Abstract
Background
Quercetin is a naturally occurring flavonol with antioxidant, anticancer and anti-ageing properties. In this study we aimed to identify genes differentially expressed in yeast cells treated with quercetin and its role in oxidative stress protection.
Methods
A microarray analysis was performed to characterize changes in the transcriptome and the expression of selected genes was validated by RT-qPCR. Biological processes significantly affected were identified by using the FUNSPEC software and their relevance in H2O2 resistance induced by quercetin was assessed.
Results
Genes associated with RNA metabolism and ribosome biogenesis were down regulated in cells treated with quercetin, whereas genes associated with carbohydrate metabolism, endocytosis and vacuolar proteolysis were up regulated. The induction of genes related to the metabolism of energy reserves, leading to the accumulation of the stress protectant disaccharide trehalose, and the activation of the cell wall integrity pathway play a key role in oxidative stress resistance induced by quercetin.
Conclusions
These results suggest that quercetin may act as a modulator of cell signaling pathways related to carbohydrate metabolism and cell integrity to exert its protective effects against oxidative stress.
Introduction
Oxidative stress is a common hallmark in the genesis of multiple age-associated diseases, such as cardiovascular diseases [1], cancer [2] and neurodegenerative [3] disorders. Oxidative stress is characterized by an imbalance between the production of reactive oxygen species (ROS) or reactive nitrogen species and cellular antioxidant defenses, resulting in the deregulation of redox homeostasis and accumulation of oxidatively damaged proteins, lipids and DNA that may lead to cell death [4]. ROS, such as hydrogen peroxide, superoxide and hydroxyl radicals, are normal by-products of mitochondrial respiration and reactions of cellular metabolism (e.g., catalyzed by cytochrome P450 and flavoprotein oxidases) or are generated from environmental insults. Reactive nitrogen species include nitric oxide (NO) produced by nitric oxide synthases, peroxinitrite generated by nonenzymatic reaction of NO with superoxide radicals, and other species such as nitrogen dioxide and dinitrogen trioxide. To maintain redox homeostasis, cells possess antioxidant defenses that neutralize reactive species in excess and repair oxidative damages [4], [5].
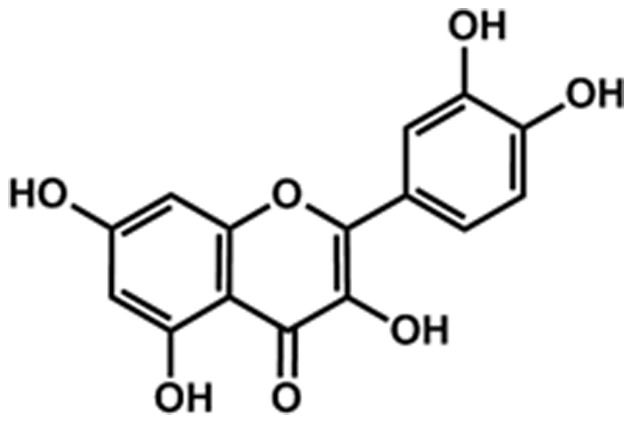
Epidemiological studies have shown an inverse correlation between the consumption of polyphenol-rich foods and oxidative stress-related chronic diseases [6]. Polyphenols are a group of plant secondary metabolites featuring more than one phenolic ring and without any nitrogen-based functional group in its structure [7]. According to their structure, polyphenols can be divided into different classes, in which flavonoids are the largest class. Quercetin (IUPAC nomenclature: 3,3′,4′,5,7-pentahydroxyflavanone) is a flavonol, a major widespread sub-class of flavonoids, being ubiquitously found in the human diet in onions, shallots, apples, berries, grapes, cappers, brassica vegetables, tea and also in red wine [8]. Quercetin has been extensively studied in many biological models, such as the nematode Caenorhabditis elegans [9], mammalian cell models [10], mice [11] and rats [12], and also to some extend in humans [13]. It is known to exhibit numerous biological and pharmacological effects, owning to its antioxidant properties [14], anti-inflammatory [15], and anticarcinogenic [16] effects. The intrinsic antioxidant activities of quercetin have been attributed to direct scavenging of ROS, through the abstraction of unpaired electrons or hydrogen atoms, or metal ions chelation that prevents the generation of hydroxyl radical through Fenton-type reactions. These properties are largely a function of the chemical structure of quercetin (Figure 1), particularly the presence and location of the hydroxyl substitutions (–OH) and the catechol-type B ring [7].
Figure 1. Chemical structure of quercetin.
More recently, it has been proposed that the mechanisms of action of polyphenols go beyond their antioxidant activities and quercetin may also exert modulatory effects on cell signaling pathways. For instance, quercetin has anti-inflammatory effects associated to the inhibition of NF-kB [17], prevents oxidant-induced liver damage by activating the mitogen-activated protein kinases (MAPK) JNK and p38, the phosphatidylinositol-3-kinase (PI3K)/Akt pathway and the Nrf2 transcription factor [18], and induces p53-dependent apoptotic cell death in HT-29 colon cancer cells by activating AMPK [19]. Quercetin binds directly to components of the ERK MAPK pathway, namely MEK1 (MAPK Kinase Kinase) and Raf1 (MAPK Kinase) [17], and to the ATP-binding site of PI3K [20]. In C. elegans, it was shown that quercetin decreases ROS levels, increases the mean lifespan and attenuates the accumulation of the aging marker lipofuscin. These beneficial effects are correlated with changes in the expression of the phase II metabolism enzyme GST-4 (glutathione S-transferase) and translocation of the DAF-16 transcription factor into the nucleus [21].
Signal transduction pathways are highly conserved during evolution [22] and the yeast S. cerevisiae has been extensively used as an eukaryotic model organism to characterize redox cell signaling and to assess in vivo the antioxidant potential of natural compounds [23]–[26]. Other studies using yeast have shown that quercetin inhibits chitin synthase II [27], the H+-translocating Mg2+-ATPase from the vacuole [28] and type-2 casein kinase, Yck2p [29], a palmitoylated plasma membrane-bound serine-threonine protein kinase that is activated by Snf3p/Rtg2p glucose sensors [30]. Quercetin also prevents the nuclear localization of the Yap1p transcription factor under oxidative stress conditions [31] and induces Oye2p and Oye3p, which are involved in the modulation of actin polymerization, oxidative stress response and cell death [32]. We have previously shown an increase in H2O2 stress resistance and chronological lifespan in yeast cells treated with quercetin [33]. In this study, we have used DNA microarrays to characterize changes in the transcriptome induced by quercetin in yeast. The effect of quercetin on carbohydrate metabolism and cell wall integrity (CWI) pathway was assessed as well as its importance for oxidative stress resistance.
Results
Microarray Analysis of Quercetin Treated Yeast Cells
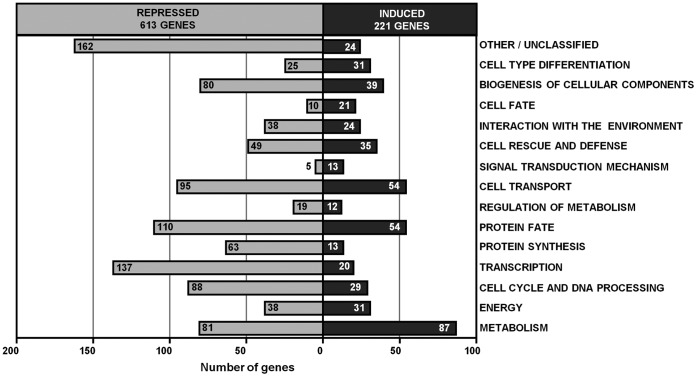
In a previous study, the analysis of cellular protection against oxidative stress in yeast exposed to quercetin for different time periods showed that a 15 min pre-treatment was sufficient to increase hydrogen peroxide resistance [33]. Aiming to characterize short-term adaptive responses triggered by quercetin and to identify cellular functions that may contribute to its protective effect against oxidative stress, changes in gene expression were analyzed by using microarrays. S. cerevisiae cells were treated with 300 µM quercetin or dimethyl sulphoxide (DMSO; control cells) during 15 min and mRNA was isolated by the hot phenol method, as described in Material and Methods. The microarray analysis showed that treatment with quercetin led to an increase of the mRNA levels of 221 genes, whereas that of 613 genes was diminished (see supplementary Tables S1 and S2). Genes differentially expressed were sorted into functional categories according to MIPS (Munich Information Center for Protein Sequences). Genes associated with transcription (22%), protein fate (18%), cell transport (15%), cell cycle and DNA processing (14%), metabolism (13%), biogenesis of cellular components (13%) and protein synthesis (10%) were down regulated, whereas genes related with metabolism (39%), protein fate (24%), cell transport (24%), biogenesis of cellular components (18%) and cell rescue and defense (16%) were up regulated by quercetin treatment (Figure 2).
Figure 2. Functional categories of genes differentially expressed in quercetin-treated cells.
S. cerevisiae BY4741 cells were grown to exponential phase and RNA was isolated as described in Materials and Methods. Genes up and down regulated in quercetin-treated cells were sorted in groups according to Munich Information Centre for Proteins Sequences (MIPS) database.
To validate the microarray data, the expression of five genes was analyzed by reverse transcription quantitative polymerase chain reaction (RT-qPCR). Four of these genes were up regulated in the microarray analysis and are functionally related with the metabolism of carbohydrates, including glycogen and trehalose (whose levels were also measured; see below): HXK1 (Hexokinase isoenzyme 1, which catalyses phosphorylation of glucose; its expression is highest during growth on non-glucose carbon sources), GPH1 (glycogen phosphorylase), TPS1 (synthase subunit of trehalose-6-phosphate synthase/phosphatase complex, which synthesizes trehalose) and TSL1 (subunit of trehalose 6-phosphate synthase/phosphatase complex). The expression of RPB5 (RNA polymerase subunit), which was down regulated in the microarray analysis, was also assessed by RT-qPCR. The mRNA levels were normalized to ACT1 (encodes for actin; internal control) and RPS6A (a second control that was chosen based on the fact that its expression was not altered by quercetin, according to the microarray data). The results obtained showed a correlation between microarray and RT-qPCR data (Figure 3).
Figure 3. Effect of quercetin (Q) on GPH1, HXK1, RPB5, TPS1 and TSL1 mRNA levels.
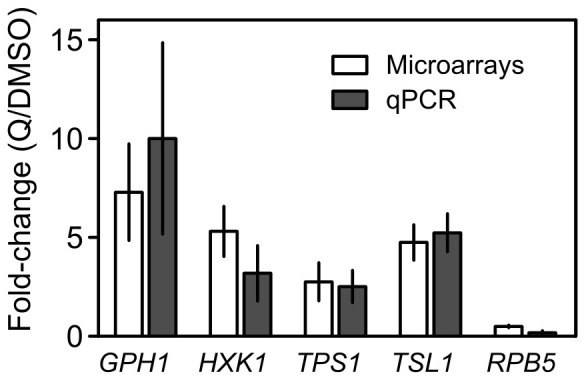
Transcription profile of GPH1 (glycogen phosphorylase), HXK1 (hexokinase isoenzyme 1), RPB5 (RNA polymerase subunit), TPS1 (synthase subunit of trehalose-6-phosphate synthase/phosphatase complex) and TSL1 (subunit of trehalose 6-phosphate synthase/phosphatase complex) were evaluated by microarrays analysis (white bars; mean ± SD of six arrays) and by qPCR (grey bars). The mean normalized fold expression by qPCR was calculated relative to the transcription of the reference genes RPS6A (component of the small (40S) ribosomal subunit) and ACT1 (actin). The results (mean and SD of triplicates) of a representative experiment (out of three independent experiments) are shown.
To identify biological processes overrepresented, gene lists were analyzed by using the FUNSPEC software [34]. The down regulated genes were mostly related with various aspects of RNA metabolism and ribosome biogenesis (Table 1). Notably, the expression of 14 genes containing a LSM (Like Sm) domain was repressed. LSM proteins form part of specific small nuclear ribonucleoproteins and are thought to be important modulators of RNA biogenesis and function [35]. The most important cellular functions induced include those associated with C-compound and carbohydrate metabolism, including metabolism of energy reserves, endocytosis and vacuolar proteolysis (Table 1).
Table 1. Functional categories overrepresented in the microarray data.
| Category | p-value | In Category from Cluster | |
| Down-regulated genes | |||
| MIPS Functional Classification | |||
| rRNA processing | 1.9E−7 | 40 GENES | |
| MIPS Subcellular Localization | |||
| nucleolus | 3.5E−6 | 50 GENES | |
| MIPS Protein Complexes | |||
| RNA polymerase II | 8.7E−6 | RPB4, RPB5, RPB8, RPB9, RPB10, RPB11, RPC10, RPO26 | |
| PFam-A Domains | |||
| LSM | 1.1E−12 | LSM2, LSM3, LSM4, LSM5, LSM6, LSM7, LSM8, MAK31, SMB1, SMD1, SMD2, SMD3, SME1, SMX2 | |
| Up-regulated genes | |||
| MIPS Functional Classification | |||
| Glycolysis and gluconeogenesis | 8.3E−7 | CDC19, ENO1, GLK1, LAT1, PDA1, PGM2, PYC1, PYC2, PYK2, ZWF1 | |
| Metabolism of energy reserves (e.g. glycogen, trehalose) | 2.4E−6 | FKS1, GDB1, GPH1, GSY2, KRE5, NTH1, PGM2, TPS1, TPS2, TPS3, TSL1 | |
| MIPS Phenotypes | |||
| Starvation sensitivity | 1.9E−5 | BCY1, IRA2, KEM1, PEP4, RVS167, SRV2, VRP1 | |
| MIPS Subcellular Localization | |||
| Transport vesicles | 8.8E−5 | APM1, COP1, RPG1, SEC7, SEC16, SEC18, SEC21, SEC24 | |
| Plasma membrane | 1.9E−4 | CSF1, CYR1, DNF1, FKS1, FUI1, GAP1, HXT2, IRA1, IRA2, IST2, KIN1, RSP5, SEC3, SED1, SSO2, YPS1, YPS3 | |
| GO Biological Process | |||
| Vacuolar protein catabolic process | 2.6E−8 | AIM3, ATG2, APE3, CAR2, CSF1, CWP1, GAP1, HPF1, IRA1, IST2, LAT1, NCR1, PDA1, PEP4, PRB1, PYC1, VPS13, VRP1, YIL169C | |
| Endocytosis | 2.7E−7 | CHC1, DNF1, EDE1, FKS1, INP53, MYO5, NEO1, PAN1, PIL1, RSP5, RVS167, SDS24, SLA1, SVL3SWH1, VRP1 | |
| GO Cellular Component | |||
| Actin cortical patch | 3.9E−6 | ABP1, BBC1, EDE1, FKS1, INP53, MYO5, PAN1, RVS167, SRV2, VRP1, | |
Quercetin Induces Glycogen Degradation and Trehalose Biosynthesis
Quercetin induced several genes related with carbohydrate metabolism that are functionally associated with the glycolytic/gluconeogenesis pathway. Most of these genes (HXK1, GLK1, PGM2, ENO1, PYK2, PYC1 and PYC2) are under catabolite repression and their expression increases in response to glucose depletion [36]–[41]. Under these conditions, they function in the gluconeogenesis pathway that is activated for glucose production. Consistent with quercetin inducing a glucose restriction-like phenotype, the HXT2 gene, which encodes a high-affinity glucose transporter induced by low levels of glucose [42], and genes that encode for the Ira1p and Ira2p GTPase-activating proteins (Table 1) were up regulated. These proteins negatively regulate RAS by converting it from the GTP- to the GDP-bound inactive form, which is required for reducing cAMP levels under nutrient limiting conditions, preventing the activation of the glucose-regulated cAMP-dependent protein kinase signaling pathway [43].
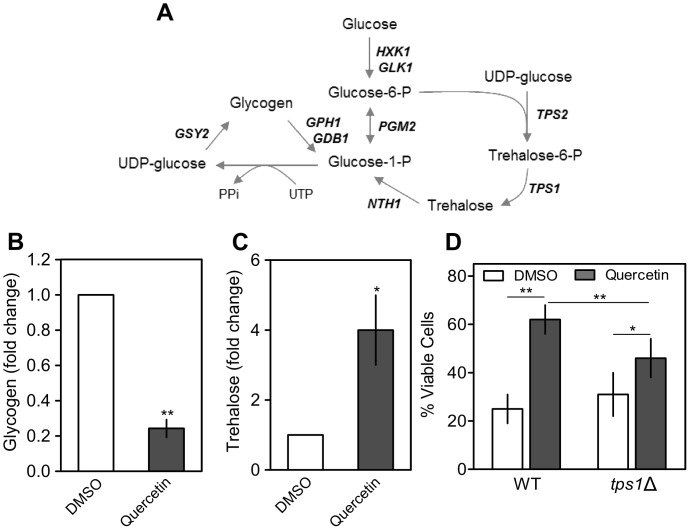
Moreover, quercetin increased the expression of the genes related to metabolism of energy reserves, such as glycogen (GSY2, GPH1 and GDB1) and trehalose (TPS1, TPS2, TSL1, TPS3 and NTH1) (Figure 4A). GSY2 encodes the majority of the glycogen synthase activity in yeast and its expression is induced by environmental stresses, nitrogen starvation and glucose deprivation [44], [45]. GPH1 and GDB1 encode for glycogen phosphorylase and debranching enzyme, respectively, which are involved in glycogen degradation to glucose-1-phosphate, and their expression is induced during late exponential growth phase and in response to various stresses [46]–[48].
Figure 4. Quercetin causes glycogen depletion and increases trehalose levels.
(A) Schematic representation of genes up regulated in cells treated with quercetin related with glycogen and trehalose metabolism. (B, C) S. cerevisiae BY4741 cells were grown to exponential phase and treated with 300 µM of quercetin or equal volume of DMSO (vehicle) for 15 min. (B) Glycogen and (C) trehalose levels were measured as described in Materials and Methods. Values are mean ± SD of three independent assays. *p<0.05; **p<0.01 (paired t-test). D) Hydrogen peroxide resistance. S. cerevisiae BY4741 (WT) and tps1Δ cells were treated with 300 µM of quercetin or equal volume of DMSO (control cells) for 15 min and subsequently exposed to 1.5 mM H2O2 for 1 h. Cell viability was determined by standard dilution plate counts and expressed as the percentage of the colony-forming units of non-stressed cells. Values are mean ± SD of at least three independent assays. **p<0.01; *p<0.05 (two-way ANOVA).
Trehalose is a disaccharide of glucose involved in responses to stress, including oxidative stress, as well as suppression of denatured protein aggregation [49]. Trehalose-6-phosphate synthase (Tps1p) and trehalose-6-phosphate phosphatase (Tps2p) are part of a complex containing also the regulatory proteins Tps3p and Tsl1p, which are co-induced under stress conditions and co-repressed by the glucose regulated Ras-cAMP pathway [50]. Notably, NTH1 gene that encodes a neutral trehalase was also up regulated in cell treated with quercetin. NTH1 is a multiple stress response gene [51]. The simultaneous increase of trehalose hydrolyzing and synthesizing enzymes also occurs during heat stress. This seemingly futile cycling of trehalose turnover during heat stress seems to be necessary for maintenance of a constant glucose concentration in the cytosol and for cellular recovery from stress [52], [53].
Changes in expression of genes related to glycogen and trehalose metabolism led us to measure the levels of these reserve carbohydrates. The results obtained show that glycogen levels decreased approximately 4-fold whereas trehalose levels increased 4-fold upon quercetin-treatment (Figure 4B–C). Overall, the gene expression results and the carbohydrate levels suggest that glycogen is metabolized to provide glucose for trehalose biosynthesis. Moreover, the protective effect of quercetin against H2O2 decreased in tps1Δ cells (Figure 4D): in the presence of this polyphenol, oxidative stress resistance increased 2.5-fold and 1.5-fold in wild type and tps1Δ cells, respectively. This suggests that trehalose production contributes to quercetin-induced H2O2 resistance.
Quercetin Activates the Cell Wall Integrity Pathway
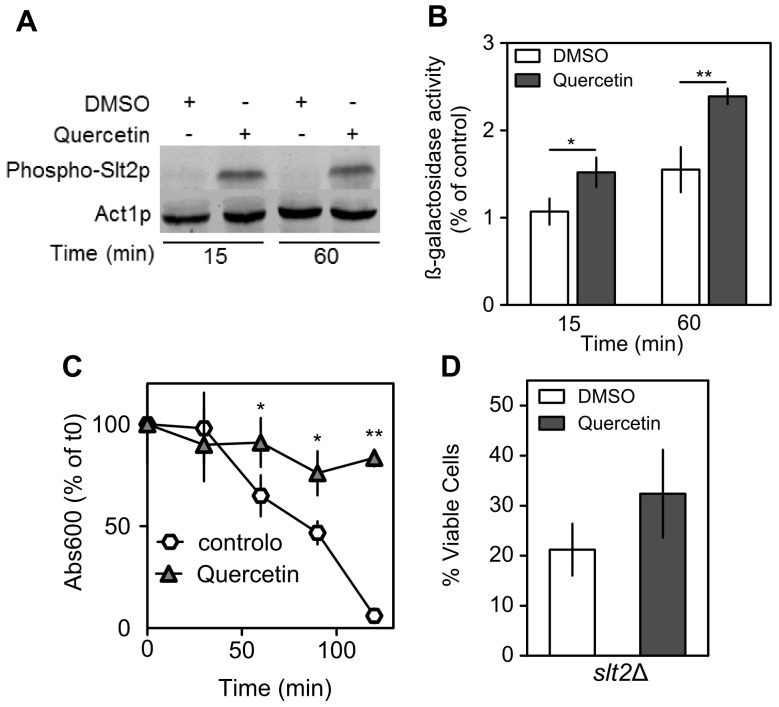
Quercetin also increased the expression of genes encoding proteins that exhibit a cortical patch membrane localization pattern (Table 1), which are involved in the regulation of actin cytoskeleton, endocytosis, cell wall biogenesis and viability following starvation or osmotic stress [54], [55]. Moreover, quercetin up regulated several genes that encode proteins related to the cell wall, such as Sed1p (cell wall glycoprotein), Yps1p and Yps3p (aspartic proteases), and Fks1p (1,3- β-D-glucan synthase) (Table 1). Cell wall biogenesis and maintenance are regulated by the CWI pathway, a Pkc1p-modulated MAPK cascade [56]. The MAPK module is composed of the MAPK kinase kinase Bck1p, a pair of redundant MAPK kinases (Mkk1p and Mkk2p), and the MAPK Slt2p/Mpk1p. To test if quercetin activates this pathway, phospho-Slt2p levels were analyzed by Western blotting, using an anti-phospho-p44/42 antibody that detects dually phosphorylated Slt2p, the active form of this MAPK. Phospho-Slt2p levels were significantly increased in cells treated with quercetin for 15 and 60 min (Figure 5A). We also characterized changes in Rlm1p, a transcription factor regulated by Slt2p [57], by measuring β-galactosidase activity in cells expressing a LacZ reporter under the control of Rlm1p (RLM1-lacZ). Consistent with an induction of the Rlm1p-reporter, β-galactosidase activity increased 50% in cells treated with quercetin (Figure 5B). The activation of the CWI pathway is important for oxidative stress resistance [58] and increases cellular resistance to cell wall perturbing agents, such as zymolyase, a lytic enzyme that degrades β1,3-glucans, the main component of cell wall. Consistent with Slt2p activation, quercetin increased zymolyase resistance (Figure 5C). Moreover, inactivation of SLT2 gene decreased the protective effect of quercetin against H2O2 (Figure 5D). These results indicate that quercetin increases H2O2 resistance through activation of the CWI pathway.
Figure 5. Quercetin activates the cell wall integrity signaling pathway.
(A) Phospho-Slt2 protein levels. S. cerevisiae BY4741 cells were grown to exponential phase and treated with 300 µM of quercetin or equal volume of DMSO for the indicated time periods. Proteins were isolated, separated by SDS-PAGE, blotted into a membrane and incubated with anti-phospho-p44/42 antibody that detects dually phosphorylated Slt2p or with anti-actin antibody (loading control), as described in Materials and Methods. A representative blot is shown. (B) Rlm1p-transcription factor activity. Exponentially growing S. cerevisiae BY4741 cells transformed with a pRLM1-LacZ reporter construct were treated with 300 µM of quercetin or equal volume of DMSO (control) at the indicated times. β-galactosidase activity was determined as described in Materials and Methods and expressed as percentage of control cells. (C) Resistance to zymolyase digestion. S. cerevisiae BY4741 cells were treated with 300 µM of quercetin or equal volume of DMSO for 15 min and incubated with 0.25 U/ml zymolyase at 37°C. Cell lysis was determined spectrophotometrically at 600 nm over time. (D) Hydrogen peroxide resistance of slt2Δ cells was assessed as described in legend to Figure 4D. Values are mean ± SD of at least three independent assays. **p<0.01; *p<0.05 (unpaired t-test).
Quercetin does not Prevent H2O2-induced Actin Polarization
Actin has also been implicated in oxidative stress resistance. Oxidation of yeast actin decreases its dynamics, causing depolarization of the mitochondrial membrane and an increase in ROS production that contributes to cell death [59]. To test if quercetin affects actin dynamics, cells were pre-treated with DMSO or quercetin for 15 min and exposed to 1.5 mM H2O2 during 1 h. To visualize actin, cells were labeled with the rhodamine-phalloidin, a high-affinity F-actin probe conjugated to the red-orange fluorescent dye, tetramethylrhodamine, and observed by fluorescence microscopy. In control cells (pre-treated with DMSO), almost 60% of the cells displayed actin depolarization after exposure to H2O2 (Figure 6). Similar results were observed in cells pre-treated with quercetin, suggesting that its protective effect is not associated with changes in actin polarization.
Figure 6. Quercetin does not affect H2O2-induced actin depolarization.
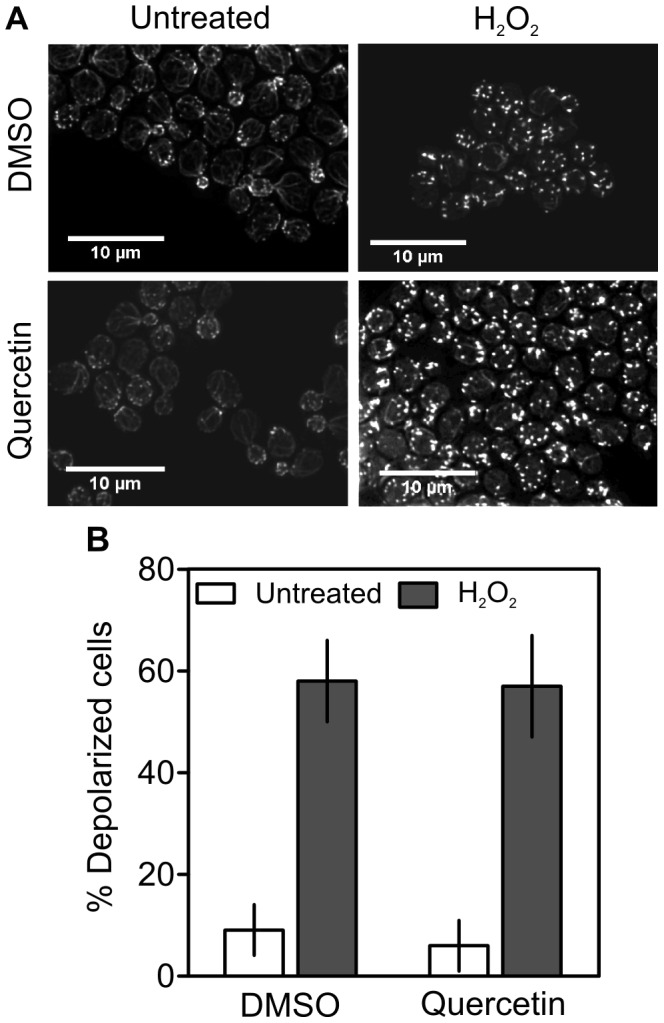
Yeast cells were grown to exponential phase and pre-treated with 300 µM of quercetin or equal volume of DMSO (control cells) for 15 min and subsequently exposed to 1.5 mM H2O2 for 1 h. Cells were incubated with rhodamine-phalloidin to stain actin and visualized on a fluorescence microscope. (A) Deconvolved and stack fluorescence images of cells with actin staining; (B) Percentage of depolarized cells obtained after counting approximately 1500 cells for each treatment condition. Data represent the mean ± SD of three independent assays.
Discussion
Oxidative stress has been implicated in aging and age-related diseases. Mounting evidence suggests that natural compounds with antioxidant properties exert health beneficial effects. The antioxidant activities of polyphenols have been attributed to ROS scavenging. However, a number of recent studies suggest that they also act through modulation of cell signaling pathways that increase cellular defense mechanisms. This phenomenon, known as xenohormesis, refers to sensing in one organism (yeast in this study) of a compound produced in another specie (plant) in response to environmental stress, leading to the induction of a defense response that increases its chances of survival [60]. We have previously shown that quercetin, the most common flavonol in the diet, increases oxidative stress resistance in yeast cells by scavenging free radicals, maintaining the redox homeostasis, and preventing protein carbonylation and lipid peroxidation [33]. Aiming to characterize genome-wide changes in gene expression induced by quercetin in yeast, a microarray analysis was performed. The results obtained show that quercetin down regulated a significant number of genes belonging to RNA metabolism and ribosome biogenesis categories. This cellular adaptation has been observed in response to multiple stress conditions [61] and is beneficial since it spares energy resources for cellular processes other than ribosome biosynthesis (the most energy consuming cellular process).
In yeast, the acquisition of oxidative stress resistance has been associated with adaptive responses that include the induction of antioxidant defences and other stress proteins, mediated by Yap1p, Skn7p and Msn2p transcription factors [62], [63]. The analysis of the promoters of the genes differentially expressed upon quercetin treatment, using YEASTRACT software [64], showed that quercetin did not affect the expression of genes known to be regulated by Yap1p or Msn2p, suggesting that these factors are not key mediators of quercetin protective effects (see supplementary Tables S3 and S4). Although a few Skn7p-target genes were up regulated, they represent 5.2% of the total number of genes directly regulated by this transcription factor. Thus, it is likely that changes in gene expression induced by quercetin are Skn7p-independent.
Notably, quercetin induced several genes belonging to carbohydrate metabolism, including metabolism of energy reserves, known to be repressed by glucose. Specifically, quercetin up regulated genes associated with gluconeogenesis, glycogenolysis, glucose uptake and trehalose biosynthesis, as well as IRA1 and IRA2 genes, which encode GTPase-activating proteins that negatively regulate the glucose-activated cAMP-dependent protein kinase signaling pathway [43]. These metabolic alterations redirect carbohydrate metabolism towards the production of trehalose, whose levels increased 4-fold after exposure to quercetin. Trehalose-6-phosphate, one of the intermediates of this pathway, and Tps1p play a major role in restricting glucose influx into glycolysis [65]. Trehalose is a disaccharide of glucose with stress-protectant functions [49]. Our results indicate that the increase in trehalose production contributes to oxidative stress resistance of cells treated with quercetin, since inactivation of TPS1 gene decreased its protective effect. Interestingly, glycogen levels also decrease whereas trehalose levels remain unchanged in stationary phase (quiescent) cells, which display a high oxidative stress resistance phenotype. Under these conditions, trehalose also functions as an energy reserve used by yeast cells upon exit from the quiescent state [66].
This adaptive, xenohormetic response to quercetin includes features observed under glucose derepression conditions, suggesting that quercetin induces glucose restriction-like phenotypes. Some of these changes were also described for mammalian cells. Quercetin has been shown to stimulate glycogenolysis [67] and to inhibit glucose uptake, insulin signaling and activation of Akt, a downstream effector of PI3K [68], [69]. Quercetin and other flavonoids may also compete with glucose for transmembrane transport [68].
Genes encoding proteins that exhibit a cortical patch membrane localization pattern were also up regulated by exposure to quercetin. These proteins are involved in the regulation of actin cytoskeleton and cellular processes such as cell wall biogenesis [54], [55]. Changes in actin polarization increases mitochondrial ROS production [59]. However, quercetin did not decrease actin depolarization induced by H2O2, suggesting that its protective effect is not associated with the modulation of actin dynamics. In contrast, our results suggest that quercetin exerts its protective effects through the activation of cell wall biogenesis and maintenance. Indeed, this flavonol up regulated several genes encoding cell wall proteins and activated components of the CWI pathway, namely the Slt2p MAPK and the Rlm1p transcription factor [57]. In addition, the increase of H2O2 resistance induced by quercetin was attenuated in slt2Δ mutant cells. These results support a role for CWI pathway in oxidative stress resistance, as previously suggested [58]. Consistent with CWI pathway activation, quercetin also increased cell resistance to zymolyase, a lytic enzyme that causes cell wall stress by degrading β1,3-glucans, the main component of cell wall. The modulation of signal transduction mediated by PKC was also observed in colon cancer cell lines exposed to quercetin [70].
In conclusion, this study shows that quercetin treatment mimics glucose restriction and modulates carbohydrate metabolism in favor of the biosynthesis of trehalose, a disaccharide with antioxidant properties. Moreover, the activation of the CWI pathway in yeast contributes to the xenohormetic activity of quercetin. The overall results suggest that quercetin exerts protective effects through the modulation of cell signaling pathways.
Materials and Methods
Reagents
All reagents and chemicals were of analytical grade. Sodium or potassium phosphates and H2O2 were purchased from Merck (VWR, Lisboa, Portugal), yeast nitrogen base from Difco (Quilaban, Sintra, Portugal), agarose from Invitrogen (Alfagene, Carcavelos, Portugal), and DMSO, quercetin and zymolyase from Sigma (Sintra, Portugal). Quercetin was dissolved in DMSO at a 200 mM stock concentration and stored at −80°C. Solutions were prepared in ultrapure water (MilliQ).
Yeast Cells and Growth Conditions
The Saccharomyces cerevisiae cells (Euroscarf, Germany) used in this study were BY4741 (Matα, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), slt2Δ (BY4741 slt2Δ::KanMX4) and tps1Δ (BY4741 tps1Δ::KanMX4). Yeast cells were grown in minimal medium [0.67% (w/v) yeast nitrogen base with ammonium sulfate without amino acids and 2% (w/v) glucose] supplemented with appropriate amino acids (40 mg His L−1, 80 mg Leu L−1, 40 mg Met L−1) and nucleotides (40 mg Ura L−1). Auxotrophic supplementation was performed before autoclaving at 110°C. Cells were grown to early exponential phase (Abs600nm = 0.6) in an orbital shaker, at 26°C and 120 rpm, with a ratio of flask volume / medium volume of 5∶1. To test the activity of the Rlm1p transcription factor, S. cerevisiae BY4741 cells were transformed by electroporation with p2xRlm1-LacZ plasmid [71] and selected in minimal medium lacking uracil.
RNA Extraction and mRNA Isolation
Exponential phase cells (90 Abs600nm units of yeast) were exposed to 300 µM quercetin (dissolved in DMSO) or equal volume of DMSO (control) by directly adding these compounds to the growth media. After 15 min of treatment, cells were collected by centrifugation at 2,900 g. Total RNA was extracted by a hot acidic phenol method [72], with a yield of approximately 1500 µg. Briefly, cells were resuspended in 500 µL acid phenol:chloroform 5∶1 pH 4.7 and the same volume of TES buffer [10 mM Tris pH 7.5 10 mM ethylenediaminetetraacetic acid (EDTA) 0.5% sodium dodecyl sulfate (SDS)], and incubated at 65°C for 1 h, with occasional, brief vortexing. After centrifugation at 10,000 g for 20 min, RNA was isolated from the aqueous phase by extractions with phenol:chloroform 5∶1 pH 4.7 (twice) and chloroform:isoamyl alcohol 25∶1, and precipitation with 100% ethanol, in the presence of sodium acetate pH 5.2. RNA was dissolved in water and its purity assessed by gel electrophoresis in 1.4% (w/v) agarose and by UV spectrophotometry, using NanoDrop ND-1000 Spectrophotometer. Only RNA samples with A260/280 ratio >1.90 were used in further experiments. For microarray studies, mRNA was isolated from 1500 µg of total RNA, as described in OligotexTM Handbook (Qiagen®), with a yield of approximately 6 µg. For hybridization quality control, mixtures of ten in vitro synthesized RNAs were added from appropriately diluted mixtures to the total RNA samples prior to mRNA enrichment.
cDNA Synthesis and Labeling
cDNA synthesis and labeling were performed as described previously [73]. Briefly, cDNA synthesis was carried out from 3 µg of mRNA enriched samples in the presence of 2-aminoallyl-dUTP (Sigma). cDNA was purified using Microcon YM-30 columns (Millipore, VWR, Lisboa, Portugal) prior to coupling to NHS ester Cy3 or Cy5 (GE Healthcare). Uncoupled dye was removed using Chromaspin-30 columns (Clontech, Enzifarma S.A., Oeiras, Portugal). The efficiency of cDNA synthesis and of dye incorporation was measured by spectrophotometry (NanoDrop). Samples with a degree of labeling (labeled nucleotides per 100 nucleotides) outside the range of 5.6 ± 1.3 were not considered for hybridization.
Microarray Hybridization
Hybridizations were carried out as previously described [73]. Samples derived from quercetin treated cells were labeled with Cy5 and were co-hybridized with control (DMSO) samples labeled with Cy3. Dye-swap hybridization replicates were performed to reduce dye-specific biases in signal intensity. For each comparison (treatment vs. control), 3 biological replicates were analyzed in 2 dye-swapped hybridizations each, in a total of 6 hybridizations. The same amount of differentially labeled treatment and control cDNA (0.30 µg) was mixed and hybridized with microarrays containing 6388 oligonucleotide probes representing the yeast ORFeome in duplicate, fabricated in the laboratory of RNA biology at University of Aveiro, Portugal. A set of 70 mer probes used to detect the spiked-in control RNA added to the total RNA sample were also included in the microarray design, in order to monitor labeling and hybridization quality. Details of the microarray composition can be found in ArrayExpress public database (http://www.ebi.ac.uk/microarray-as/ae/) under the accession number A-MEXP-1185.
Image Acquisition and Data Analysis
Microarray images were obtained with the Agilent G2565BA microarray scanner. Raw data was extracted using QuantArray v3.0 software (PerkinElmer). Pre-processing and normalization of the data was performed using the Biometric Research Branch (BRB)-ArrayTools v3.4.0 software. Briefly, manually flagged bad spots were eliminated and the local background was subtracted prior to averaging of replicate features on the array. Log2 intensity ratios (M values) were adjusted by global Lowess normalization. The MIAME (Minimum Information About a Microarray Experiment) compliant data was deposited in ArrayExpress (http://www.ebi.ac.uk/miamexpress/) under the accession number E-MEXP-3523.
Treatment and control samples were compared using Significance Analysis for Microarrays (SAM) [74] implemented in TM4 Microarray Software Suite (MeV) 4.3 [75]. Significance analysis, with a false discovery rate (90th percentile) of 0.016%, resulted in a common set of 1687 significant genes, from which 834 were selected based on an average fold change of more than 2.0. Statistical analysis of overrepresentation of functional groups was performed by using FUNSPEC [34]. All available databases were addressed by using probability cut-off of 10−3 and the Bonferroni correction for multiple testing. Genes differentially expressed in quercetin-treated cells were sorted in functional groups according to Munich Information Centre for Protein Sequences (MIPS) database (Tables S1 and S2).
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Glycogen phosphorylase (GPH1), hexokinase I (HXK1), RNA polymerase subunit B (RPB5), trehalose-6-phosphate synthase (TPS1) and trehalose synthase subunit (TSL1) transcription profile was validated by RT-qPCR. For each analysis RPS6A and ACT1 were used as internal normalizers. Two micrograms of DNase I-treated (Promega, VWR, Lisboa, Portugal) total RNA were transcribed with the iScriptTM Select cDNA Synthesis Kit (Bio-Rad, Amadora, Portugal), using the random primers supplied and following the manufacturer’s recommendations. Control PCRs were performed with RT-untreated RNA to confirm the absence of contaminating DNA in the RNA preparations. qPCR amplifications were performed using 2 µL of template cDNA, 10 µL of SYBR Green Supermix (Bio-Rad) and 0.2 µM of each primer (Table 2). Reactions were carried out in the iCycler iQ5 Real-Time PCR detection system (Bio-Rad). Cycling conditions were as follows: 95°C for 3 min; 40 cycles of 95°C for 30 s, 56°C or 57°C (according to the set of primers used) for 60 s and 72°C for 30 s. Standard dilutions of the cDNA were used to check the relative efficiency and quality of the primers. Negative controls (no template cDNA) were included in all qPCR. A melting curve analysis was performed at the end of each qPCR assay to exclude the formation of non-specific products. The obtained data was analyzed using the software provided by the supplier (iQ5 Optical System Software v2.1, Bio-Rad) and relative expression of the selected genes in quercetin-treatment vs. control cells was determined using the Pfaffl method [76].
Table 2. Primers used in real-time PCR.
| Primera | Systematic Name | Sequence |
| ACT1_S | YFL039C | TGGATTCTGAGGTTGCTGCTTT |
| ACT1_AS | YFL039C | CCGACGATAGATGGGAAGACAG |
| RPS6A_S | YPL090C | TCGGTCAAGAAGTCGATGGTGA |
| RPS6A_AS | YPL090C | CACCTTGCTTCATTGGGAAACC |
| GPH1_S | YPR160W | ACATGGCTGCTTATGAAGCTGC |
| GPH1_AS | YPR160W | TCCAAAGCCCTACCCATCAAA |
| HXK1_S | YFR053C | AGGTCCAAAGAAACCACAGGCT |
| HXK1_AS | YFR053C | AACCGGGAATCATTGGAATGTT |
| RPB5_S | YBR154C | TCAAGAGGAAGTCGAATTGCCA |
| RPB5_AS | YBR154C | TGGATTTGCCTGGAAGGACA |
| TPS1_S | YBR126C | CGATGAGAAGGATCAGGTGAGG |
| TPS1_AS | YBR126C | GATGGTAATGGAATAACGGCCA |
| TSL1_S | YML100W | CGCCAATCCAACAGCAACAG |
| TSL1_AS | YML100W | GAAGTAGCCGCCGAAGTAGG |
aS, sense; AS, antisense.
Trehalose and Glycogen Assays
The content of intracellular trehalose and glycogen was determined in cells (30 Abs600nm units of yeast) treated with quercetin or DMSO, as described above, by measuring glucose released by trehalase or amyloglucosidase, respectively [77]. The amount of glucose released from trehalose and glycogen was determined using glucose (GO) assay kit (Sigma), according to manufacturer’s recommendations. Results are expressed as fold change relative to control (DMSO-treated) cells.
Oxidative Stress Resistance
Yeast cells (2×107/mL) were pre-treated with quercetin or DMSO for 15 min, as described above, and subsequently exposed to 1.5 mM H2O2 for 1 h. Cell viability was determined by standard dilution plate counts on YPD medium containing 1.5% (w/v) agar. Colonies were counted after growth at 26°C for 3 days. Viability was expressed as the percentage of the colony-forming units.
Actin Staining with Rhodamine-phalloidin
Yeast cells (3 Abs600nm units) were pre-treated with quercetin or DMSO for 15 min and subsequently exposed to 1.5 mM H2O2 for 1 h, as described above. Cells were then fixed by adding directly to the culture 3% formaldehyde for 10 min, washed twice with phosphate-buffered saline (PBS) and fixed in 10% formaldehyde/PBS for 1 h with agitation at room temperature. Cells were washed twice in PBS and permeabilized in 1% tritonX100/PBS for 3 min and washed again with PBS. For actin stain, cells were incubated in the dark for 1 h with 200 U/ml rhodamine-phalloidin/PBS (Invitrogen) and washed one time with PBS before resuspending in 20 µl of anti-bleaching solution (Vectashield, Vector Lab. Inc.) and mounted on agarose beds. All centrifugations were performed at 1,600 g 5 min. Z-stacks images were acquired using a AxioImager Z1® fluorescence microscope (Axiovision 4.7® acquisition software) equipped with a 43HE filter set for rhodamine-phalloidin, a 40x oil-immersion objective and an Axiocam MR ver.3.0® (Carl Zeiss). Z-stack images were deconvolved using Huygens® Essential software. Image stacks and cell counting were performed using Image J/Fiji® software. Cells with an irregular and sparse distribution pattern of rhodamine-phalloidin fluorescence were considered depolarized.
Western Blotting
Yeast cells (30 Abs600nm units) were exposed to quercetin or DMSO for 15 or 60 min, as described above, and collected by centrifugation at 2,900 g. Yeast extracts were prepared in 50 mM potassium phosphate buffer (pH 7.0) containing protease inhibitors (Complete, Mini, EDTA-free Protease Cocktail Inhibitor Tablets; Roche Applied Science, Amadora, Portugal) and phosphatase inhibitors (50 mM sodium fluoride, 5 mM sodium pyrophosphate, 1 mM sodium orthovanadate), by vigorous shaking of the cell suspension in the presence of glass beads for 5 min. Short pulses of 1 min were used, with 1 min intervals on ice. Cell debris was removed by centrifugation at 10,000 g for 15 min at 4°C and protein content was determined by the Lowry method, using bovine serum albumin as a standard. Proteins (50 µg) were separated by SDS-PAGE using 10% polyacrylamide gels and blotted onto a nitrocellulose membrane (Hybond-C, GE Healthcare). The membrane was incubated with rabbit anti-phospho-p44/42 MAPK (Cell Signaling Technology, Izasa, Lisboa, Portugal) at a 1∶2,000 dilution or rabbit anti-actin (Sigma) at a 1∶200 dilution as primary antibodies to detect phospho-Slt2p and actin (loading control), respectively, as previously described [78]. Subsequently, the membrane was incubated with the secondary antibody, goat anti-rabbit IgG-peroxidase (Sigma), at a 1∶5,000 dilution. Immunodetection was performed by chemiluminescence, using a kit from GE Healthcare (RPN 2109). Quantification of band intensities was performed by densitometry using Quantity One package (Bio-Rad).
β-galactosidase Activity
Yeast cells (30 Abs600nm units) expressing the p2xRlm1-LacZ reporter or pLGΔ178 were treated with quercetin or DMSO for 15 or 60 min, as described above, and collected by centrifugation at 2,900 g. The β-galactosidase activity was measured as previously described [33].
Cell Wall Stress Assay
For the analysis of zymolyase sensitivity, yeast cells (2×107/mL) were treated with quercetin or DMSO for 15 min, as described above, washed twice, and resuspended in 10 mM Tris-HCl pH 7.5. Cells were then incubated with 0.25 U/ml zymolyase at 37°C and absorbance at 600 nm followed overtime. A decrease in Abs600nm indicates cell lysis due to degradation of β1,3-glucans, the main component of cell wall.
Statistical Analysis
Data are expressed as mean values ± standard deviation of independent experiments. Values were compared by Student’s t-test or Two-way ANOVA using GraphPad Prism Software v5.01 (GraphPad Software).
Supporting Information
Genes up regulated in quercetin treated cells.
(PDF)
Genes down regulated in quercetin treated cells.
(PDF)
Transcription factors with documented direct regulation of genes induced by exposure to quercetin.
(PDF)
Transcription factors with documented direct regulation of genes repressed by exposure to quercetin.
(PDF)
Acknowledgments
We are grateful to the National Facility for DNA Microarrays at University of Aveiro for DNA microarray analysis, Paula Magalhães (CCGen) for excellent technical assistance, and David E. Levin (Goldman School of Dental Medicine, Boston University, Boston, USA) for generously providing the p2xRlm1-LacZ plasmid used in this study.
Funding Statement
This work was financially supported by FEDER (Fundo Europeu de Desenvolvimento Regional) through the program “Programa Operacional Factores de Competitividade-COMPETE” and by FCT (Fundação para a Ciência e Tecnologia) through the projects FCOMP-01-0124-FEDER-007422-PTDC/QUI/65501/2006 and FCT/FEDER/PTDC/BIA-BCM/64745/2006. R.V. was supported by iBeSa (Instituto de Bebidas e Saúde) and V.M. by FCT (fellowship PTDC/QUI/65501/2006). M.V.M. was supported by “Programa Ciência 2007” sponsored by POPH (Programa Operacional Potencial Humano) QREN (Quadro de Referência Estratégico Nacional) Tipologia 4.2 Promocão do Emprego Científico program and co-financed by the European Social Fund and Portuguese national funds from the MCTES (Ministério da Ciência, Tecnologia e Ensino Superior). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alameddine FM, Zafari AM (2002) Genetic polymorphisms and oxidative stress in heart failure. Congest Heart Fail 8: 157–164, 172. [DOI] [PubMed]
- 2. Acharya A, Das I, Chandhok D, Saha T (2010) Redox regulation in cancer: a double-edged sword with therapeutic potential. Oxid Med Cell Longev 3: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Radak Z, Zhao Z, Goto S, Koltai E (2011) Age-associated neurodegeneration and oxidative damage to lipids, proteins and DNA. Mol Aspects Med 32: 305–315. [DOI] [PubMed] [Google Scholar]
- 4. Halliwell B (1999) Antioxidant defence mechanisms: from the beginning to the end (of the beginning). Free Radic Res 31: 261–272. [DOI] [PubMed] [Google Scholar]
- 5. Trujillo M, Ferrer-Sueta G, Radi R (2008) Peroxynitrite detoxification and its biologic implications. Antioxid Redox Signal 10: 1607–1620. [DOI] [PubMed] [Google Scholar]
- 6. Prior RL (2003) Fruits and vegetables in the prevention of cellular oxidative damage. Am J Clin Nutr 78: 570S–578S. [DOI] [PubMed] [Google Scholar]
- 7. Quideau S, Deffieux D, Douat-Casassus C, Pouysegu L (2011) Plant Polyphenols: Chemical Properties, Biological Activities, and Synthesis. Angew Chem Int Ed Engl 50: 586–621. [DOI] [PubMed] [Google Scholar]
- 8. Erlund I (2004) Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr Res 24: 851–874. [Google Scholar]
- 9. Pietsch K, Saul N, Menzel R, Sturzenbaum SR, Steinberg CE (2009) Quercetin mediated lifespan extension in Caenorhabditis elegans is modulated by age-1, daf-2, sek-1 and unc-43. Biogerontol 10: 565–578. [DOI] [PubMed] [Google Scholar]
- 10. Zhou W, Kallifatidis G, Baumann B, Rausch V, Mattern J, et al. (2010) Dietary polyphenol quercetin targets pancreatic cancer stem cells. Int J Oncol 37: 551–561. [DOI] [PubMed] [Google Scholar]
- 11. Kobori M, Masumoto S, Akimoto Y, Oike H (2011) Chronic dietary intake of quercetin alleviates hepatic fat accumulation associated with consumption of a Western-style diet in C57/BL6J mice. Mol Nutr Food Res 55: 530–540. [DOI] [PubMed] [Google Scholar]
- 12. Park HK, Kim SJ, Kwon do Y, Park JH, Kim YC (2010) Protective effect of quercetin against paraquat-induced lung injury in rats. Life Sci 87: 181–186. [DOI] [PubMed] [Google Scholar]
- 13. Shoskes DA, Zeitlin SI, Shahed A, Rajfer J (1999) Quercetin in men with category III chronic prostatitis: a preliminary prospective, double-blind, placebo-controlled trial. Urology 54: 960–963. [DOI] [PubMed] [Google Scholar]
- 14. Meyers KJ, Rudolf JL, Mitchell AE (2008) Influence of dietary quercetin on glutathione redox status in mice. J Agric Food Chem 56: 830–836. [DOI] [PubMed] [Google Scholar]
- 15. Lee EJ, Ji GE, Sung MK (2010) Quercetin and kaempferol suppress immunoglobulin E-mediated allergic inflammation in RBL-2H3 and Caco-2 cells. Inflamm Res 59: 847–854. [DOI] [PubMed] [Google Scholar]
- 16. Vargas AJ, Burd R (2010) Hormesis and synergy: pathways and mechanisms of quercetin in cancer prevention and management. Nutr Rev 68: 418–428. [DOI] [PubMed] [Google Scholar]
- 17. Lee KW, Kang NJ, Heo YS, Rogozin EA, Pugliese A, et al. (2008) Raf and MEK protein kinases are direct molecular targets for the chemopreventive effect of quercetin, a major flavonol in red wine. Cancer Res 68: 946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weng CJ, Chen MJ, Yeh CT, Yen GC (2011) Hepatoprotection of quercetin against oxidative stress by induction of metallothionein expression through activating MAPK and PI3K pathways and enhancing Nrf2 DNA-binding activity. New Biotechnol 28: 767–777. [DOI] [PubMed] [Google Scholar]
- 19. Kim HJ, Kim SK, Kim BS, Lee SH, Park YS, et al. (2010) Apoptotic effect of quercetin on HT-29 colon cancer cells via the AMPK signaling pathway. J Agric Food Chem 58: 8643–8650. [DOI] [PubMed] [Google Scholar]
- 20. Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, et al. (2000) Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol Cell 6: 909–919. [DOI] [PubMed] [Google Scholar]
- 21. Kampkotter A, Timpel C, Zurawski RF, Ruhl S, Chovolou Y, et al. (2008) Increase of stress resistance and lifespan of Caenorhabditis elegans by quercetin. Comp Biochem Physiol B Biochem Mol Biol 149: 314–323. [DOI] [PubMed] [Google Scholar]
- 22. Chen RE, Thorner J (2007) Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae . Biochim Biophys Acta 1773: 1311–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu MJ, O'Doherty PJ, Fernandez HR, Lyons V, Rogers PJ, et al. (2011) An antioxidant screening assay based on oxidant-induced growth arrest in Saccharomyces cerevisiae . FEMS Yeast Res 11: 379–387. [DOI] [PubMed] [Google Scholar]
- 24. Martorell P, Forment JV, de Llanos R, Monton F, Llopis S, et al. (2011) Use of Saccharomyces cerevisiae and Caenorhabditis elegans as model organisms to study the effect of cocoa polyphenols in the resistance to oxidative stress. J Agric Food Chem 59: 2077–2085. [DOI] [PubMed] [Google Scholar]
- 25. Jimenez A, Lisa-Santamaria P, Garcia-Marino M, Escribano-Bailon MT, Rivas-Gonzalo JC, et al. (2010) The biological activity of the wine anthocyanins delphinidin and petunidin is mediated through Msn2 and Msn4 in Saccharomyces cerevisiae . FEMS Yeast Res 10: 858–869. [DOI] [PubMed] [Google Scholar]
- 26. Dani C, Bonatto D, Salvador M, Pereira MD, Henriques JA, et al. (2008) Antioxidant protection of resveratrol and catechin in Saccharomyces cerevisiae . J Agric Food Chem 56: 4268–4272. [DOI] [PubMed] [Google Scholar]
- 27. Hwang EI, Ahn BT, Lee HB, Kim YK, Lee KS, et al. (2001) Inhibitory activity for chitin synthase II from Saccharomyces cerevisiae by tannins and related compounds. Planta Med 67: 501–504. [DOI] [PubMed] [Google Scholar]
- 28. Uchida E, Ohsumi Y, Anraku Y (1985) Purification and properties of H+-translocating, Mg2+-adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae . J Biol Chem 260: 1090–1095. [PubMed] [Google Scholar]
- 29. Meggio F, Grankowski N, Kudlicki W, Szyszka R, Gasior E, et al. (1986) Structure and properties of casein kinase-2 from Saccharomyces cerevisiae. A comparison with the liver enzyme. Eur J Biochem 159: 31–38. [DOI] [PubMed] [Google Scholar]
- 30. Moriya H, Johnston M (2004) Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci U S A. 101: 1572–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bednarska S, Leroy P, Zagulski M, Bartosz G (2008) Efficacy of antioxidants in the yeast Saccharomyces cerevisiae correlates with their effects on protein thiols. Biochimie 90: 1476–1485. [DOI] [PubMed] [Google Scholar]
- 32. Amari F, Fettouche A, Samra MA, Kefalas P, Kampranis SC, et al. (2008) Antioxidant small molecules confer variable protection against oxidative damage in yeast mutants. J Agric Food Chem 56: 11740–11751. [DOI] [PubMed] [Google Scholar]
- 33. Belinha I, Amorim MA, Rodrigues P, de Freitas V, Moradas-Ferreira P, et al. (2007) Quercetin increases oxidative stress resistance and longevity in Saccharomyces cerevisiae . J Agric Food Chem 55: 2446–2451. [DOI] [PubMed] [Google Scholar]
- 34. Robinson MD, Grigull J, Mohammad N, Hughes TR (2002) FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kufel J, Allmang C, Petfalski E, Beggs J, Tollervey D (2003) Lsm Proteins are required for normal processing and stability of ribosomal RNAs. J Biol Chem 278: 2147–2156. [DOI] [PubMed] [Google Scholar]
- 36. Rodriguez A, De La Cera T, Herrero P, Moreno F (2001) The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae . Biochem J 355: 625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oh D, Hopper JE (1990) Transcription of a yeast phosphoglucomutase isozyme gene is galactose inducible and glucose repressible. Mol Cell Biol 10: 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boy-Marcotte E, Perrot M, Bussereau F, Boucherie H, Jacquet M (1998) Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae . J Bacteriol 180: 1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cohen R, Yokoi T, Holland JP, Pepper AE, Holland MJ (1987) Transcription of the constitutively expressed yeast enolase gene ENO1 is mediated by positive and negative cis-acting regulatory sequences. Mol Cell Biol 7: 2753–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boles E, Schulte F, Miosga T, Freidel K, Schluter E, et al. (1997) Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J Bacteriol 179: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brewster NK, Val DL, Walker ME, Wallace JC (1994) Regulation of pyruvate carboxylase isozyme (PYC1, PYC2) gene expression in Saccharomyces cerevisiae during fermentative and nonfermentative growth. Arch Biochem Biophys 311: 62–71. [DOI] [PubMed] [Google Scholar]
- 42. Ozcan S, Johnston M (1999) Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev 63: 554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Colombo S, Ronchetti D, Thevelein JM, Winderickx J, Martegani E (2004) Activation state of the Ras2 protein and glucose-induced signaling in Saccharomyces cerevisiae . J Biol Chem 279: 46715–46722. [DOI] [PubMed] [Google Scholar]
- 44. Ni HT, LaPorte DC (1995) Response of a yeast glycogen synthase gene to stress. Mol Microbiol 16: 1197–1205. [DOI] [PubMed] [Google Scholar]
- 45. Parrou JL, Enjalbert B, Francois J (1999) STRE- and cAMP-independent transcriptional induction of Saccharomyces cerevisiae GSY2 encoding glycogen synthase during diauxic growth on glucose. Yeast 15: 1471–1484. [DOI] [PubMed] [Google Scholar]
- 46. Hwang PK, Tugendreich S, Fletterick RJ (1989) Molecular analysis of GPH1, the gene encoding glycogen phosphorylase in Saccharomyces cerevisiae . Mol Cell Biol 9: 1659–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Teste MA, Enjalbert B, Parrou JL, Francois JM (2000) The Saccharomyces cerevisiae YPR184w gene encodes the glycogen debranching enzyme. FEMS Microbiol Lett 193: 105–110. [DOI] [PubMed] [Google Scholar]
- 48. Sunnarborg SW, Miller SP, Unnikrishnan I, LaPorte DC (2001) Expression of the yeast glycogen phosphorylase gene is regulated by stress-response elements and by the HOG MAP kinase pathway. Yeast 18: 1505–1514. [DOI] [PubMed] [Google Scholar]
- 49. Gancedo C, Flores CL (2004) The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res 4: 351–359. [DOI] [PubMed] [Google Scholar]
- 50. Winderickx J, de Winde JH, Crauwels M, Hino A, Hohmann S, et al. (1996) Regulation of genes encoding subunits of the trehalose synthase complex in Saccharomyces cerevisiae: novel variations of STRE-mediated transcription control? Mol Gen Genet 252: 470–482. [DOI] [PubMed] [Google Scholar]
- 51. Zahringer H, Burgert M, Holzer H, Nwaka S (1997) Neutral trehalase Nth1p of Saccharomyces cerevisiae encoded by the NTH1 gene is a multiple stress responsive protein. FEBS Lett 412: 615–620. [DOI] [PubMed] [Google Scholar]
- 52. Hottiger T, Schmutz P, Wiemken A (1987) Heat-induced accumulation and futile cycling of trehalose in Saccharomyces cerevisiae . J Bacteriol 169: 5518–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nwaka S, Kopp M, Holzer H (1995) Expression and function of the trehalase genes NTH1 and YBR0106 in Saccharomyces cerevisiae . J Biol Chem 270: 10193–10198. [DOI] [PubMed] [Google Scholar]
- 54. Donnelly SF, Pocklington MJ, Pallotta D, Orr E (1993) A proline-rich protein, verprolin, involved in cytoskeletal organization and cellular growth in the yeast Saccharomyces cerevisiae . Mol Microbiol 10: 585–596. [DOI] [PubMed] [Google Scholar]
- 55. Friesen H, Colwill K, Robertson K, Schub O, Andrews B (2005) Interaction of the Saccharomyces cerevisiae cortical actin patch protein Rvs167p with proteins involved in ER to Golgi vesicle trafficking. Genetics 170: 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Levin DE (2005) Cell wall integrity signaling in Saccharomyces cerevisiae . Microbiol Mol Biol Rev 69: 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim KY, Truman AW, Levin DE (2008) Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol Cell Biol 28: 2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vilella F, Herrero E, Torres J, de la Torre-Ruiz MA (2005) Pkc1 and the upstream elements of the cell integrity pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, are required for cellular responses to oxidative stress. J Biol Chem 280: 9149–9159. [DOI] [PubMed] [Google Scholar]
- 59. Gourlay CW, Carpp LN, Timpson P, Winder SJ, Ayscough KR (2004) A role for the actin cytoskeleton in cell death and aging in yeast. J Cell Biol 164: 803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Howitz KT, Sinclair DA (2008) Xenohormesis: Sensing the Chemical Cues of Other Species. Cell 133: 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, et al. (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rodrigues-Pousada CA, Nevitt T, Menezes R, Azevedo D, Pereira J, et al. (2004) Yeast activator proteins and stress response: an overview. FEBS Lett 567: 80–85. [DOI] [PubMed] [Google Scholar]
- 63. de la Torre-Ruiz MA, Mozo-Villarias A, Pujol N, Petkova MI (2010) How budding yeast sense and transduce the oxidative stress signal and the impact in cell growth and morphogenesis. Curr Protein Pept Sci 11: 669–679. [DOI] [PubMed] [Google Scholar]
- 64. Monteiro PT, Mendes ND, Teixeira MC, d'Orey S, Tenreiro S, et al. (2008) YEASTRACT-DISCOVERER: new tools to improve the analysis of transcriptional regulatory associations in Saccharomyces cerevisiae . Nucleic Acids Res 36: D132–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Noubhani A, Bunoust O, Rigoulet M, Thevelein JM (2000) Reconstitution of ethanolic fermentation in permeabilized spheroplasts of wild-type and trehalose-6-phosphate synthase mutants of the yeast Saccharomyces cerevisiae . Eur J Biochem 267: 4566–4576. [DOI] [PubMed] [Google Scholar]
- 66. Shi L, Sutter BM, Ye X, Tu BP (2010) Trehalose is a key determinant of the quiescent metabolic state that fuels cell cycle progression upon return to growth. Mol Biol Cell 21: 1982–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gasparin FR, Salgueiro-Pagadigorria CL, Bracht L, Ishii-Iwamoto EL, Bracht A, et al. (2003) Action of quercetin on glycogen catabolism in the rat liver. Xenobiotica 33: 587–602. [DOI] [PubMed] [Google Scholar]
- 68. Strobel P, Allard C, Perez-Acle T, Calderon R, Aldunate R, et al. (2005) Myricetin, quercetin and catechin-gallate inhibit glucose uptake in isolated rat adipocytes. Biochem J 386: 471–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nomura M, Takahashi T, Nagata N, Tsutsumi K, Kobayashi S, et al. (2008) Inhibitory mechanisms of flavonoids on insulin-stimulated glucose uptake in MC3T3-G2/PA6 adipose cells. Biol Pharm Bull 31: 1403–1409. [DOI] [PubMed] [Google Scholar]
- 70. van Erk MJ, Roepman P, van der Lende TR, Stierum RH, Aarts JM, et al. (2005) Integrated assessment by multiple gene expression analysis of quercetin bioactivity on anticancer-related mechanisms in colon cancer cells in vitro. Eur J Nutr 44: 143–156. [DOI] [PubMed] [Google Scholar]
- 71. Jung US, Sobering AK, Romeo MJ, Levin DE (2002) Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol Microbiol 46: 781–789. [DOI] [PubMed] [Google Scholar]
- 72.Ausubel FA, Brent R, Kingston D, Moore D, Seidman JG, et al. (1998) Current Protocols in Molecular Biology, New York: John Wiley and Sons.
- 73. van de Peppel J, Kemmeren P, van Bakel H, Radonjic M, van Leenen D, et al. (2003) Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep 4: 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Saeed AI, Sharov V, White J, Li J, Liang W, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378. [DOI] [PubMed] [Google Scholar]
- 76. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Parrou JL, Francois J (1997) A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem 248: 186–188. [DOI] [PubMed] [Google Scholar]
- 78. Bermejo C, Rodriguez E, Garcia R, Rodriguez-Pena JM, Rodriguez de la Concepcion ML, et al. (2008) The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol Biol Cell 19: 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes up regulated in quercetin treated cells.
(PDF)
Genes down regulated in quercetin treated cells.
(PDF)
Transcription factors with documented direct regulation of genes induced by exposure to quercetin.
(PDF)
Transcription factors with documented direct regulation of genes repressed by exposure to quercetin.
(PDF)