bio-mathematical balance of codons, according to the amino acids they codify. Here, I demonstrate that when using the conventional or classic circular genetic code, a clearer pattern for the human codon usage per amino acid and per genome emerges. The most used human codons per amino acid were the ones ending with the three hydrogen bond nucleotides: C for 12 amino acids and G for the remaining 8, plus one codon for arginine ending in A that was used approximately with the same frequency than the one ending in G for this same amino acid (plus *). The most used codons in man fall almost all the time at the rightmost position, clockwise, ending either in C or in G within the circular genetic code. The human codon usage per genome is compared to other organisms such as fruit flies (Drosophila melanogaster), squid (Loligo pealei), and many others. The biosemiotic codon usage of each genomic population or ‘Theme’ is equated to a ‘molecular language’. The C/U choice or difference, and the G/A difference in the third nucleotide of the most used codons per amino acid are illustrated by comparing the most used codons per genome in humans and squids. The human distribution in the third position of most used codons is a 12-8-2, C-G-A, nucleotide ending signature, while the squid distribution in the third position of most used codons was an odd, or uneven, distribution in the third position of its most used codons: 13-6-3, U-A-G, as its nucleotide ending signature. These findings may help to design computational tools to compare human genomes, to determine the exchangeability between compatible codons and amino acids, and for the early detection of incompatible changes leading to hereditary diseases.
Keywords: codon usage, compatible genomics, triplet, genome, adaptability, adaptation, variation, biomathematics, bioinformatics, computational biology
Introduction
I was searching for the reverse translation from proteins to nucleotides in bioinformatics when I read that different organisms often have radically different codon preferences, a fact profoundly affecting their capability to express different proteins (Wishart and Fortin, 2001).
Then, I decided to visit the earliest references on the subject, finding a surprising consistency of choices among genes of the same or similar genomes, a very precise bio-system for choosing between codons. Each gene in a genome tends to conform to its ‘species’ usage “of the codon catalog” (Grantham et al., 1980).
With these antecedents I decided to compare the human codon usage, focused first on the most used codons per amino acid, and second on the most used codons per genome. Earlier I found that the rotating circular genetic code is useful to distinguish the rules of variation as applied to the amino acid exchange (Castro-Chavez, 2010). Then, I noticed a precise positional bio-mathematical balance of codons per amino acid within the classic representation of the circular genetic code (Castro-Chavez, 2012a).
So, I decided to use the circular genetic code as the representation of choice to visualize the pattern of distributions for the most used codons in humans, while focusing my attention on the human molecular language (Ikemura, 1985), according to the most used codons per amino acid and per genome, discovering a clear example of the C/U choice or difference for the third or ‘wobbled’ nucleotide of the codons (Grantham et al., 1986) 2 and of the G/A choice, also for the third nucleotide by comparing codon preferences between humans as the chosen prototype for vertebrates and squids, the chosen prototype for invertebrates.
Since the earliest sequence analysis of ∼39 genes, these C or G third-position codon-preferences per amino acid in humans were clearly noticed (Ikemura, 1985), together with the finding that the richness of G or C bases at the third position in vertebrate genes could not be a consequence of their genome-base composition, since their overall genome G+C percentage is known to range from 40% to 45%, while codon usages per amino acid for each vertebrate converged in an essentially identical pattern, with the possible exception of arginine (Arg, R); this pattern was noticeably different from the pattern of higher plants and “from the dialect of E. coli or yeast” (Ikemura, 1985). This early work demonstrated the existence of a constraint in vertebrates to keep the third position of codons G+C rich; Ikemura (1985) concluded that the possible factor responsible for this converging pattern was related to the tRNA populations within vertebrates.
I want to stop here to emphasize the remarkable and early use of the word “dialect” by Ikemura (1985), used by him in relation to the specific codon usage pattern of an organism. In my previous related article and before reading the article by Ikemura (1985), I defined a group of organisms that have compatible genomes as a single ‘Theme’, the genomic species or population capable to speak the same molecular language through different accents, “with each variety within a theme being a different version of the same book” (Castro-Chavez, 2012a). Ikemura (1985) was also referring to the same molecular language; what he called the molecular “dialect” in 1985, I considered it as “different accents” in 2011, being such ‘accents’ the varieties within a ‘Theme’.
In a way similar to Ikemura (1985), earlier Garel (1982) proposed that apparently the quantitative adaptation of tRNAs to the codon content of mRNA seemed to be optimizing the efficiency of translation, influencing both the mean elongation rate and the fidelity. However, Grantham opposed both by declaring that “apart from leaving unexplained the origin of differential tRNA concentrations, which, in fact, poses the same problem as does codon usage, we have just seen that this cannot be the case with biased C/U choice”, finding that both codon and tRNA distributions varied enormously between species. Grantham then asked the question: “why does the bias exist?” His literal response was that such question was so difficult to treat scientifically that in effect it remained philosophical (Grantham et al., 1986).
In the present article, I have confirmed the bias in the C/U choice by comparing the most used codons per amino acid between man and squid; by doing this comparison I also found a clear bias in the G/A choice.
My hope is that these preliminary and by no means exhaustive comparisons will be able to further our understanding for the research on compatible genomics (Castro-Chavez, 2012a), and at the same time to further the practical application of my theoretical discoveries to produce and preserve biodiversity, and for the early detection of hereditary genetic diseases.
Experimental Section
The rotating circular genetic code is used here as the engine for the comparison of sequences (Castro-Chavez, 2010 and 2012a), coupled to the most useful database of codon usage available thus far (Nakamura et al., 2000), URL: http://www.kazusa.or.jp/codon [Data source: NCBI-GenBank, flat file database release 160.0 of June 15 2007]. Nakamura's human database contained 40,662,582 codons when these analyses were done. The codons present in the database for other organisms will be presented in the next section.
Results and Discussion
The results of most used codons per amino acid in humans are shown in Table 1.
Table 1.
Most used codons in humans per amino acid. The percentage represents the most used codon within each group. [End of codons in bold, the Arg % similarity underlined].
| 12 most used codons end in C | 8 most used codons end in G | 2 most used codons end in A |
|---|---|---|
| His CAC, 58 % | Met AUG, 100 % | Stop UGA, 47 % |
| Tyr UAC, 56 % | Trp UGG, 100 % | Arg AGA, 21 % |
| Phe UUC, 54 % | Gln CAG, 73 % | |
| Cys UGC, 54 % | Glu GAG, 58 % | |
| Asp GAC, 54 % | Lys AAG, 57 % | |
| Asn AAC, 53 % | Val GUG, 46 % | |
| Ile AUC, 47 % | Leu CUG, 40 % | |
| Ala GCC, 40 % | Arg AGG, 21 % | |
| Thr ACC, 36 % | ||
| Gly GGC, 34 % | ||
| Pro CCC, 32 % | ||
| Ser AGC, 24 % |
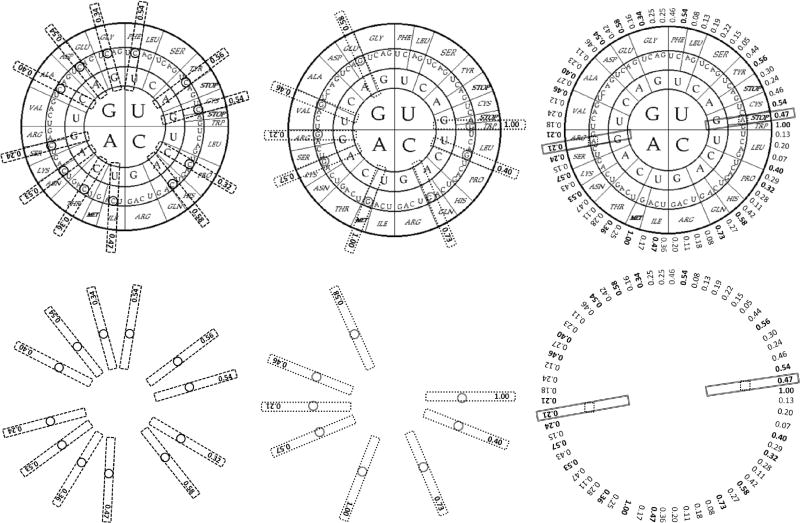
When I applied the rules of variation (Castro-Chavez, 2010) to the human codon usage, I did see that the most used codons per amino acid were able to keep the same relative positions within quadrants in the circular genetic code.
The first aspect to be recognized when using the classic circular genetic code is that its most used codons can be found either at the rightmost, or last, position for 13 amino acids, or in the second position (clockwise) for 5 amino acids, while 2 of them: Trp and Met in all organisms, only have one codon that ended in G. Arg is almost equally divided in its codon usage for two codons: Its last codon, clockwise, ending in G, and its companion codon to the left ending in A.
In Fig. 1, the figures in the lower row underlie the relative positions of most used codons, showing that if we roughly divide the circular genetic code into three parts, we obtain the asymmetric distribution of 3 × 4 codons, each with a C at its third nucleotide (Fig. 1, left); if we divide it into two parts, we obtain the asymmetric distribution of 2 × 4 codons, each with a G at the third nucleotide (Fig. 1, center), or a symmetric one (1 × 2), with one codon with A at the third nucleotide (Fig. 1, right).
Figure 1.
Most used codons per amino acid in the human genome as illustrated by the circular genetic code. Left. 12 of the most used codons per amino acid end in C, and are (clockwise): Phe UUC, Tyr UAC, Cys UGC, Pro CCC, His CAC, Ile AUC, Thr ACC, Asn AAC, Ser AGC, Ala GCC, Asp GAC, Gly GGC. Center. 8 of the most used codons end in G, and are: Trp UGG, Leu CUG, Gln CAG, Met AUG, Lys AAG, Arg AGG, Val GUG, Glu GAG. Right. 2 most used codons end in A: Stop UGA and Arg AGA. The figures below underlie their relative positions. The human distribution of most used codons is a 12-8-2, C-G-A nucleotide ending signature. Based on the human table at http://www.webcitation.org/63F2wuwCY
If we use more decimals, from the ‘Codon Frequency’ output style, GCG Wisconsin Package™ (Thompson, 2003), the total genomic use instead of the use per amino acid, and averaged per 1K of nucleotides, we have Arg AGA 12.17%, while Arg AGG is 11.96%, indicating a small 0.21% global increase for the Arg codon AGA (Nakamura et al., 2000; database update of 2007), making Arg AGA the only exception, even if small, to the human (and vertebrates in general) rule of the 3-Hydrogen bond G-C dominance at the third position.
If we reorder all of the 64 codons according to their relative abundance per genome and not per amino acid usage, as measured by the average of every thousand (1K) codifying nucleotides, which is the standard way presented by Nakamura et al. (2000), we obtain Table 2.
Table 2.
Most used codons in humans per genome; average use of all 64 codons per thousand (1K) codifying nucleotides. The number within the brackets is the relative position of each codon within the list, if the brackets are bold, that indicates that those codons correspond to a cluster of similar usage (when two contiguous members in the cluster are separated by 0.02% from each other), being the cluster number the number in italics at the left side of the brackets. There are 16 clusters and 14 non-clustered codons.
| Amino Acid, Codon, % | Amino Acid, Codon, % | Amino Acid, Codon, % |
|---|---|---|
| 1 [1] Leu/L CUG 3.96 | 5 [22] Phe/F UUU 1.76 | 11 [43] Ser/S AGU 1.21 |
| 1 [2] Glu/E GAG 3.96 | 5 [23] Pro/P CCU 1.75 | 11 [44] Arg/R AGA 1.2 |
| [3] Gln/Q CAG 3.42 | 6 [24] Asn/N AAU 1.7 | 11 [45] Arg/R AGG 1.2 |
| [4] Lys/K AAG 3.19 | 6 [25] Pro/P CCA 1.69 | [46] Arg/R CGG 1.14 |
| [5] Glu/E GAA 2.9 | 7 [26] Gly/G GGA 1.65 | 12 [47] Val/V GUU 1.1 |
| [6] Val/V GUG 2.81 | 7 [27] Gly/G GGG 1.65 | 12 [48] His/H CAU 1.09 |
| [7] Ala/A GCC 2.77 | 8 [28] Ile/I AUU 1.6 | 12 [49] Gly/G GGU 1.08 |
| [8] Asp/D GAC 2.51 | 8 [29] Ala/A GCA 1.58 | 12 [50] Cys/C UGU 1.06 |
| [9] Lys/K AAA 2.44 | 9 [30] Tyr/Y UAC 1.53 | 12 [51] Arg/R CGC 1.04 |
| 2 [10] Gly/G GGC 2.22 | 9 [31] Ser/S UCU 1.52 | 13 [52] Leu/L UUA 0.77 |
| 2 [11] Met/M/Start AUG 2.2 | 9 [32] His/H CAC 1.51 | 13 [53] Ile/I AUA 0.75 |
| 2 [12] Asp/D GAU 2.18 | 9 [33] Thr/T ACA 1.51 | 13 [54] Ala/A GCG 0.74 |
| [13] Ile/I AUC 2.08 | [34] Val/V GUC 1.45 | 13 [55] Leu/L CUA 0.72 |
| [14] Phe/F UUC 2.03 | 10 [35] Trp/W UGG 1.32 | 13 [56] Val/V GUA 0.71 |
| 3 [15] Pro/P CCC 1.98 | 10 [36] Leu/L CUU 1.32 | 13 [57] Pro/P CCG 0.69 |
| 3 [16] Leu/L CUC 1.96 | 10 [37] Thr/T ACU 1.31 | 14 [58] Arg/R CGA 0.62 |
| 3 [17] Ser/S AGC 1.95 | 10 [38] Leu/L UUG 1.29 | 14 [59] Thr/T ACG 0.61 |
| 4 [18] Asn/N AAC 1.91 | [39] Cys/C UGC 1.26 | 15 [60] Arg/R CGU 0.45 |
| 4 [19] Thr/T ACC 1.89 | 11 [40] Gln/Q CAA 1.23 | 15 [61] Ser/S UCG 0.44 |
| [20] Ala/A GCU 1.84 | 11 [41] Ser/S UCA 1.22 | [62] STOP UAG 0.16 |
| 5 [21] Ser/S UCC 1.77 | 11 [42] Tyr/Y UAU 1.22 | 16 [63] STOP UAA 0.1 |
| 16 [64] STOP UGA 0.08 |
It is noteworthy that when comparing the actual usage percent of the genetic code instead of the most used codons per amino acid, we found that amino acids that only have two codons dominate the higher percentages. For example, Gln GAA occupies the 5th place in abundance below Leu CUG, Glu GAG, Gln CAG (its brother codon), and Lys AAG. Leu CUG and Glu GAG, the leaders, are represented with the same dominant 3.96%. The next one that also has two codons, Lys AAA, appears just below the amino acids: Val GUG, Ala GCC and Asp GAC in position number 9 (with its sister codon Lys AAG occupying the 4th position). Then, it is Asp GAU which appears in position number 12 (with its cognate codon Asp GAC occupying the 8th position), just below Gly GGC and Met AUG codons. Leu CUC appears, after Ile AUC, Phe UUC, and Pro CCC, in position number 16 (with its cognate codon Leu CUG sharing the 1st place) (Table 2). There is a cluster of 10 codons before the most used Tyr (Y) UAC that appears in position 30. These observations demonstrate that the most used codons per amino acid are not the same as the actual set of most used codons in the averaged nucleotides (by 1Ks) from the complete codifying genome. His CAC is in position 32, Trp UGG in 35, Cys UGC in 39, while the two Arg: AGA and AGG occupy the global positions of 44 and 45, respectively.
In Table 2 we also see 16 clusters when two contiguous members in the cluster are separated by 0.02% from each other, meaning that those codons are more or less used with a similar intensity, ranging from two to six, i.e., Leu CUG and Glu GAG, both at the top of the list (cluster number 1), with an identical frequency of 3.96%, while Leu UUA, Ile AUA, Ala GCG, Leu CUA, Val GUA and Pro CCG, range from 0.77% to 0.69% (cluster number 13).
Table 2 also shows that the three most used codons for hydrophobic amino acids are: Leu CUG 3.96%, Val GUG 2.81%, and Ala GCC 2.77%; the three most used codons for acidic amino acids are: Glu GAG 3.96%, Glu GAA 2.9%, and Asp GAC 2.51%; the most used codons for basic amino acids are: Lys AAG 3.19% and Lys AAA 2.44%, while the most used amides are: Gln CAG 3.42% and Asn AAC 1.91%, etc. On the other side, the three less used codons for hydrophobic amino acids are: Val GUA 0.71%, Leu CUA 0.72%, Ala GCG 0.74%; the three less used codons for basic amino acids are: Arg CGU 0.45%, Arg CGA 0.62%, Arg CGC 1.04%; the less used codons for signal-transduction related amino acids are: Ser UCG 0.44%, Thr ACG 0.61%; globally, the less used codons are the stop codons: UAG 0.16%, UAA 0.1%, UGA 0.08%, etc.
If we stop a minute to consider dinosaurs that died in the first, coming from the outer space, water flood and glacial age, the 2008 NOVA program entitled “Arctic Dinosaurs” presented some results related to two places from the North Slope of Alaska:
the Liscomb Bonebed, and
the Colville River, describing fossils of dinosaurs found in the Arctic pole, including: Pachyrhinosaurus, Gorgosaurus, Troodon, Thescelosaurus, and Edmontosaurus, a Hadrosaur, “with >6000 specimens collected”, assemblage that “resulted from mass mortality” (Gangloff and Fiorillo, 2010), and they even found evidence for the presence of a Pterosaur (Fiorillo et al., 2009).
Related findings have been made in the Russian Arctic, where we may add the Dromaeosaurid and Ankylosaurian teeth (Godefroit et al., 2009); these authors included pictures of fossilized specimens of tropical and subtropical plants: Ginkgo ex. gr. adiantoides, Elatocladus smittiana, Sequoia minuta, Encephalartopsis vassilevskajae, Nilssonia serotina, Cissites kautajamensis, Carpolithes ceratops, Corylus ageevii, Celastrinites septentrionalis, Platanus rarinervis, Trochodendroides grossidentata, etc.
These plants and other pieces of evidence are indicative that at the highest latitudes in the far past, the climates were warm enough, possibly with a more even day length through the whole year (Douglas and Williams, 1982).
In the Antarctic, fossils have been found for at least 21 dinosaur groups (including the Leaellynasaurus mentioned in NOVA), plus birds (‘aves’), amphibians, crocodilia, turtle, marsupialia, monotremes, multituberculata, tritylodontid, placentalia (?), etc… (Rich et al., 2002); there, we read: “the large size of some of its trees [fossilized in southeastern Australia] far exceed that found in such cold environments today”.
Next we will read some NOVA's “Arctic Dinosaurs” excerpts:
“Picture yourself standing on this lake shoreline. It's warm, sunny; there are herds of duck-billed dinosaurs and other kinds of plant-eating dinosaurs along the lake, eating vegetation like horsetails, gingkoes, the flowering plants; an abundance of flying insects and crawling insects on the shoreline…” Steve Hasiotis.
“… we have at least 15 of these, from this particular quarry. It's as if somebody took 15 Pachyrhinosaurus and dumped them in a blender for 30 seconds and then poured all the mess out into a large batch of concrete and let it solidify… Everything is completely jumbled about. Bones are oriented in all sorts of different directions, and nothing's connected” Ron Tykoski.
“This is the most diverse assemblage of fossil…of dinosaur bones here on the North Slope. It's got a variety of kinds of animals… There's 14 individuals of the horned dinosaur (Pachyrhinosaurus) in this little pit so far… A density of bone so high that it's easy to envision this being a jam… a bone jam… It challenges everything that we think we know about dinosaurs” Anthony Fiorillo.
“I don't see [in the Hadrosaur] the growth rings that I would have expected in an animal that would have been affected by seasonal climatic conditions… this animal was very well adapted to the environment in which it lived” Anusuya Chinsamy.
After the first glacial age (Custance, 1989), the planets and their satellites started moving, while the stars in the sky, including our sun, were ‘turned on’ once more (Bullinger, 1893, 1922; Seiss, 1882; Rolleston, 1862; Wierwille, 1982), while life emerged and diversified, culminating with men (Stock, 1878; Schoenheit, 2004)http://fdocc.ucoz.com/fdocc-eternity.pdf. After the second and partial glacial age/global flood, there was a dramatic reduction in the human population. Here, a consanguineous genetic bottleneck, with its consequent health and longevity-reduction problems, is proposed, maybe for the loss of some alleles (Hunter and Gibbs, 2007).
Evidence from the second polar or partial glacial age, coupled to a global flood, is the study of human petrified bones, where minerals replaced bone tissue; examples from Texas were taken by the author (Fig. 2).
Figure 2.

Images of what appears to be petrified human bones. A1 and A2 show what seems to be a petrified skull covered with a thin and brown peeling layer of what seems to be petrified skin; A2 also shows the detail of a Y-shaped crack in the skull, between the 11 and 11.5 inches mark. B1 and B2 show a petrified tooth and what seems to be bone and medulla remains of a petrified lower leg, the red color within the center of the bigger piece of medulla and of its surroundings indicates a high presence of iron. B1 shows the petrified tooth between the marks of centimeters 2 and 3.5, while B2 shows the bigger petrified medulla between centimeters 4 and 6. These images were taken in Texas.
Evidence of both glacial ages and of waters arriving from outside the Earth to drawn it, remaining on it most specially the ones from the second flooding, I think, are the waters found in the dark side of the moon (Colaprete et al., 2010), where its dark pole is located (Duke, 2002), and in the south pole of mars (Mackenzie, 2002), plus its dried rivers and lakes (Kerr, 2000), water in Io (Salama et al., 1990), and in other moons of Jupiter and of other planets (Sammonds, 2006), and both in the Kuiper belt (Jewitt and Luu, 2004), and in the Oort comet cloud (Hsieh and Jewitt, 2006) in the outermost part of the Solar System mostly composed of gigantic frozen ice-cubes like Eris (Bertoldi et al., 2006; Sicardy et al., 2011), Ceres (Thomas et al., 2005), Pluto (Ridpath, 1978), now removed from the status of planet (Spahr, 2006), and many others (Grundy et al., 2009); H2O also in the colorful nebula/nebulae (Miranda et al., 2001) and in comets (Meier et al., 1998), meteors/meteorites (Jenniskens and Mandell, 2004), asteroids (Rivkin and Emery, 2010), and regoliths (Seshadri et al., 2008), etc., being the water in them the one that evaporates, forming their characteristic long comet-tails, making them smaller and smaller every time… As a matter of fact, the presence of water all over the outer space beyond our atmosphere is so overwhelming that it deserves its own and careful review.
Even when several more regional or localized ice periods may have occurred, two appear to be the most prominent world-wide, with the last one still ongoing on the poles of the Earth.
Due to their importance to understand fossils and strata, a group of quotations related to those two historical and global glacial ages will be posted here (Hambrey and Harland, 2011):
“In the Broken Hill area of New South Wales, two glacial periods are represented... In the Amadeus and Ngalia basins of central Australia evidence of two main glacial periods is again preserved... Further to the northwest, in the Kimberley area, the same picture of two main glacial periods prevails...” (p. 945);
“The two glacial periods represented in East Greenland...” (p. 947);
“Divided into two glacial substages” (p. 415);
“The palaeogeographic setting of the two glacial valley sequences... is not well-understood. At Alligator Peak...” (p. 235);
“In Shaba (Zaire) two glacial events...” (p. 944);
“Yancowinna Sub-Group... may indicate two glacial events, the youngest event being more intense” (p. 536);
“Two glacial episodes can be recognized in the stratal record of the Yudnamutana Subgroup” (p. 541);
“Nearly all workers agree that the two tillite divisions are evidence of two glacial episodes separated by an interglacial period...” (p. 780);
“They indicate a recurrence between two glacial phases of a less severe climate more favourable to life” (p. 103);
“In central Africa... two glacial episodes are recognized, the second being characterized by several short fluctuations in position of the ice front” (p. 81).”
Flint (1947) declared:
“That Iceland yields evidence of at least two glacial stages has long been known.” (p. 347).
“In Severnaya Zemlya (Siberia) two stages are recorded …in the Chukotsk Peninsula two stages, of which the later is identified with the Wisconsin stage in Alaska …and at least two stages in the Sayan Mountains” (p. 360).
“In the Caucasus region two glacial stages are established.” (p. 363).
“In several parts of the South Island (New Zealand) two glacial stages have been identified on evidence furnished by moraines and outwash.” (p. 376).
“In Indian Tibet the Pang-gong Tso and Pangur Tso basins contained contemporaneous lakes during at least two glacial ages, the later generation having been smaller than the earlier.” (p. 480).
“The two glacial stages on the high mountains of East Africa are correlated with the Last Pluvial and Great Pluvial stages.” (p. 480).
“…in the mountains of western United States only two glacial expansions of considerable magnitude occurred during the Wisconsin age…” “the other expansions recognized in Central North America are not represented in the west”, view “advocated by Antevs (1945)… The several Wisconsin drifts reported from the Colorado Rockies do not necessarily carry weight… it has not been demonstrated that all of them have substage value. Some may represent glacier fluctuations of lesser magnitude.” (p. 301).
Additional references dealing with the two main or most important historical ice ages are:
“In no arctic locality has clear evidence of more than two glacial ages yet come to light… There is clear evidence of at least two glacial ages in the frozen ground in the Yukon River basin in central Alaska.” (Flint, 1953).
“Concerning the ice ages, the majority of the studies (de Martonne, 1907; Phleps, 1914; Niculescu et al. 1960) bring arguments in favour of the two glacial ages, Riss, when the maximal expansion is reached, and Würm, of less intensity (Valsan river and Fagaras Mountains, Romania).” (Marcu, 2011).
“The two glacial ages of particular importance to the physiographic development of Ogle County were the Illinois Episode and the more recent Wisconsin Episode” (Ogle County Planning & Zoning Department, 2008).
“Francois Matthes, in an interesting “detective story” written in the form of a geological essay, discusses Moraine Dome extensively. He deduced, using three examples, that the moraines around the dome were the product of at least two glacial ages — a notion contrary to the thinking of the time. The morainal till of the last glacial age characterizes the underfooting of the descent into Little Yosemite Valley.” (Morey et al., 2005).
“The California marine section calls for two glacial ages before the deformation and uplift…” (Grant and Gale, 1931).
“Some evidence of two glacial stages in the Klamath Mountains in California… the older glacial deposits and the associated rock gorges… represent a distinct stage of glaciation of a pre-Wisconsin age.” (Hershey, 1903).
“…the simplest hypothesis of age requires two glacial ages to account for the facts.” (Deevey, 1949; see more recently, Cooper, 2002), etc.
If we move back to the living systems, we can see that recently, Svante Paabo dodged twice the “species” question by declaring “I think discussion of what is a species and what is a subspecies is a sterile academic endeavour”, “why take a stand on it when it will only lead to discussions and no one will have the final word”.
However, Svante's colleague and co-author Jean-Jacques Hublin stated that many consider Neandertals a species separate from modern humans because the anatomical and developmental differences are “an order of magnitude higher than anything we can observe between extant human populations”, and that Mayr's “species” concept doesn't hold up because “there are about 330 closely related species of mammals that interbreed, and at least a third of them can produce fertile hybrids” (Gibbons, 2011); however, if we include birds, reptiles, amphibians, fishes, insects, plants, etc. the number will increase tremendously.
The 100% of the original components of petrified bones, such as proteins and nucleic acids (DNA and RNA), were completely replaced with the minerals present in the soil and salty waters where they were deposited; something similar happened to petrified wood, but being the bones thinner and more fragile, it is well known that the original shape of petrified bones is easily distorted by the surrounding humidity and changing temperatures of the soil.
Sometimes, the polymorphisms can experience a non-random reversion by a process currently barely understood (Castro-Chavez, 2012a); even harmful mutations after the overload of the quality control mechanism of cells and organisms seem to follow a non-random pattern concentrated in the mutational “hot-spots” (Volpato and Pelletier, 2009; Perrot et al., 2009; Namekawa et al., 2006), a process that seems to be reversible under certain conditions (Zhou et al., 2003).
The genomic variation to adapt to a changing environment is a slow process that starts working selflessly for at least two generations before the inheritance of a more adaptively advantageous polymorphism, perhaps similar to the time it takes for a hereditary disease to appear in the offspring once the damage has been done in the ancestors (Fig. 3).
Figure 3.
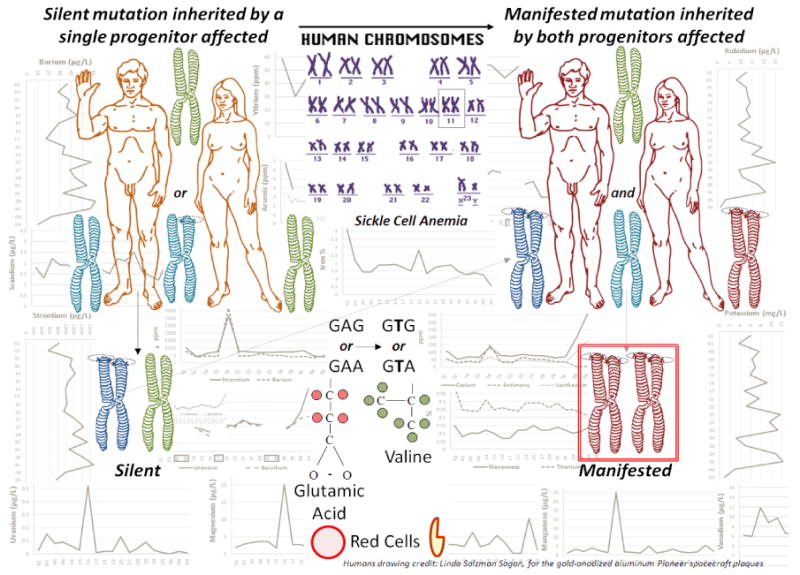
Representation of the origin of hereditary diseases initially affecting the gametic genome of only one progenitor; it starts by a silent damage on nucleotide(s) of one single arm of one chromosome; however, if inherited, it will be duplicated to both arms of such first generation chromosome, but it will continue being silent, even if transferred to the offspring, being covered by the still healthy genes of an unrelated spouse; it will only be manifested by consanguinity, if close relatives of the first affected donor intermarry, i.e., cousins; also people affected by the same and extreme contaminants increase the risk of manifesting hereditary diseases in earlier generations (i.e., Chernobyl, Fukushima, the 9/11 World Trade Center (WTC) at N.Y., with graphics in the background discussed in detail by Castro-Chavez (2012a), etc.) The example shows the sickle cell anemia, but the principle illustrated here applies to any hereditary disease and maybe as well to the beneficial adaptive changes. Credits: Humans drawn by Linda Salzman Sagan, for the gold-anodized aluminum Pioneer spacecraft plaques.
Fig. 3 illustrates the possible origin of hereditary diseases, a process in need of several generations to express itself, apparently similar to the time needed for selfless and favorable adaptations to appear; the background of the figure shows the pattern of atomic expression on the 9/11 World Trade Center (WTC) toxic dust.
If we focus here only in strontium (Sr) and barium (Ba), we will discover that, in at least three different studies, the coordinated pattern of Sr contamination showed to be more abundant than Ba, starting with the analysis using two different methodologies performed by the U.S.G.S., where an average in the soluble leachate of 1.1 mg/L for Sr, and of 34 μg/L for Ba was found; here, Ba represented only 3 % when compared to Sr being 100%; X-ray fluorescence was also performed on the dry dust, finding a coordinated 726.6 ppm of Sr and 533.38 ppm of Ba, representing 73.4 % when compared to Sr (100%) for this methodology (Clark et al., 2001); to see the six mishandlings of the 9/11 WTC reporting of strontium (Sr) by the U.S.G.S., go to: http://www.webcitation.org/63YyJEzsi
A second study that analyses only three samples, found the next averages (in mg/g dry weight): 630.2 mg/g for Sr and 380.6 mg/g for Ba, which represented the 60.3 % when compared to Sr (100%) for this methodology (Lioy et al., 2002); here, beta radiation was twice the background level in these few samples of the 9/11 WTC toxic dust analyzed. Jenkins called into question the 9/11 WTC pH values reported by this group (Jenkins, 2007), one of the groups against which she raised her complaint.
A third study analyzing eight and five 9/11 WTC dust samples by two different methodologies corroborated in general the patterns presented by the U.S.G.S. analysis; i.e., the average values for the eight samples (mg/g of solid sample) was of 0.7 mg/g for Sr and 0.2 mg/g for Ba, which represented the 28.6 % when compared to Sr (100%) (McGee et al, 2003). These authors wrote that “further detailed analyses and toxicity testing of the WTC PM (Particulate Matter)2.5 fraction may be necessary to evaluate other components found in the bulk WTC fallout dust, which may be enriched in the smaller inhalable particle sizes, and which have known long-term health consequences. WTC PM that infiltrated into buildings is most likely of smaller particle sizes and more of an inhalation hazard than that found outdoors” (McGee, 2003). The discrepancy between the different studies may be due to the different methodologies used and/or also to a possible decay due to different timing in the analyses.
In my last minute update, I found that Miller (2012) presented his complaint related to the sloppiness of the most recent pulmonary 9/11 WTC research done by the "WTC Medical Monitoring and Treatment Program (MMTP)" centered at Mount Sinai on the 9/11 first responders: "It is disappointing that authors from an institution so wealthy in both WTC and sarcoidosis patients have not provided fuller information on their cases of SLGPD ["Sarcoid Like" Granulomatous Pulmonary Disease ]", and even the scant pulmonary function testing done in their 38 reported cases "was highly deficient and limited to spirometry (itself lacking in a third of the patients)", additionally, "decreased exercise tolerance and its equivalent, shortness of breath, were not graded or described any more specifically. No mention was made of indications for or response to therapy". Meanwhile, Kahn et al (2011) compared the poor reproducibility for a superficial 9/11 dust analysis based on the measurement of the concentration of slag wool fibers, considered to be an indicator of WTC dust, finding that in the five laboratories deemed as qualified for such analysis, "their ability to distinguish between the batches was somewhat mixed"; and finally, Naik (2011) published on the disturbing high lack of reproducibility within the biomedical field: 64.2% publications were non-replicable, 11.9% were partially replicable, and only a 20.9% of research studies were fully replicable [http://gcalhoun.files.wordpress.com/2011/12/11-12-02-wsj-scientists-elusive-goal-e28093-reproducing-study-results.pdf]; one of the main reasons for such lack of reproducibility could be partly attributed to the inhumane pressures exerted by the grant-seeking PIs over their students, which I think corners the honest researchers on trying to find a different activity for their lives.
On Sept. 11, 2001 (9/11) at the WTC, N.Y., the earliest explosion apparently happened underground at 08:46:26 AM and was registered (ML = 0.9) by the earthquake monitoring station in Palisades, N.Y. (Kim et al, 2001). Mike Pecoraro and his colleague, who were below the site of this underground explosion, remembers ascending from where they were to the basement's C level seeing the equipment, including a 50 Ton hydraulic press, reduced to rubble, while their co-workers were practically ‘evaporated’ (Vito Deleo and David Williams); “as they ascended to the B Level, one floor above, they were astonished to see a steel and concrete fire door that weighed about 300 pounds, wrinkled up “like a piece of aluminum foil” and “lying on the floor” (Pecoraro et al, 2001).3 By comparison, the 1993 explosion in the basement's parking garage under WTC1 was not detected by any earthquake monitoring station (Tahil, 2006); according to this author, the concentration of Zn had a direct linear relationship to the concentration of Ba, lead (Pb), and other elements.
The timing for the other 9/11 underground explosions as reported at Palisades, N.Y., are 09:02:54 (ML = 0.7), 09:59:04 (ML = 2.1, before or at the fall of WTC2), 10:28:31 (ML = 2.3, before or at the fall of WTC1). The WTC ground shook before the fall of both twin towers [http://www.webcitation.org/63YyikkM2], and at 11:01:07, 11:15:04, 11:29:46…, etc., … 5:20:33 (ML = 0.6), being this one very important because this timing was just immediately before the fall of WTC7, just when the east mechanical penthouse fell to the ground [http://www.ldeo.columbia.edu/LCSN/Eq/20010911_wtc.html], a precise timing also mentioned by FEMA's McAllister et al. (2002). The WTC7, also called the ‘Salomon Brothers’ building, had its east Penthouse falling first all way down through the core few seconds (5 sec?) before the fall of the complete building, also in a matter of seconds [http://www.youtube.com/watch?v=iOLIlKHDd78].
“Much higher subsurface temperatures were inferred from the lower portions of removed steel beams glowing red. The sub-surface of the collapse piles remained hot for months despite use of massive amounts of water to cool them, with the last spontaneous surface fire occurring in mid-December” (Cahill et al, 2004); Cahill also declared in the same article that such extreme heat was “shown by the melting of metals”. In another article, he said that “the air from Ground Zero was laden with extremely high amounts of very small particles, probably associated with high temperatures in the underground debris pile”; “normally, in New York City and in most of the world, situations like this just don't exist” (C. A. Editors, 2002). Those volatile elements produced for months in the WTC smoldering pit were persistent and more dangerous than the ones present in the 9/11 dust when the two towers fell (Cahill, to Jenkins, 2007). The 9/11 WTC toxic dust sickening trajectory of Doug Copp, CEO of the “American Rescue Team International” has been documented by himself.4
A “toxic eyesore” that was in front of WTC7, the “Fiterman Hall, contaminated with WTC dust” (Kaysen, 2005a), was later demolished; also demolished was the Deutsche Bank (WTC4) at 130 Liberty St. (Brion, 2006) and at four (4) Albany St. (Kaysen, 2005b), which were “two 9/11-damaged structures on the south side of the WTC”; “…a negative pressure region is maintained inside the building to contain any contaminated dust while the area is cleared”; workers under such extreme hazards wore haz-mat suits (Brion, 2006); however, very few select persons did wear those suits at the WTC on 9/11 (Finkelstein, 2001; Lombardi, 2006).
Several of the 9/11 “meteorites” have been presented [http://www.youtube.com/watch?v=hWHvnprvSbc http://www.youtube.com/watch?v=1WWAnCd_6lg], some of them seem to be made of iron and concrete, like chunks of ‘lava’ solidified when the temperatures dropped. “One of the oddest shapes is called the compression. It's so valuable that it's been locked inside a tent within the hangar. It looks like a meteorite and nobody knows which tower it came from says Charles Gargano, vice chairman of the Port Authority of New York and New Jersey, which owns the hangar and the Trade Center Site. Gargano says the compression is actually 4 stories that have been crushed into a jagged object four feet high” (Fertig, 2006).5 Rich Garlock declared that in WTC 6 the debris past the columns was red-hot, molten and “running” [http://www.webcitation.org/63YzRIRlu, http://www.webcitation.org/63Yzcgmed]; molten concrete engulfed metal materials, guns, etc.
“Sample one (From WTC 7) showed “evidence of a severe high temperature corrosion attack on the steel, including oxidation and sulfidation with subsequent intergranular melting”, “readily visible in the near-surface microstructure”. Sample two (From WTC 1 or WTC 2) showed that its “thinning of the steel occurred by high temperature corrosion due to a combination of oxidation and sulfidation”. “The severe corrosion and subsequent erosion of Samples 1 and 2 are a very unusual event. No clear explanation for the source of the sulfur has been identified”, “a detailed study into the mechanisms of this phenomenon is needed” (Barnett et al., 2002). However, “for more than three months, structural steel from the World Trade Center has been and continues to be cut up and sold for scrap”, “such crucial evidence is on the slow boat to China, perhaps never to be seen again in America until you buy your next car” (Manning, 2002).
These important pieces of evidence for the use of highly energetic materials to destroy the WTC on 9/11 were removed and/or destroyed, even when the extreme human damage to the exposed individuals is still present today (Castro-Chavez, 2012a), not only through the bio-available and soluble Sr but also through tiny carbon nano-tubes (Wu et al., 2010). A special foaming agent called Pyrocool FEF was used to “absorb the high-energy emissions”, and to “re-emit the energy at a longer, lower-energy wavelength”; “One target was the large Freon tanks that had served the WTC air-conditioning system and might have exploded” (Beard, 2001).”
For the critical consideration of the reader, I leave the next: At Google images, while searching for the words: 40ft pothole 9/11 WTC, I found a fascinating conformation that was carelessly destroyed (see the left side of the ‘pothole’ that was facing the U.S. Post Office building): http://www.webcitation.org/63QAeQMSu (Dunlap, 2008a). The formation closer-to-the-surface was found near to where the WTC2 stood, flanking the glacier ‘pothole’ that was under the Deutsche Bank (WTC4) that was located at the right side of WTC2. In the next image, we see the destruction of this peculiar structure similar to ‘Inwood marble’, a sugar-like crystal with white or blue-grey colors. Due to its high light-reflectance, some have called it the 9/11 “manhatite”: http://www.webcitation.org/63QAteMPn here seen in the opposite view to the previous image (Dunlap, 2008b). The now disappeared ‘pothole’ was deemed to be the product of the ‘Ice Age’ by Cheryl J. Moss and Anthony Pontecorvo, from ‘Mueser Rutledge Consulting Engineers’ (mrce.com); their news report ended with their statement that: “The pothole and other features are being covered, filled in or blasted away. ‘It’s nice to look at, but it’s all got to go’” (Reina et al., 2008).6 Next, saved images of the last steps in the fast destruction of this 9/11 ‘manhatite’ formation, a part of a unique and highly valuable, yet destroyed, rock formation under the WTC: http://www.webcitation.org/63SjjeA5F , http://www.webcitation.org/63Su9rRVw , http://www.webcitation.org/63QBQCq6C (WTC4 image mislabeled in its original website as WTC3). This is but a brief recount of 9/11 and its damaging health implications for the first responders (Castro-Chavez, 2012a), In Memory of Lynn Margulis (Barrett, 2011a,b). More diverse details can be found at: Khalezov (2011a,b,c).
The vital role of the relatives of the victims has been blotted out from history, even from the TIME magazine’s timeline: http://www.time.com/time/magazine/article/0,9171,2092218-3,00.html, the same as the omission of the WTC7 falling to its footprint at 5:20 PM on 9/11: http://www.time.com/time/magazine/article/0,9171,2092218-1,00.html (?), and from TIME’s 9/11 tenth anniversary number [Beyond 9/11, Special Commemorative Issue, Sept. 19, 2011, 64 p.], among other omissions, i.e., Architects and Engineers for Truth, advanced aircraft flying before, during and after the 9/11 attacks (i.e., E-4Bs, Gulfstreams, advanced choppers and UAVs, etc.), the nuclear atomic profile of the dust (Castro-Chavez, 2012a), the 9/11 scripted beforehand TV news, the active involvement of foreign intelligence operatives, as well as the important role of proficient writers dissenting of the official story (i.e., David Ray Griffin, Mark H. Gaffney, Christopher Bollyn, etc.), etc. See here two movies made by those 9/11 suffering relatives, now victims of oblivion: “Press for Truth”: http://www.youtube.com/watch?v=iMT2CHSvyGw,“In Their Own Words”: http://video.google.com/videoplay?docid=4399917864007973679
Recently, I found that a high similarity in the frequency of two nucleotides of the LDLrap-1 gene was present between families with Mexican and Japanese ancestry (Castro-Chavez, 2012a); archaeological findings of figurines of men sculpted over jade by the Mexican Olmec culture agree with the statistical findings: http://fdocc.ucoz.com/olmec-eyes.pdf
Between the extremes of global fitness (Bergeron, 2011; Maslen and Mitchell, 2006; Edwards et al., 1986; Custance, 1980; Talley, 198?; Wierwille, 1980) and, as Hublin above said (Gibbons, 2011), of an unfitness barely human (Culotta, 1999; Mayell, 2003; Underdown, 2008)http://www.webcitation.org/63YujQxF9, most normal and natural humans will fall near the center of a Gaussian graphic.
In relation to the gene flow, I found recently the clip: “Human Degeneracy into Neandertals, The Forgotten Gene Flow” [http://youtu.be/Osw1ChnSlhY], where we hear David Reich: “…The other explanation is ancient structure… the human population was sub-structured into three groups… exchanging genes but quite differentiated, like human populations today, and then what happened is: Population one went off and became Neandertals…” His article says: “Although we detect gene flow only from Neandertals into modern humans, gene flow in the reverse direction may also have occurred. Scenario 4 represents old substructure in Africa that persisted from the origin of Neandertals until the ancestors of non-Africans left Africa. This scenario is also compatible with the current data” (Green et al., 2010).
Now, if we apply the codon usage logic per genome to other organisms: chimpanzee, dolphin, elephant, llama and Canis (Castro-Chavez, 2012a), and to cow, mouse, hen, fruit fly, zebrafish, etc., we obtain similar positional results for the global frequency of codon usage (average of codifying sequences with a length of 1K) according to the circular genetic code. The relative proportions for each codon change dramatically; however, the proportional global abundance of codon usage, when compared to humans, is maintained with only slight exceptions characteristic of the organism being studied.
For example, when analyzing the chimpanzee (Pan troglodytes), the relative usage of codons remained the same as in humans, except that chimpanzee had almost the same most used codon percent per amino acid for two Ser codons: Ser UCC (24.07%), and Ser AGC (24.2%), being the last one the most used in humans, having these most used codons for Ser in chimpanzee a minimum difference of 0.13% (the ratio of the analyzed human/chimpanzee codons was 40,662,582/328,023).
The dolphin (Tursiops truncatus) had a very striking similarity to the most used codons in humans per amino acid, being the only minor difference the dominant codon for Arg (AGG 27.51%), while the next ‘Args’ had almost the same frequency: Arg AGA 18.34%, and Arg CGG and Arg CGC, both with a 17.69% usage, etc. (the ratio of the analyzed human/dolphin codons was 40,662,582/6,771).
A similar finding was done in elephants (Elephas maximus). The dominant codon for Arg also was AGG 28.27%, while the next ones were: Arg CGC 27.23%, Arg CGG 19.63%, and Arg AGA 13.61%, etc. (the ratio of the analyzed human/elephant codons was 40,662,582/2,124).
Similarly, the llama (Lama glama) had only one minor change in Arg when compared to the most used codons in humans per amino acid: The most used Arg in llamas was AGG 32.71%, followed by Arg AGA 23.06%, etc. (the ratio of the analyzed human/llama codons was 40,662,582/3,871).
The difference for most used codons in mouse (Mus musculus) when compared to humans, is that the most frequently used codons for Pro are CCU (30.62%) and CCC (30.28%), which is a small 0.34% difference, being the last one the most used codon for Pro in humans and dogs (the ratio of the analyzed human/mouse codons was 40,662,582/24,533,776).
The slight exception for the dog (Canis familiaris) was that its two most frequently used codons for Arg are: AGG (as in humans), but also Arg CGG, instead of Arg AGA, which is the one we find as the most used in humans. For the most used codifying codons for Arg in dogs, the percentage differences are as follows: Arg AGG 21.06%, Arg CGG 20.87% (a small difference of 0.19%), Arg CGC 20.11%, Arg AGA 19.92%, etc.; being the most used, the ones ending in a three hydrogen bond nucleotide (G or C), being the difference among each of them of less than 1% (the ratio of the analyzed human/dog codons was 40,662,582/559,501).
The wolf (Canis latrans), genetically compatible to the dog (Castro-Chavez, 2012a), had as its only difference the main preference for Arg CGC 29.38%, when compared to dogs or to humans, being followed by Arg CGG 23.54%, both ending in a three hydrogen bond network (the ratio of the analyzed dog/wolf codons was 559,501/2,710).
Similar to wolves, the difference of the most used codons in pigs, when compared to humans, was in the most used codons for Arg: CGC 21.84%, Arg CGG 21.48% (a small 0.36% difference), Arg AGG 20.58% and Arg AGA 18.59% (the ratio of the analyzed human/pig codons was 40,662,582/1,168,059).
The difference in cows (Bos taurus) was that the most used codon for Arg is CGG, instead of either AGA or AGG, which are the equally most used codons in humans for Arg. The percentage difference for the most used Arg codons in cows was: Arg CGG 22.05%, Arg AGG 20.11%, Arg CGC 19.58%, Arg AGA 18.87%, etc. (the ratio of the analyzed human/cow codons was 40,662,582/5,198,458).
In the hen (Gallus gallus), there was again a very slight difference in the Arg codon usage, where the most used codon was AGA 22.30%, followed by AGG 21.39%, with a percent difference <1% among them (the ratio of the analyzed human/hen codons was 40,662,582/2,719,057).
It was surprising to find, while studying the most used codons per amino acid in the bread mold (Neurospora crassa), that its only differences when compared to humans were: Val GUC 41.68%, Leu CUC 32.33%, Arg CGC 28.48%, and Ser UCC 24.48%. Notice that even when the most used codons are different, they preserve the three hydrogen bond nucleotide C at the end (The ratio of the analyzed human/bread mold codons was 40,662,582/2,048,035).
The blue whale (Balaenoptera acutorostrata) exhibited differences when compared to the most used codons in humans in two amino acids: Ser UCC 30.43%, followed by Ser AGC 26.28% (the most used by humans), and in Arg, where the most used codon was Arg CGG 21.92%, followed by an approximately similar percentage for Arg AGA 21.14% and Arg AGG 20.74% (a difference of 0.40%, a small difference for the two that are most used in humans). The most used stop codon in whales was UAA 56.52% (the ratio of the analyzed human/blue whale codons was 40,662,582/4,726).
The fruit fly (Drosophila melanogaster) also exhibited differences with the most used codons in humans for the next codons, corresponding to two amino acids: Asp GAU 52.87% and Arg CGC 32.85%, Arg CGU 16.06%, Arg CGA 15.33% and Arg CGG 14.96%, etc. (the ratio of the analyzed human/fruit fly codons was 40,662,582/21,945,319).
Due to the extremely high amount of fruit fly varieties in the Nakamura's (2000) database when compared to any other organism (in its 2007 update), we explored in more detail their differences.
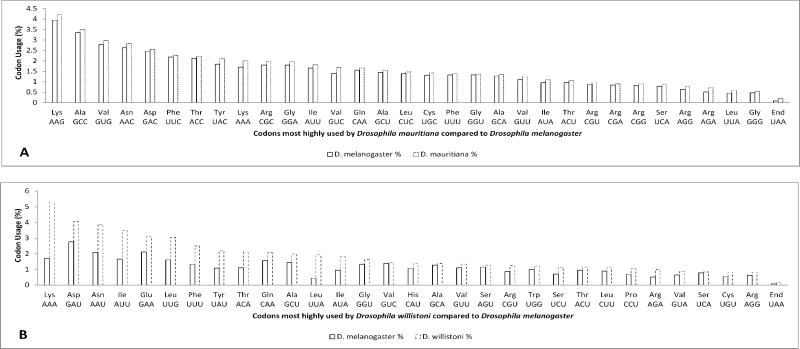
When the total usage of codons for the genomes of fruit fly varieties was compared, the ones expressing the less difference were: D. melanogaster and D. mauritiana (average of standard deviations: 0.10), while the highest difference in usage was between D. melanogaster and D. willistoni (average of standard deviations: 0.48), see Figure 4.
Figure 4.
Codons most frequently used by Drosophila mauritiana (A) and by Drosophila willistoni (B) when compared to Drosophila melanogaster. D. melanogaster is the fruit fly with most codons in the database. A. D. melanogaster is compared to its closest match for total codon usage: D. mauritiana (averaged standard deviation per codons (ĀSD): 0.10 for this comparison of its 32 most expressed codons). B. D. melanogaster is compared to its most distant match for total codon usage: D. willistoni (ĀSD: 0.48 for this comparison of its 31 most expressed codons).
The only amino acids that did not increase their total codon usage in D. mauritiana when compared to D. melanogaster were Met, Pro, His, Trp, Glu, and Ser in its AG_ side, some of the most functionally unique or specialized amino acids (Met starts, Pro bends and His catalyzes). All the amino acids were increased in their total codon usage for D. willistoni except for Met, the protein starter. When comparing D. melanogaster to D. mauritiana and to D. willistoni for the unique codon which codes for Met, the next were their percentages: 2.361%, 2.365% and 1.895%, respectively.7
In Fig. 4, the ratio of codons analyzed for D. melanogaster/D. mauritiana was 21,945,319/27,319, while the ratio for D. melanogaster/D. willistoni was 21,945,319/43,315. Interestingly, the Student's t-test comparison gives p = ∼0.5 when the percentages for all the 64 codons are compared for both the examples presented in Fig. 4A and Fig. 4B; however, when the Student's t-test of all the 64 codons are compared for only the standard deviations, the p = 1.22 × 10-9. However, when only the codons presented in images Fig. 4A and Fig. 4B are compared, the Student's t-test for Fig. 4A (32 codons) provides a p = 3 × 10-12, while for Fig. 4B (31 codons), a p = 7.7 × 10-6. The Student's t-test for the comparison of standard deviations for the first 31 codons of images Fig. 4A and Fig. 4B resulted in a p = 7.6 × 10-5 (excluding the Stop codon UAA of Fig. 4A to make an even comparison).
Moving on to an aquatic vertebrate, the zebrafish (Danio rerio) presented major changes in its most used codons per amino acid: Its most used codon for Cys was UGU 54.3%, followed by Cys UGC 45.7%, which is the most used codon for the terrestrial vertebrates analyzed; zebrafish most used codons for proline were: Pro CCU 31.2% and Pro CCA 29.5%, instead of the most used CCC 23.9% for terrestrial vertebrates; then, the zebrafish most used codon for Thr was ACA 30.85%, followed by Thr ACC 29.4% (the most used by land animals), while Thr ACU had a use of 26.32%. Another difference in zebrafish was the most used codon for Ala GCU (31.86%), followed by Ala GCC 29.73% (the most used in land animals), while Ala GCA had a use of 25.3%. The zebrafish most used Gly codon was GGA 34.46%, followed by Gly GGC 27.56% (the most used Gly in land animals), while Gly GGU had 21.96% of usage. The 7th zebrafish difference (that was also seen in land animals), was that the most used codon for Arg was AGA 26.34%, followed by Arg AGG 18.78%, and by Arg CGC 17.68%, having the rest of its Arg codons a use of ∼12.4% each (the ratio of the analyzed human/zebrafish codons was 40,662,582/8,042,248).
From this point on, the differences increase dramatically if we compare the most used codons in humans per amino acid to an amphibian (frog Xenopus laevis) or a reptile (cobra Naja naja) [data not shown]; and even more abundant differences were seen when comparing the most used codons in humans to the most used codons of a plant (Arabidopsis thaliana, which will be compared elsewhere within this journal, in my topic of the quantum workings of the genetic code) (Castro-Chavez, 2011), except for the most abundant Lys AAG 51.5% and Arg AGA 35.19%. Unexpectedly, the differences between humans and bacteria (E. coli K12), in relation to the most used codons, are less notable than the ones for the last three mentioned organisms (frog, cobra and plants). The archaea Methanosarcina acetivorans C24A had slightly more differences than E. coli.
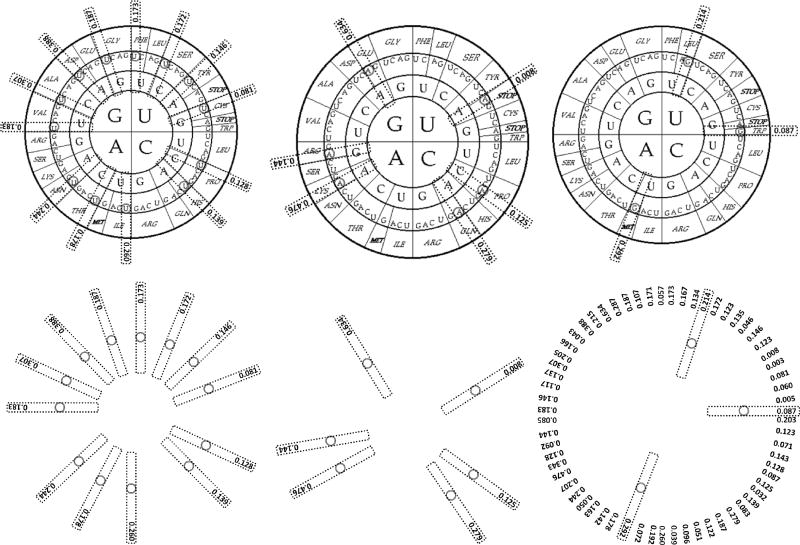
Other two organisms with the most radical differences when compared to the most used codons in humans were the squid (Loligo pealei, see Fig. 5), and the octopus (Octopus vulgaris) [data not shown], except for only one of the Arg AGA codons: Arg 26.82% (Loligo pealei) and Arg 35.8% (Octopus vulgaris).
Figure 5.
Most used codons per amino acid in the genome of the squid as illustrated by the circular genetic code. Left. 13 of the most used codons end in U and are (clockwise): Phe UUU, Ser UCU, Tyr UAU, Cys UGU, Pro CCU, His CAU, Ile AUU, Thr ACU, Asn AAU, Val GUU, Ala GCU, Asp GAU, Gly GGU. Center. 6 of the most used codons end in A and are: Stop UAA, Pro CCA, Gln CAA, Lys AAA, Arg AGA, Glu GAA. Right. 3 of the most used codons end in G: Leu UUG, Trp UGG and Met AUG. The figures below underlie their relative positions in the circular genetic code. The ratio of the analyzed human/squid codons was 40,662,582/25,614. Squid had a mostly odd or uneven distribution of most used codons, 13-6-3, U-A-G as their nucleotide ending signature.
Fig. 5 shows clearly the “biased C/U choice” described by Grantham et al. (1986) in 12 of the squid's most frequently used codons, plus a biased G/A choice for 4 of the most used codons in squid; 12 of the most used amino acid codons that end in C in humans, here in the squid end in U, while 4 of the most used amino acid codons that end in G in humans, in squid end in A. Only one codon changed from G to U in the squid (the G/U rare choice): Val GUG/U. The amino acids with only one codon (Trp, Met), remained the same. Leu remained the same at its third nucleotide (G), while changing its first nucleotide: Leu CUG/UUG. In squid, the two codons equally used were those for Pro: CCU 34.41% (already included in the previous sum of 12 Us for the squid) and CCA 33.6%, with a 0.81% of difference among them, while in humans, as mentioned earlier, the codons equally used were two for Arg: AGA 12.17% and AGG 11.96% (with a 0.21% difference).
In squids there was no most used codon ending in C, while in humans there was no most used codon ending in U. Humans had an even distribution of most used codons, a 12-8-2, C-G-A nucleotide ending signature (Fig. 1), while in squid the distribution was mostly uneven: 13-6-3, U-A-G nucleotide ending signature (Fig. 5).
The comparison of the most used codons between humans and squids was done to demonstrate the changes experienced among some of the most diametrically opposed organisms and their different third nucleotide choices.
The nematode Caenorhabditis elegans, which, except for the Phe codon UUC 50.64% and Arg AGA 29.33%, it had a different most used codon for the rest of the amino acids (the ratio of the analyzed human/nematode codons was 40,662,582/11,197,796).
Table 3 compares the most used codons in humans versus squid (human/squid), clearly illustrating these independent choices for their third nucleotide: 12 C/U, 3 G/A, and 1 G/U. The codon for Leu changed in its first nucleotide only, from CUG to UUG. The amino acids that only have one codon remained the same. The Pro CCA is considered an addition in squid, rather than a change, similar to the Arg AGG highly used in humans but not in squid.
Table 3.
Differences in the third nucleotide between men and squid.
| Codons ending in C/U* | Codons ending in G/A | Codons ending in A |
|---|---|---|
| His CAC/U | #Arg AGG/A [similar % to #] | #Arg AGA [only in humans] |
| @Pro CCC/U [similar % to @] | Gln CAG/A | @Pro CCA [only in squid] |
| Phe UUC/U | Glu GAG/A | |
| Cys UGC/U | Lys AAG/A | Codons ending in G/G |
| Asp GAC/U | Leu CUG/UUG | |
| Asn AAC/U | Codons ending in G/U | Trp UGG/UGG |
| Ile AUC/U | Val GUG/U | Met AUG/AUG |
| Ala GCC/U | ||
| Thr ACC/U | Codons ending in A/A | |
| Gly GGC/U | Stop UGA/UAA | |
| Tyr UAC/U | ||
| Ser AGC/UCU |
Human (Homo sapiens)/squid (Loligo pealei).
Obviously, the most stable comparison will be with the genomic codon usage per amino acid. In the examples reported above, except for humans, the results are of the globally expressed genes. Some organisms had fewer sequenced codons in the database, such as the wolf, the elephant and the llama in the above examples. The results presented here corroborate the early findings that “the usage of codons is not random and differs between organisms and genes” (Widmann et al., 2008) being useful, especially as a point of reference for the global codon usage in humans, and also to track the changes experienced by individual proteins within individual varieties, ethnic groups or breeds, which are the molecular languages or dialects, between these proteins compared to the global codon usage per amino acid, and between the genomic composition of the genetic DNA versus the punctual post-transcriptional changes due to the mRNA editing, to learn about its regulatory purposes. For example, “slow translation rate correlates with the high content of rare codons” (Pedersen, 1984) and vice-versa, a ribosome translating a strand of 9 codons in which some, or all of them, are low-usage “will move more slowly than over a comparable stretch of message containing no low-usage codons” (Zhang et al., 1994).
Codon frequencies are not random among codons specifying a given amino acid as some may be common while some may be rare. Usage biases varying within and between genomes help to regulate the rate of protein synthesis. “If the transfer RNA that matches a codon is rare, then transcription of genes including that codon will be slowed. For some proteins there is evidence that such pace-setting codons help ensure correct folding of the amino acid chain” (Hayes, 2004).
When comparing bacterial genes versus genes of phages, Grantham declared with a great surprise: “single-stranded DNA phages fabricate messenger sequences that use the translation apparatus and tRNA of their bacterial hosts, yet make different choices from the host among synonymous codons” (Grantham, 1980); in general, “viruses do not closely imitate the use of the codon catalogue by the host” (Grantham et al., 1986); in the same article Grantham and collaborators stated that species could be characterized by the slight variations in their coding strategies.
Grantham declared that “each kind of transcription product (mRNA, rRNA and tRNA) have a rather limited range of G + C content that is most often different (and in animals, at least, generally higher) than that of the whole genome”, then he posed the questions: “Why, for example, do α- and β-globin mRNAs make such different third base choices when they are translated at the same time and at similar abundances in the same cell?”; “The same question can be asked regarding C and V segments of immunoglobulin mRNA. Here the situation is even worse since the two kinds of segments are incorporated into the same messenger”; “Especially difficult to understand is the large variation in individual human genes, in which percentage (G + C) in codon position III runs from around 40 to over 90 per cent” (Grantham et al., 1986).
These frequencies can be useful for developing tools for bioinformatics to compare codon usage in individual peptides and proteins, and to determine their bias from the general trend within a genomic population or ‘Theme’ (Castro-Chavez, 2012a).
Conclusions and Outlook
The codon usage follows a precise pattern clearly defined in the human codifying genome analyzed. The general principle is that the most used codons have a C or a G in their third position. The discrepancy with other vertebrate organisms is small, mostly focused in the Arg codons (dogs, cows, dolphins, elephant, llama, wolf, pig), in the Ser codons (chimpanzee), in both (blue whale), or in the Pro codons (mouse).
By determining the genomic preference of codons per amino acid and/or per total amino acid usage in any organism we may have a fast way to determine genomic compatibilities, and design computational tools to specifically compare human genomes, their compatible codons and amino acids, and the early detection of incompatible changes leading to hereditary diseases, such as the potential damage of interbreeding for the 9/11 responders.
These findings can be clearly differentiated from methodological artifacts reported elsewhere (Castro-Chavez, 2005, 2012b), “which are products of the methodologies used, such as the host-vector interactions leading to an artificial rearrangement-splicing” (Castro-Chavez, 2004).
Acknowledgments
Joel D. Morrisett reviewed an earlier version of this article; Donald R. Forsdyke posted online the Grantham’s works; Ellen M. Carroll and Tracy L. Duncan did help in preparing this manuscript, partly supported by NIH grant ROI-HL-63090 and T32 HL-07812. To Janet Kay Wooley and the Texas Workforce Commission for helping me financially to complete the writing of these two unpaid works, and thanks to S. Tarlaci, and Tidjani Negadi, guest Editor of NeuroQuantology, for inviting me to write this article. The author declares no conflict of interests. In memory of Lynn Margulis (1938-2011). Thanks to Estefano Macedo Reyes for his interest in the research of ancient human petrified bones. 9/11 Is/was rather elusive, cribbing mineral allelisms…
Last update 03/07/2012.
Footnotes
Abbreviations: Amino acids: A: alanine (Ala), V: valine (Val), I: isoleucine (Ile), L: leucine (Leu), M: methionine (Met), F: phenylalanine (Phe), W: tryptophan (Trp), D: aspartic acid (Asp), N: asparagine (Asn), E: glutamic acid (Glu), Q: glutamine (Gln), R: arginine (Arg), K: lysine (Lys), S: serine (Ser), T: threonine (Thr), G: glycine (Gly), P: proline (Pro), H: histidine (His), C: cysteine (Cys), Y: tyrosine (Tyr). Nucleotides: U: uracil, C: cytosine, A: adenine, G: guanine, T: thymine.
Note: It seems that the underground explosions under the twin towers, registered by the earthquake monitors, were aimed to be coupled to the serious damages exerted upon the higher floors of both towers; falling elevators and ‘jet fuel’ are not capable to destroy 50 ton hydraulic presses, twist like aluminum foil a 300 pound fire door, and to “evaporate” humans, and still burning for months the underground where the WTC buildings stood. A similar conclusion was reached by Lynn Margulis (2010).
GARGANO: This is metal that has been compressed again as the result of the collapse of these extremely tall buildings. And that is made up, composition of different materials steel, concrete and other materials and you can see how that was compressed.
Amazingly, it's possible to make out individual objects. There are bathroom tiles, a pipe, and blackened pieces of paper carbonized by the heat. It smells like charcoal. Peter Gatt, who's one of the preservationists here, points to the spine of what looks like a corporate report.
GATT: This is a book. You can find a book inside. You can read letters on these little papers and for some reason they're still here.
BETH (Fertig): You can actually make out words on some of these tiny pieces of carbonized paper. The word representative, action, employment. [sic]
GATT: we checking every inches of this piece and we photographing every inches.
Gatt has been watching for any signs of deterioration. Cracks emerged over a year ago (2005)* and they're now marked with tiny green pins, to monitor the slightest change. The humidity and temperature in this room are tightly controlled.
[* Related to other kind of 9/11 WTC cracks: “a crack opened up on the [Liberty] street overnight (Sep., 2001)”, “[Peter] Rinaldi recalled”, Halbfinger (2006).]
Related example: “A laser scan of the boat as the excavation continues”: http://www.webcitation.org/63YwB2GrL and ‘Laser mapping’, a clip from “Relics From the Rubble”: http://www.youtube.com/watch?v=qQgEwD14Fpw“It could have been incorporated into the memorial or it could have been a tourist attraction — it could have been many things other than buried in concrete...” — Risa“Are we the only ones that think it should be preserved as part of the memorial site? ...Why is this not a bigger deal?” — anna“This is just more evidence that something else may have gone on. To think the builders of WTC did not take ground sonar or know about this (if it was there before) is turning a blind eye to reality...” — Ken
Pictures of these 3 fruit flies: D. melanogaster: http://www.webcitation.org/63Z01cSBc; D. mauritiana, http://www.webcitation.org/63Z06ZdKy; D. willistoni, http://www.webcitation.org/63Z0BMgtD. There is almost no visual difference between them (Ratnasingham and Hebert, 2007).
References
- Antevs EV. Correlation of Wisconsin glacial maxima. Amer J Sci. 1945;243-A:1–39. [Google Scholar]
- Barnett J, Biederman RR, Sisson RD., Jr Limited Metallurgical Examination. Appendix C. (13 p.) World Trade Center Building Performance Study. FEMA. 2002:403. URL: http://www.fema.gov/pdf/library/fema403_apc.pdf, saved at: http://www.webcitation.org/60zPXGotQ.
- Barrett K, Keogh J, Ranzz M, et al. Lynn Margulis: 1938-2011. 911blogger 2011a [Google Scholar]
- Barrett K. Lynn Margulis, Truth Seeker, 1938-2011. Veterans Today 2011b [Google Scholar]
- Beard J. Ground Zero's fires still burning. New Scientist. 2001 URL: http://www.newscientist.com/article/dn1634-ground-zeros-fires-still-burning.html [also see p. 7 of: http://www.epa.gov/gcc/pubs/docs/award_entries_and_recipients1998.pdf, saved at: http://www.webcitation.org/60zQ7VkEp]
- Bergeron JW. The crucifixion of Jesus: Review of hypothesized mechanisms of death and implications of shock and trauma-induced coagulopathy. J Forensic Leg Med. 2011:4. doi: 10.1016/j.jflm.2011.06.001. in press. [DOI] [PubMed] [Google Scholar]
- Bertoldi F, Altenhoff W, Weiss A, Menten KM, Thum C. The trans-neptunian object UB313 is larger than Pluto. Nature. 2006;439(7076):563–564. doi: 10.1038/nature04494. DOI: 10.1038/nature04494. [DOI] [PubMed] [Google Scholar]
- Brion PM. The Deconstruction of Deutsche Bank. Structural Engineers Association of New York. 2006;11(1) URL: http://www.seaony.org/publications/article.php?id=16, saved at: http://www.webcitation.org/60zNRAl5j. [Google Scholar]
- Bullinger EW. The witness of the stars. Eyre and Spottiswoode; London: 1893. [Accessed date: June 27, 2011]. p. 204. (plus chart) URL: http://www.archive.org/download/witnessofthestar00bulluoft/witnessofthestar00bulluoft.pdf, http://philologos.org/__eb-tws. [Google Scholar]
- Bullinger EW. The Companion Bible. 1922 [See Appendix 12] http://www.archive.org/details/CompanionBible.Bullinger.1901-Haywood.2005.
- C. A., editor. Trade Center air laden with very fine particles, DELTA scientists find. California Agriculture. 2002;56(3):84–84. URL: http://www.webcitation.org/60zLCYDQC. [Google Scholar]
- Cahill TA, Cliff SS, Perry KD, et al. Analysis of aerosols from the World Trade Center Collapse Site, New York, October 2 to October 30, 2001. Aerosol Science Technology. 2004;38(2):165–183. URL: http://www.webcitation.org/5wtNQUkwN, Personal Site: http://www.webcitation.org/5wYA07Vmt. [Google Scholar]
- Castro-Chavez F. Some Implications for the Study of Intelligent Design Derived from Molecular and Microarray Analysis. [Accessed date: June 27, 2011];PCID. 2004 3.1 URL: http://www.iscid.org/pcid/2004/3/1/chavez_microarray_analysis.php. [Google Scholar]
- Castro-Chavez F. Palindromati. [Accessed date: June 27, 2011];PCID. 2005 4.2 URL: http://www.iscid.org/pcid/2005/4/2/chavez_palindromati.php. [Google Scholar]
- Castro-Chavez F. The rules of variation: amino acid exchange according to the rotating circular genetic code. J Theor Biol. 2010;264(3):711–21. doi: 10.1016/j.jtbi.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Chavez F. The rules of variation expanded, implications for the research on compatible genomics. Biosemiotics. 2012a;5(1):121–145. doi: 10.1007/s12304-011-9118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Chavez F. Escaping the cut by restriction enzymes through single-strand self-annealing of host-edited 12-bp and longer synthetic palindromes. DNA Cell Biol. 2012b;31(2):151–163. doi: 10.1089/dna.2011.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Chavez F. The quantum workings of the rotating 64-grid genetic code. NeuroQuantology. 2011;9(4):728–746. doi: 10.14704/nq.2011.9.4.499. http://www.neuroquantology.com/index.php/journal/article/view/499 [current issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RN, Green RO, Swayze GA, et al. Environmental Studies of the World Trade Center area after the September 11, 2001 attack. United States Geological Survey. 2001 saved URL: http://www.webcitation.org/5wtEMG1Hp.
- Colaprete A, Schultz P, Heldmann J, et al. Detection of water in the LCROSS ejecta plume. Science. 2010;330(6003):463–468. doi: 10.1126/science.1186986. [DOI] [PubMed] [Google Scholar]
- Copper P. Silurian and Devonian reefs: 80 million years of global greenhouse between two ice ages. In: Kiessling W, Flugel E, Golonka J, editors. Phanerozoic Reef Patterns. Society of Economic Paleontologists and Mineralogists; 2002. pp. 181–238.pp. 72 [DOI] [Google Scholar]
- Culotta E. Neanderthals Were Cannibals, Bones Show. Science. 1999;286(5437):18–19. doi: 10.1126/science.286.5437.18b. [DOI] [PubMed] [Google Scholar]
- Custance AC. The Seed of the Woman. Doorway Publications; Ontario: 1980. [Accessed date: June 27, 2011]. pp. 210–232.pp. 254–267. URL: http://www.custance.org/Library/SOTW/Index.html. [Google Scholar]
- Custance AC. Without form and void. Doorway Publications; Ontario: 1989. p. 211. [Also by: Classic Reprint Press, 2008, 292 p.] http://www.webcitation.org/63oLDi4mo. [Google Scholar]
- Deevey ES., Jr Biogeography of the Pleistocene. Part I: Europe and North America. [Accessed date: June 27, 2011];GSA Bulletin. 1949 60(9):1315–1416. URL: http://bulletin.geoscienceworld.org/cgi/content/abstract/60/9/1315. [Google Scholar]
- de Martonne E. Recherches sur l’évolution morphologique des Alpes de Transylvanie (Karpates méridionales) 1906-1907. Rev Geogr Annuelle. 1907 Paris, 1:286. vezi şi, Lucrări geografice despre România”, Edit. Academiei 1:271, Bucureşti, 1981. [Google Scholar]
- Douglas JG, Williams GE. Southern polar forests: The Early Cretaceous floras of Victoria and their palaeoclimatic significance. Palaeogeogr Palaeoclimatol Palaeoecol. 1982;39(3-4):171–185. doi: 10.1016/0031-0182(82)90021-9. [DOI] [Google Scholar]
- Duke MB. Lunar polar ice deposits: scientific and utilization objectives of the Lunar Ice Discovery Mission proposal. Acta Astronaut. 2002;50(6):379–383. doi: 10.1016/s0094-5765(01)00184-9. [DOI] [PubMed] [Google Scholar]
- Dunlap DW, Tamaro GJ, Merguerian C, Horenstein S, Racanelli J. At Ground Zero, scenes from the Ice Age. The New York Times. 2008a Sep 22;:B4. http://www.nytimes.com/2008/09/22/nyregion/22rocks.html.
- Dunlap DW, Lobel M, Woolhead J, Moore CP. City Room: Geologic Time at Ground Zero. The New York Times. 2008b Oct 6; Slide Show: Image 5 (06rock_5) http://www.nytimes.com/slideshow/2008/10/06/nyregion/06rock_5.html. [Google Scholar]
- Edwards WD, Gabel WJ, Hosmer FE. On the physical death of Jesus Christ. [Accessed date: June 27, 2011];JAMA. 1986 255(11):1455–1463. URL: http://jama.ama-assn.org/content/255/11/1455.short. [PubMed] [Google Scholar]
- Fertig B. Preserving The WTC Wreckage. A Visit to Hangar 17. WNYC (93.9 FM, AM 820) New York Public Radio 2006. URL: http://www.wnyc.org/articles/wnyc-news/2006/jun/14/preserving-the-wtc-wreckage, saved at: http://www.webcitation.org/60zOzyCmZ. see also: Fertig B. Artifacts from Ground Zero Await Memorial Site. NPR. 2006 Jun 14; http://www.npr.org/templates/story/story.php?storyId=5484916, saved at: http://www.webcitation.org/60zQn1bdd.
- Finkelstein KE. 40 Hours in Hell. American Journalism Review. 2001;23(9):19. U. of Maryland also has a 2005 reprint. http://www.ajr.org/Article.asp?id=2381, saved at: http://www.webcitation.org/60zOIhQiV. [Google Scholar]
- Fiorillo AR, Hasiotis ST, Kobayashi Y, Tomsich CS. A pterosaur manus track from Denali National Park, Alaska Range, Alaska, United States. PALAIOS. 2009;24:466–472. doi: 10.2110/palo.2008.p08-129r. [DOI] [Google Scholar]
- Flint RF. Glacial Geology and the Pleistocene Epoch. Wiley; New York, NY: 1947. p. 589. [Google Scholar]
- Flint RF. Wetmore A. Annual report of the Board of Regents of the Smithsonian Institution. Publ. 4111. Showing the Operations, Expenditures, and Conditions of the Institution for the Year Ended June 30 1952. Washington: 1953. The Ice Age in the North American Arctic (pp 251-252) p. 461. [Google Scholar]
- Gangloff RA, Fiorillo AR. Taphonomy and paleoecology of a Bonebed from the Prince Creek Formation, North Slope, Alaska. PALAIOS. 2010;25:299–317. doi: 10.2110/palo.2009.p09-103r. From NOVA: http://www.pbs.org/wgbh/nova/nature/arctic-dinosaurs.html. [DOI] [Google Scholar]
- Garel JP. The silkworm, a model for molecular and cellular biologists. Trends Biochem Sci. 1982;7:105–108. doi: 10.1016/0968-0004(82)90158-X. [DOI] [Google Scholar]
- Gibbons A. Anthropology. The species problem. Science. 2011;331(6016):394. doi: 10.1126/science.331.6016.394. [DOI] [PubMed] [Google Scholar]
- Gilsanz R, DePaola EM, Marrion C, Nelson HB. WTC7 (Chapter 5), 5.5.4 Sequence of WTC 7 Collapse, pp. 5-23 (32 p.) In: McAllister T, editor. World Trade Center Building Performance Study: Data Collection, Preliminary Observations, and Recommendations. FEMA; Washington, DC; 2002. [Accessed date: June 27, 2011]. p. 403. http://www.fema.gov/pdf/library/fema403_ch5.pdf. [Google Scholar]
- Godefroit P, Golovneva L, Shchepetov S, Garcia G, Alekseev P. The last polar dinosaurs: high diversity of latest Cretaceous arctic dinosaurs in Russia. Naturwissenschaften. 2009;96(4):495–501. doi: 10.1007/s00114-008-0499-0. [DOI] [PubMed] [Google Scholar]
- Grant US, IV, Gale HR. Memoirs of the San Diego Society of Natural History. Vol. 1. San Diego, CA: 1931. Catalogue of the marine Pliocene and Pleistocene mollusca of California and adjacent regions (p. 71) p. 1037. [Google Scholar]
- Grantham R. Workings of the genetic code. [Accessed date: June 27, 2011];Trends Biochem Sci. 1980 5:327–331. http://post.queensu.ca/∼forsdyke/granth01.htm. [Google Scholar]
- Grantham R, Gautier C, Gouy M, Mercier R, Pavé A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980;8(1):r49–r62. doi: 10.1093/nar/8.1.197-c. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC327256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R, Perrin P, Mouchiroud D. Patterns in codon usage of different kinds of species. [Accessed date: June 27, 2011];Oxf Surv Evol Biol. 1986 3:48–81. URL: http://post.queensu.ca/∼forsdyke/granth01.htm. [Google Scholar]
- Green RE, Krause J, Briggs AW, et al. A draft sequence of the Neandertal genome (Fig. 6) Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy WM, McKinnon WB, Ammannito E, et al. SBAG Community White Paper. Exploration Strategy for the Ice Dwarf Planets 2013-2022. Delivered to the Primitive Bodies Decadal Panel, 9/5/09. NASA. 2009 8 p. [Google Scholar]
- Halbfinger C. WTC slurry wall a symbol of survival. LoHud.com. 2006 Saved at: http://www.webcitation.org/60zO0Mh02.
- Hambrey MJ, Harland WB. Earth’s Pre-Pleistocene Glacial Record. Cambridge University Press; 2011. p. 1022 p. [Google Scholar]
- Hayes B. Ode to the code [A closer look at the question of whether the code is optimal in some sense] [accessed date: June 27, 2011];Am Sci. 2004 92:494–499. URL: http://bit-player.org/bph-publications/AmSci-2004-11-Hayes-genetic-code.pdf, both links. http://www.americanscientist.org/issues/id.3441,y.0,no.,content.true,page.1,css.print/issue.aspx. [Google Scholar]
- Hershey OH. Some evidence of two glacial stages in the Klamath Mountains in California. Am Geologist. 1903;31:139–156. [Google Scholar]
- Hsieh HH, Jewitt D. A population of comets in the main asteroid belt. Science. 2006;312(5773):561–563. doi: 10.1126/science.1125150. [DOI] [PubMed] [Google Scholar]
- Hunter ML, Jr, Gibbs JP. Ch 5. Genetic Diversity. Processes that diminish genetic diversity, (p. 99-104) 3rd. Blackwell; Malden, MA: 2007. Fundamentals of conservation biology; p. 497. [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. [Accessed date: June 27, 2011];Mol Biol Evol. 1985 2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. URL: http://mbe.oxfordjournals.org/content/2/1/13.long. [DOI] [PubMed] [Google Scholar]
- Jenkins C. Request for an investigation by the senate subcommittee on superfund and environmental health into the falsification of pH corrosivity data for World Trade Center dust. Journal of 911 Studies. 2007;12:134. URL: http://www.webcitation.org/5wZljPqYr. [Google Scholar]
- Jenniskens P, Mandell AM. Hydrogen emission in meteors as a potential marker for the exogenous delivery of organics and water. Astrobiology. 2004;4(1):123–134. doi: 10.1089/153110704773600285. [DOI] [PubMed] [Google Scholar]
- Jewitt DC, Luu J. Crystalline water ice on the Kuiper belt object (50000) Quaoar. Nature. 2004;432(7018):731–733. doi: 10.1038/nature03111. [DOI] [PubMed] [Google Scholar]
- Kahn HD, Rosati JA, Bray AP. Statistical evaluation of data from multi-laboratory testing of a measurement method intended to indicate the presence of dust resulting from the collapse of the World Trade Center. Environ Monit Assess. 2011 Nov 18; doi: 10.1007/s10661-011-2426-7. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Kaysen R. Fiterman Hall's last days may finally be in sight. Downtown Express. 2005a;17(52) URL: http://www.downtownexpress.com/de_106/fitermanhalllast.html, saved at: http://www.webcitation.org/60zNAAqfA. [Google Scholar]
- Kaysen R. Demolition work begins at 4 Albany St. Downtown Express. 2005b;17(51) URL: http://www.downtownexpress.com/de_105/demolitionworkbegins.html, saved at: http://www.webcitation.org/60zNErcyS. [Google Scholar]
- Kerr RA. Planetary science. A dripping wet early Mars emerging from new pictures. Science. 2000;290(5498):1879–1880. doi: 10.1126/science.290.5498.1879. [DOI] [PubMed] [Google Scholar]
- Khalezov D. Bio-terrorism and anthrax. Veterans Today. 2011a http://www.veteranstoday.com/author/khalezov.
- Khalezov D. Ground Zero. Veterans Today 2011b [Google Scholar]
- Khalezov D. Chernobyl and Manhattan. Veterans Today 2011c [Google Scholar]
- Kim WY, Sykes LR, Armitage JH, et al. Earthquake Monitoring Station, Palisades: Lamont-Doherty Earth Observatory, University of Columbia; 2001. Seismic Waves Generated by Aircraft Impacts and Building Collapses at World Trade Center, New York City; p. 9. URL: http://www.ldeo.columbia.edu/LCSN/Eq/20010911_WTC/WTC_LDEO_KIM.pdf. [Google Scholar]
- Lioy PJ, Weisel CP, Millette JR, et al. Characterization of the Dust/Smoke Aerosol that Settled East of the World Trade Center (WTC) in Lower Manhattan after the Collapse of the WTC 11 September 2001. Environ Health Perspect. 2002;110(7):703–714. doi: 10.1289/ehp.02110703. URL: http://www.ncbi.nlm.nih.gov/pubmed/12117648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi K. Death by Dust: The Frightening Link between the 9-11 Toxic Cloud and Cancer. Village Voice. 2006:9. URL: http://www.villagevoice.com/2006-11-21/news/death-by-dust/5, saved at: http://www.webcitation.org/60zOdxWdm.
- Mackenzie D. Planetary science. And an icy patch at Mars's south pole. Science. 2002;298(5600):1866–1867. doi: 10.1126/science.298.5600.1866b. [DOI] [PubMed] [Google Scholar]
- Manning W. $elling out the investigation. Fire Engineering. 2002:4. Saved at: http://www.webcitation.org/60zPm5Jx2.
- Marcu MF. Glacial and periglacial relief in the Făgăraş Mountain, with special focus on the Vâlsan river basin. [Accessed date: June 27, 2011];Forum Geografic. 2011 10(1):27–34. URL: http://forumgeografic.ro/wp-content/uploads/2011/07/Marcu.pdf. [Google Scholar]
- Margulis L. Two hit, three down—the biggest lie. Rock Creek Free Press; 2010. http://rockcreekfreepress.tumblr.com/post/353434420/two-hit-three-down-the-biggest-lie. [Google Scholar]
- Maslen MW, Mitchell PD. Medical theories on the cause of death in crucifixion. J R Soc Med. 2006;99(4):185–188. doi: 10.1258/jrsm.99.4.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayell H. Neandertals Not Our Ancestors, DNA Study Suggests. [Accessed date: June 27, 2011];National Geographic News. 2003 URL: http://news.nationalgeographic.com/news/2003/05/0514_030514_neandertalDNA.html.
- McGee JK, Chen LC, Cohen MD, et al. Chemical Analysis of World Trade Center Fine Particulate Matter for Use in Toxicological Assessment. Environ Health Perspect. 2003;11(7):972–980. doi: 10.1289/ehp.5930. URL: http://www.ncbi.nlm.nih.gov/pubmed/12782501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier R, Owen TC, Matthews HE, et al. A determination of the HDO/H2O ratio in comet C/1995 O1 (Hale-Bopp) Science. 1998;279(5352):842–844. doi: 10.1126/science.279.5352.842. [DOI] [PubMed] [Google Scholar]
- Miller A. "Sarcoid like" granulomatous pulmonary disease (SLGPD) in WTC responders. Am J Ind Med. 2012 Feb;55(2):189. doi: 10.1002/ajim.21005. [DOI] [PubMed] [Google Scholar]
- Miranda LF, Gómez Y, Anglada G, Torrelles JM. Water-maser emission from a planetary nebula with a magnetized torus. Nature. 2001;414(6861):284–286. doi: 10.1038/35104518. [DOI] [PubMed] [Google Scholar]
- Morey K, White M, Corless S, Winnett T. Sierra North: Backcountry Trips in California's Sierra Nevada (p 181) 9th. Wilderness Press; Berkeley, CA: 2005. p. 344. [Google Scholar]
- Naik G. Scientists’ Elusive Goal: Reproducing Study Results. Wall Street Journal. 2011 Dec 2;55:A1–A16. URL: http://www.webcitation.org/65p4UiQfi. [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. [Accessed date: June 27, 2011];Nucleic Acids Res. 2000 28(1):292. doi: 10.1093/nar/28.1.292. Software URL: http://www.kazusa.or.jp/codon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa M, Nelson I, Ribai P, et al. A founder effect and mutational hot spots may contribute to the most frequent mutations in the SPG3A gene. Neurogenetics. 2006;7(2):131–132. doi: 10.1007/s10048-006-0028-2. [DOI] [PubMed] [Google Scholar]
- Niculescu G, Nedelcu E, Iancu S. Nouvelle contribution a l’étude de la morphologie glaciaire des Carpates roumaines, în Recueil’études géographiques concernant le territoire de la République Populaire Roumaine, Edit. Academiei Bucureşti. 1960:29–43. [Google Scholar]
- Ogle County Planning & Zoning Department. Topography and Physiography (p 27) Ogle County, IL: 2008. [Accessed date: June 27, 2011]. Amendatory Comprehensive Plan “2k8 Update”; p. 116. URL: http://www.oglecounty.org/zoning/Comprehensive%20Plan/Comprehensive%20Plan%202K8%20Update.pdf. [Google Scholar]
- Pecoraro M, et al. We Will Not Forget. The Chief Engineer. 2001 URL: http://www.chiefengineer.org/article.cfm?seqnum1=1029, saved at: http://www.webcitation.org/60zIMCzVf.
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984;3(12):2895–8. doi: 10.1002/j.1460-2075.1984.tb02227.x. URL: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC557784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot A, Hussein S, Ruppert V, et al. Identification of mutational hot spots in LMNA encoding lamin A/C in patients with familial dilated cardiomyopathy. Basic Res Cardiol. 2009;104(1):90–99. doi: 10.1007/s00395-008-0748-6. [DOI] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert PDN. BOLD : The Barcode of Life Data System. Molecular Ecology Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2006.01678.x. www.barcodinglife.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina RB, Moss CJ, Pontecorvo A. Mueser Rutledge Consulting Engineers, and Mail Foreign Service. Pictured: The 40ft ‘pothole’ that shows Ground Zero was once the site of an Ice Age glacier. Mail Online, UK. 2008 http://www.dailymail.co.uk/news/article-1060043/Pictured-The-40ft-pothole-shows-Ground-Zero-site-Ice-Age-glacier.html.
- Rich TH, Vickers-Rich P, Gangloff RA. Paleontology. Polar dinosaurs. Science. 2002;295(5557):979–980. doi: 10.1126/science.1068920. [DOI] [PubMed] [Google Scholar]
- Ridpath I. Pluto—Planet or Imposter? Astronomy. 1978:6–11. http://myweb.tiscali.co.uk/ianridpath/Pluto.pdf.
- Rivkin AS, Emery JP. Detection of ice and organics on an asteroidal surface. Nature. 2010;464(7293):1322–1323. doi: 10.1038/nature09028. [DOI] [PubMed] [Google Scholar]
- Rolleston F. Mazzaroth; or, The Constellations. (Including Planisphere of Dendera and Zodiac of Esne) Rivingtons; London: 1862. [Accessed date: June 27, 2011]. p. 257. for URL: http://books.google.com/books/about/Mazzaroth_or_The_constellations_by_F_Rol.html?id=hTABAAAAQAAJ, http://philologos.org/__eb-mazzaroth. [Google Scholar]
- Salama F, Allamandola LJ, Witteborn FC, Cruikshank DP, Sandford SA, Bregman JD. The 2.5-5.0 micrometers spectra of Io: evidence for H2S and H2O frozen in SO2. Icarus. 1990;83(1):66–82. doi: 10.1016/0019-1035(90)90006-u. [DOI] [PubMed] [Google Scholar]
- Sammonds PR. Planetary science. Creep and flow on the icy moons of the outer planets. Science. 2006;311(5765):1250–1251. doi: 10.1126/science.1123985. [DOI] [PubMed] [Google Scholar]
- Schoenheit JW. The Christian’s Hope: The Anchor of the Soul. 2nd Ed. CES; Indianapolis, IN: 2004. 287 p. http://www.truthortradition.com/modules.php?name=News&file=article&sid=317. [Google Scholar]
- Seiss JA. The Gospel in the Stars. Claxton & Comp.; Philadelphia: 1882. [Accessed date: June 27, 2011]. p. 154. URL: http://www.archive.org/download/JoesphA.Seiss-TheGospelInTheStars/J_A_Seiss-Gospel_in_the_stars.pdf. [Google Scholar]
- Seshadri S, Chin KB, Buehler MG, Anderson RC. Using electrical impedance spectroscopy to detect water in planetary regoliths. Astrobiology. 2008;8(4):781–792. doi: 10.1089/ast.2007.0180. [DOI] [PubMed] [Google Scholar]
- Sicardy B, Ortiz JL, Assafin M, et al. A Pluto-like radius and a high albedo for the dwarf planet Eris from an occultation. Nature. 2011;478(7370):493–496. doi: 10.1038/nature10550. DOI: 10.1038/nature10550. [DOI] [PubMed] [Google Scholar]
- Spahr TB. ‘MPEC 2006-R19 : EDITORIAL NOTICE’. Minor Planet Center 2006-09-07, 17:35 UT. http://www.minorplanetcenter.net/iau/mpec/K06/K06R19.html. [Google Scholar]
- Stock E. Heaven not our home but the renovated earth the eternal abode of the redeemed saints. Elliot Stock; London: 1878. 299 pp. http://books.google.com/books?id=NgkDAAAAQAAJ. [Google Scholar]
- Tahil W. Ground Zero. 2006:178. URL: http://nucleardemolition.com/GZero_Report.pdf, Preserved at: http://www.webcitation.org/5wZdXDneQ.
- Talley PT. Another Study in Biblical Accuracy. American Christian Press; Ohio: [Accessed date: June 27, 2011]. Jesus Christ, The Last Adam (1 Cor. 15:45, 49) p. 9. n.d. URL: http://www.reocities.com/fdocc3/christ.htm. [Google Scholar]
- Thomas PC, Parker JW, McFadden LA, Russell CT, Stern SA, Sykes MV, Young EF. Differentiation of the asteroid Ceres as revealed by its shape. Nature. 2005;437:224–226. doi: 10.1038/nature03938. DOI: 10.1038/nature03938. [DOI] [PubMed] [Google Scholar]
- Thompson SM. Constructing and refining multiple sequence alignments with PileUp, SeqLab, and the GCG suite. Curr Protoc Bioinformatics. 2003;Chapter 3(Unit 3.6) doi: 10.1002/0471250953.bi0306s00. [DOI] [PubMed] [Google Scholar]
- Underdown S. A potential role for transmissible spongiform encephalopathies in Neanderthal extinction. Med Hypotheses. 2008;71(1):4–7. doi: 10.1016/j.mehy.2007.12.014. [DOI] [PubMed] [Google Scholar]
- Volpato JP, Pelletier JN. Mutational ‘hot-spots’ in mammalian, bacterial and protozoal dihydrofolate reductases associated with antifolate resistance: sequence and structural comparison. Drug Resist Updat. 2009;12(1-2):28–41. doi: 10.1016/j.drup.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Widmann M, Clairo M, Dippon J, Pleiss J. Analysis of the distribution of functionally relevant rare codons. BMC Genomics. 2008;9:207. doi: 10.1186/1471-2164-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierwille VP. Jesus Christ our Passover. American Christian Press; Ohio: 1980. [Accessed date: June 27, 2011]. p. 527. URL: http://tinyurl.com/3sz99bn. [Google Scholar]
- Wierwille VP. Jesus Christ Our Promised Seed. American Christian Press; Ohio: 1982. [Accessed date: June 27, 2011]. p. 306. URL: http://tinyurl.com/tsemach. [Google Scholar]
- Wishart DS, Fortin S. The BioTools Suite. A comprehensive suite of platform-independent bioinformatics tools. Mol Biotechnol. 2001;19(1):59–77. doi: 10.1385/MB:19:1:059. [DOI] [PubMed] [Google Scholar]
- Wu M, Gordon RE, Herbert R, et al. Case report: Lung disease in World Trade Center responders exposed to dust and smoke: carbon nanotubes found in the lungs of World Trade Center patients and dust samples. Environ Health Perspect. 2010;118(4):499–504. doi: 10.1289/ehp.0901159. URL: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2854726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Goldman E, Zubay G. Clustering of low usage codons and ribosome movement. J Theor Biol. 1994;170(4):339–54. doi: 10.1006/jtbi.1994.1196. [DOI] [PubMed] [Google Scholar]
- Zhou NY, Bates SE, Bouziane M, Stary A, Sarasin A, O'Connor TR. Efficient repair of cyclobutane pyrimidine dimers at mutational hot spots is restored in complemented Xeroderma pigmentosum group C and trichothiodystrophy/xeroderma pigmentosum group D cells. J Mol Biol. 2003;332(2):337–351. doi: 10.1016/S0022-2836(03)00793-9. [DOI] [PubMed] [Google Scholar]