Abstract
In the bone marrow cavity, hematopoietic stem cells (HSC) have been shown to reside in the endosteal and subendosteal perivascular niches, which play specific roles on HSC maintenance. Although cells with long-term ability to reconstitute full hematopoietic system can be isolated from both niches, several data support a heterogenous distribution regarding the cycling behavior of HSC. Whether this distinct behavior depends upon the role played by the stromal populations which distinctly create these two niches is a question that remains open. In the present report, we used our previously described in vivo assay to demonstrate that endosteal and subendosteal stromal populations are very distinct regarding skeletal lineage differentiation potential. This was further supported by a microarray-based analysis, which also demonstrated that these two stromal populations play distinct, albeit complementary, roles in HSC niche. Both stromal populations were preferentially isolated from the trabecular region and behave distinctly in vitro, as previously reported. Even though these two niches are organized in a very close range, in vivo assays and molecular analyses allowed us to identify endosteal stroma (F-OST) cells as fully committed osteoblasts and subendosteal stroma (F-RET) cells as uncommitted mesenchymal cells mainly represented by perivascular reticular cells expressing high levels of chemokine ligand, CXCL12. Interestingly, a number of cytokines and growth factors including interleukin-6 (IL-6), IL-7, IL-15, Hepatocyte growth factor (HGF) and stem cell factor (SCF) matrix metalloproteases (MMPs) were also found to be differentially expressed by F-OST and F-RET cells. Further microarray analyses indicated important mechanisms used by the two stromal compartments in order to create and coordinate the “quiescent” and “proliferative” niches in which hematopoietic stem cells and progenitors reside.
Keywords: bone marrow, trabecular bone, microenvironment, osteoblast, niche
Introduction
Hematopoietic stem cells (HSCs) are able to self-renew and differentiate into lympho-hematopoietic cells through a finely controlled balance between intrinsic signals and microenvironment derived factors (Wilson and Trumpp, 2006; Morrison and Spradling, 2008). In order to maintain this balance, HSCs reside and lodge in highly specialized niches – endosteal and perivascular – composed by stromal cells that modulate HSCs behavior – self-renewal, quiescence, proliferation, and commitment (Heissig et al., 2002; Calvi et al., 2003; Zhang et al., 2003; Arai et al., 2004; Visnjic et al., 2004; Kiel et al., 2005; Kollet et al., 2006; Sugiyama et al., 2006; Bourke et al., 2009; Butler et al., 2010; Robey 2011). The role of endosteal osteoblasts on HSC maintenance and self-renewal was first proposed in vitro by Taichman and Emerson (1994a; 1994b; 1996; 1998) and later in vivo evidence was acquired by others (Calvi et al., 2003; Zhang et al., 2003; Visnjic et al., 2004). An increased number of osteoblasts in the marrow cavity lead to an increased number of long-term HSC, without affecting any other hematopoietic subpopulation in the bone marrow (Calvi et al., 2003; Zhang et al., 2003). Furthermore, osteoblast ablation from the marrow cavity results in a loss of HSCs (Visnjic et al., 2004). These data provide evidence that osteoblasts play a crucial role in HSC maintenance and behavior.
HSCs also reside in the perivascular niche (Arai et al., 2004; Kiel et al., 2005; Sugiyama et al., 2006), in the abluminal side of bone marrow sinusoids, interacting with the endothelial and perivascular reticular cells. Sugiyama et al.9 observed that HSCs residing in the perivascular niche were in close association with reticular cells, which express high levels of CXCL12, a chemokine required for HSC maintenance and lodging (Jung et al., 2006). In situ observation demonstrated that most of hematopoietic stem cells are concentrated in the trabecular zone of the marrow cavity, which also harbors high numbers of niche osteblasts, sinusoids, and CXCL12-positive reticular cells (Calvi et al., 2003; Zhang et al., 2003; Sugiyama et al., 2006). Functional assays indicated that HSC maintenance by both endosteal and perivascular niches are, at least in part, mediated by Jagged-Notch (Calvi et al., 2003) and angiopoietin-1-Tie2 interactions (Arai et al., 2004).
Several studies demonstrate that fast-cycling HSCs preferentially reside in vascular niche, also referred as to “the proliferative niche”, whereas quiescent/slow-cycling HSCs are found in close association with endosteal osteoblasts, which create a “quiescent niche” (Zhang et al., 2003; Arai et al., 2004; Wilson and Trumpp, 2006; Parmar et al., 2007; Robey 2011). This is evidenced in in vivo myelosuppressive models, in which HSCs colonizing the vascular niches in the subendosteal region are mostly ablated. Although still controversial, and depending on the ablation protocol used, almost all HSCs in contact with endosteal osteoblasts are preserved (Arai et al., 2004; Kiel et al., 2007). Whether this heterogeneous distribution and/or HSC cycling behavior regarding the two niches depend upon the role played by the two stromal populations is a question that remains open.
Different markers, such as Osteopontin and Sca-1, have been used to distinguish and isolate osteoblasts from other stromal cells (Mayack and Wagers, 2008; Winkler et al., 2010). However, most of the markers tested are not only expressed by osteoblasts, but also mesenchymal cells at distinct stage of differentiation. In our previous work, we established a protocol to isolate and culture separately endosteal osteoblasts (F-OST) and subendosteal reticular cells (F-RET) from the marrow cavity of murine long bones (Balduino et al., 2005) and suggested that F-OST and F-RET fractions mainly comprised osteoblasts and undifferentiated mesenchymal cells, respectively. In the present study, our main objective was to elucidate the in vivo potential of these two closely associated stromal populations, which are responsible for HSC niche creation in the marrow cavity. Despite the close range, an in vivo ectopic ossicle formation assay (Taichman et al., 2010) allowed us to demonstrate that cells from the two fractions are totally distinct regarding the commitment to the skeletal lineage differentiation cascade. Following in vivo assays, global gene analyses establishes the differences between these two stromal populations and also give us directions to better understand the possible roles played by the stromal cells in the creation of HSC niches.
Methods
Animals and Cells
Pathogen-free C57BL/6 and BALB/c mice were purchased from Jackson Laboratories (Bar Harbor, ME) and Charles River Laboratories (Wilmington, MA), respectively, and housed in environmentally controlled rooms with a 12h light-dark cycle according to the procedures outlined in the "Guide for the Care and Use of Laboratory Animals" [NIH publication no. 86-23, 1985]. Cells were harvested from 8 – 12 week old mice, as previously described (Balduino et al., 2005). Briefly, femurs were removed, cleaned of all connective tissue, and digested in trypsin and collagenase (1:1 – 5×30 min; both from Sigma, St. Louis, MO, USA). Epiphyses were removed and marrows were flushed with DMEM supplemented with 10% FBS. Bones were sectioned and washed, and fragments were digested twice with 0.1% collagenase, for 30 minutes each. Cells were harvested separately, suspended and plated in DMEM 10% FBS. Bone fragments were plated together with cells harvested from the second collagenase digestion. Cells harvested from the first and second collagenase treatments were named F-RET and F-OST, respectively. All experiments on animals were performed following institutional guidelines.
Scanning Electron Microscopy
Femurs were excised and marrows were flushed. A few femurs underwent collagenase digestion. All bones were fixed, post-fixed, and dehydrated as previously described. Using a scalpel, femurs were cut longitudinally (half-pipe-like). Bone inner face was metalized and examined with ZEISS DSM 940A scanning electron microscope at 15 kV.
In Vivo Assessment of Multi-lineage Activity
Flushed bone marrow (BM), subendosteal (F-RET), and endosteal (F-OST) populations were tested for in vivo multipotency (Taichman et al., 2010). Cells were isolated from animals as described above. In some cases the donor mice were treated with 5-FU (American Pharmaceutical Partners, Schaumburg, IL) administrated intravenously via the lateral tail vein at a dose of 150 mg/kg body weight in 100 µL PBS five days prior to marrow harvest. Control animals were injected with an equal volume of 0.9% sodium chloride vehicle solution. Cell/scaffold constructs were transplanted subcutaneously into 5-week -old male mice (NIH-bg-nu-xid BR, Harlan Sprague Dawley, Indianapolis, IN), as previously described (Wang et al., 2006). The implants were harvested 5 weeks post surgery, fixed, and analysed in an EVS corp. µCT scanner (London, Ontario, Canada). GEMS MicroView® software was used to make a 3-D reconstruction from the set of scans. For histology, the specimens were decalcified for 3 days in 10% formic acid, embedded in paraffin and 5-µm serial sections prepared and stained with hematoxylin and eosin.
Microarray Analyses
Flushed bone marrow (BM), F-RET, and F-OST populations were isolated as previously described (Balduino et al., 2005), and cultured in DMEM 10% FBS (Invitrogen Life-Technologies, Carlsbad, CA) up to 90% confluence. Hematopoietic cell distributions on the three niches were evaluated by flow cytometry (Supplemental Figure 1). CD45+ Lin+ cells were magnetically depleted (Miltenyi Biotec Inc., Auburn, CA). Stromal cells were analyzed in triplicate. Microarray and Real Time RT-PCR were achieved as previously described (Lustig et al., 2009). Functional grouping was performed on the Ingenuity Pathways Analysis tool (https://analysis.ingenuity.com/pa/login/login.jsp). Further distance-based gene selection was done on the Microarray Data Analysis website of the National Human Genome Research Institute (http://arrayanalysis.nih.gov).
Real Time RT-PCR
One microgram of RNA was used to generate cDNA. The SYBR Green I assay and the GeneAmp 5700 Sequence Detection System (PE Applied Biosystems) were used for the detection of real-time PCR products as previously described (Lustig et al., 2009). Primers are listed in Table 1.
Table 1.
Primer sequences for the real time reverse transcriptase polymerase chain reaction
| Gene | Gene Name | Primer set (5’ – 3’) |
|---|---|---|
| β-actin | actin, β |
S CCTAAGGCCAACCGTGAAAA 3’ A CCATCACAATGCCTGTGGTA |
| CD164l1 | CD164 sialomucin-like 1 |
S ATGAGTGCCAGATTGCTGGT A AGCTGATACCATCTGCCTCAA |
| CD34 | CD34 antigen |
S CACATCTAGCATCCCTGGAA A TTGGGGAAGTCTGTGGTTGT |
| Cdh13 | cadherin 13 |
S GGAGTGGATCAAGACCCTAAA A ACTGGCCCTTCCAGAGTTTT |
| Cebpb | CCAAT/enhancer binding protein (C/EBP), beta |
S GTTTCGGGACTTGATGCAAT A ACCCCGCAGGAACATCTTTA |
| Cebpd | CCAAT/enhancer binding protein (C/EBP), delta |
S CCTTTGAGACTCTGAACGACCT A ATAGCTTCTCTCGCAGTCCA |
| Ctgf | connective tissue growth factor |
S ATCTCCACCCGAGTTACCAA A GACAGGCTTGGCGATTTTAG |
| Fzd6 | frizzled homolog 6 (Drosophila) |
S ACCCTGTCGGAAATTGTGTG A AAGCTCTGTGTGTGGATGAGAA |
| Fzd9 | frizzled homolog 9 (Drosophila) |
S AACCCCGAGAAGTTCCAGTA A AGGAAGGTGAACACGGTGAA |
| Gpc1 | glypican 1 |
S TATTGCCGAAATGTGCTCAA A TGAGTGTGTCCTTGTTGTCCT |
| Hgf | hepatocyte growth factor |
S CCCATTACTGAAGATTAAAACCAAA A CCACTTGACATACTATTGAAAGGA |
| Ibsp | integrin binding sialoprotein (bone sialoprotein) |
S GAAAATGGAGACGGCGATAG A TGGAAAGTGTGGCGTTCTCT |
| IL11ra1 | interleukin 11 receptor, alpha chain 1 |
S AGCCCTGATGAAGGCACTTA A TGGACTCCAAGTACAGGAGAAG |
| IL15 | interleukin 15 |
S ATCCTGCTGTGTTTGGAAGG A GCTGACATGGGTTTCTGTGTT |
| IL6 | interleukin 6 |
S CGATGATGCACTTGCAGAAA A TGTTCTTCATGTACTCCAGGTAGC |
| Kitl | stem cell factor |
S TCTCAAATATTTCTGAAGGCTTG A CAGGAGTAAAGGATCTAGTTTCTGG |
| Mgp | matrix gamma-carboxyglutamate (gla) protein |
S TCAACAGGAGAAATGCCAAC A ATCTCGTAGGCAGGCTTGTT |
| Mmp13 | matrix metalloproteinase 13 |
S AGTGCCTGATGTGGGTGAAT A TGGTGAAATTCAGTGGTGTCA |
| Mmp3 | matrix metalloproteinase 3 |
S TGATGCATAAGCCCAGGTGT A CAAAGCTTTTTCAATGGCAGA |
| Osmr | oncostatin M receptor |
S AATCAGGGCTCTGGTAGATGA A ATGGTGACATTGGAGCCTTC |
| Osx | Osterix (Osx-pending) |
S TTCTCTCCATCTGCCTGACT A AGCGTATGGCTTCTTTGTGC |
| Otc | Bone gamma-carboxyglutamate protein 1 (Osteocalcin) |
S AAGCAGGAGGGCAATAAGGT A AGGCGGTCTTTAAGCCATAC |
| Pdgfa | platelet derived growth factor, alpha |
S TCCCATGCCATTAACCATGT A ATCAGGAAGTTGGCCGATGT |
| Pdgfra | platelet derived growth factor receptor, alpha polypeptide |
S GCTTCAACGGAACCTTCA A TCGTTGTTAAAGACGGCACA |
| Runx2 | runt related transcription factor 2 |
S GCCGGGAATGATGAGAACTA A TGGGGAGGATTTGTGAAGAC |
| Sdc4 | syndecan 4 |
S GGGATGACATGTCCAACAAA A TCGTAACTGCCTTCGTCCTT |
| Sfrp1 | secreted frizzled-related sequence protein 1 |
S TGCTCAAATGTGACAAGTTCC A TGCACAGAGATGTTCAATGATG |
| Sfrp2 | secreted frizzled-related sequence protein 2 |
S ACGACAACGACATCATGGAA A CAGCACGGATTTCTTCAGGT |
| Tgfb3 | transforming growth factor, beta 3 |
S TTTCGTTTCAATGTGTCCTCA A ATGTAGCGCTGCTTGGCTAT |
| Timp3 | tissue inhibitor of metalloproteinase 3 |
S ACATTCACACGGAAGCCTCT A CCTCTCCACAAAGTTGCACA |
| Wisp1 | WNT1 inducible signaling pathway protein 1 |
S TCGGATCTCTAACGTCAATGC A GTACACAGCCAGGCATTTCT |
| Wisp2 | WNT1 inducible signaling pathway protein 2 |
S TGTGATGACGGTGGTTTCAC A AGGAGTGACAAGGGCAGAAA |
| Wnt10b | wingless related MMTV integration site 10b |
S TTCTCTCGGGATTTCTTGGA A CCCTCCAACAGGTCTTGAAT |
| Wnt4 | wingless-related MMTV integration site 4 |
S AACGGAACCTTGAGGTGATG A CTGCTGAAGAGATGGCGTAT |
Statistical Analysis
For the microarray analysis, the Z score statistical analysis method (Cheadle et al., 2003) was used. Genes presenting a 1.5 fold Z-Score difference (positive or negative) were included. For RT-RT-PCR and bone density analysis significant results were determined by Students’s t test. P value less than 0.05 was considered statistically significant.
Results
Isolation of Stromal Cells from the Trabecular Bone Zone
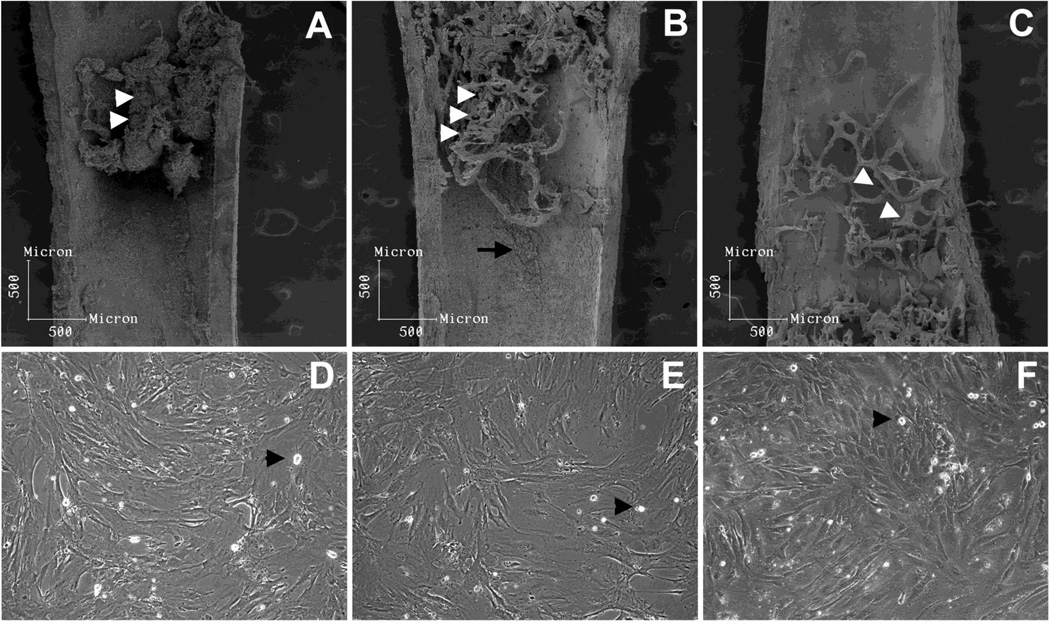
Several studies indicate that most of HSC are concentrated in the trabecular zone, the same region in which CXCL12-reticular perivascular cells preferentially reside, as described by Sugiyama et al. (2006). We observed that, after the bone marrow was flushed, most of the cells that remained attached to the bone surface were in the trabecular zone of the metaphysis and epiphysis (Figure 1A). As previously described, after the first round of collagenase digestion, subendosteal cells (F-RET) were extracted, as osteoblasts (F-OST) remained attached to the bone surface (Figure 1B). Osteoblasts were extracted after a second round of collagenase digestion (Figure 1C). When in culture, flushed bone marrow (Figure 1D) and F-RET cells (Figure E) presented myofibroblast pattern of growth in vitro. In contrast, endosteal osteoblasts presented in a cuboidal shape and maintain good contact inhibition, as previously observed (Figure 1F). Macrophages were found in all three primary stromal cells culture (Figures 1D, E, and F, arrow heads). When we compared hematopoietic cells in the three niches, we observed that a higher frequency of myeloid cells, mostly granulocytes, reside in the subendosteal and endosteal regions, as compared to flushed central marrow (Supplemental Figure 1). Conversely, B-lymphocytes frequency decreases slightly in the subendosteal and endosteal regions when compared to central flushed marrow. No differences were observed for the T lymphocyte cell distribution in the three niches. In addition, nonadherent cells were removed (at least, the majority of them) after three days of incubation. Only a few myeloid cells remained attached in all three stromal cultures.
Figure 1. Analysis of the inner surface of femurs and morphology of the stromal populations isolated.
After bone marrow was flushed, subendosteal and endosteal stromal cells which remained attached were mainly distributed in the trabecular region (A, white arrow heads) at the metaphysis and epiphysis. After the first round of collagenase digestion procedure, subendostal stromal cells were removed (B) and osteoblasts remained attached (B, white arrow heads and arrow), being retrieved only after the second collagenase digestion (C, white arrow heads pointing to osteoblast-free trabecular bone). When in culture, flushed bone marrow (D) and subendosteal stromal cells (E) presented similar myofibroblastic morphology. Conversely, endosteal osteoblats (F) presented a cuboidal morphology, as expected. A few macrophage-like cells were observed in all three primary cultures (arrow heads). Primary cells were cultured to confluence in DMEM 10% FBS.
In Vivo Multipotency Detained by Subendosteal Stromal Cells
In our previous work, we suggested that F-OST and F-RET populations comprised stromal cells with distinct potential regarding the skeletal differentiation cascade (Balduino et al., 2005). Using the same techniques, we found that the isolated the F-OST populations were quite homogenous regarding their committment to the osteogenic lineage, and all cells expressed high levels of bone sialoprotein, osteopontin and alkaline phosphatase. On the other hand, all F-RET cells were also positive for alkaline phosphatase, osteonectin, and osteopontin. Even though immunocytochemistry isn’t a quantitative assay, no differences on the expression levels of these markers was observed among all cells on the F-RET population (Balduino et al., 2005)..
To define stromal cells potency, all three fractions were isolated combined to a scaffold, and subcutaneously implanted following the method described in Taichman et al., 2010. After 5 weeks of implantation, cells from BM and F-RET fractions gave rise to significant amount of cortical bone, with a recruited hematopoietic marrow. Cells from F-OST fraction, on the other hand, only gave rise to compact bone ossicles, with no bone marrow. These data establish that stromal cells from F-OST fraction are fully committed osteoblasts. Conversely, stromal cells distributed in the subendosteal region contain uncommitted mesenchymal cells. These observations further validated the method used to isolate separately bone marrow endosteal and subendosteal fractions. These data support that, once implanted in vivo, multipotent mensenchymal cells will give rise to bone tissue, hematopoiesis supporting stroma and marrow adipocytes. All these cells/tissue could be observed when FRET cells were implanted. On the other hand, committed osteoblasts (FOST), upon implantation, gave rise only to bone tissue and no hematopoietic supporting stroma or adipocytes were observed. Based on this in vivo data, we support the potential of each population, and that while the in vitro studies examing the differentiating potential of these isolated cells, we feel that such assays would not add any major new perspective to the present work.
Gene Expression Profiling
We identified 749 known genes differentially expressed by stromal cells from subendosteal and endosteal fractions: 430 and 319 transcripts were up-regulated in the F-RET and F-OST cells, respectively. In order to confirm microarray data, several genes were tested on real time RT-PCR analyses (Table 2). As expected, endosteal cells expressed higher levels of osteogenic lineage and bone extracellular matrix related genes. Subendosteal stromal cells expressed higher levels of MMP3, MMP13, and TIMP1, and osteoblasts expressed higher levels of MMP2, MMP14, and TIMP3 (Figure 3).
Table 2.
Comparison between microarray and Real-Time RT-PCR analysis. F-RET/F-OST ratio.
| LogRatio (F-RET/F-OST) | |||
|---|---|---|---|
| Entrez Gene ID | Gene | Microarray | RT-RT-PCR |
| NM_054042 | CD164l1 | 0.53 | 0.60 |
| NM_133654 | CD34 | 0.52 | 0.61 |
| NM_019707 | Cdh13 | 0.65 | 0.83 |
| NM_009883 | Cebpb | 0.35 | 0.47 |
| NM_007679 | Cebpd | 0.66 | 0.84 |
| NM_010217 | Ctgf | −0.43 | −0.35 |
| NM_008056 | Fzd6 | −0.26 | −0.37 |
| AK021164 | Fzd9 | 0.23 | 0.53 |
| NM_016696 | Gpc1 | −0.34 | −0.46 |
| X82046 | Hgf | 1.12 | 2.15 |
| NM_008318 | Ibsp | −0.31 | −0.61 |
| NM_010549 | IL11ra1 | 0.51 | 0.48 |
| NM_008357 | IL15 | 0.40 | 1.04 |
| NM_031168 | IL6 | 0.66 | 1.53 |
| BC009120 | Mgp | 1.11 | 1.12 |
| NM_008607 | Mmp13 | 0.25 | 0.24 |
| NM_010809 | Mmp3 | 1.89 | 2.74 |
| NM_011019 | Osmr | 0.48 | 0.49 |
| NM_130458 | Osx | −0,18 | −0.72 |
| BC005483 | Otc | −0.41 | −0.45 |
| NM_008808 | Pdgfa | −0.23 | −0.27 |
| NM_011058 | Pdgfra | 1.13 | 1.40 |
| D14636 | Runx2 | −0.18 | −0.72 |
| NM_011521 | Sdc4 | 0.22 | 0.48 |
| NM_013834 | Sfrp1 | 0.73 | 1.07 |
| NM_009144 | Sfrp2 | 0.69 | 1.49 |
| NM_009368 | Tgfb3 | −0.25 | −0.27 |
| NM_011595 | Timp3 | −0.71 | −0.74 |
| NM_018865 | Wisp1 | −0.75 | −0.89 |
| NM_016873 | Wisp2 | 0.59 | 0.59 |
| NM_011718 | Wnt10b | −0.28 | −0.86 |
| NM_009523 | Wnt4 | −0.31 | −0.67 |
Real Time RT-PCR was used to verify array results. Thirty microarray-present samples were tested and results (log ratio) were compared to microarray data.
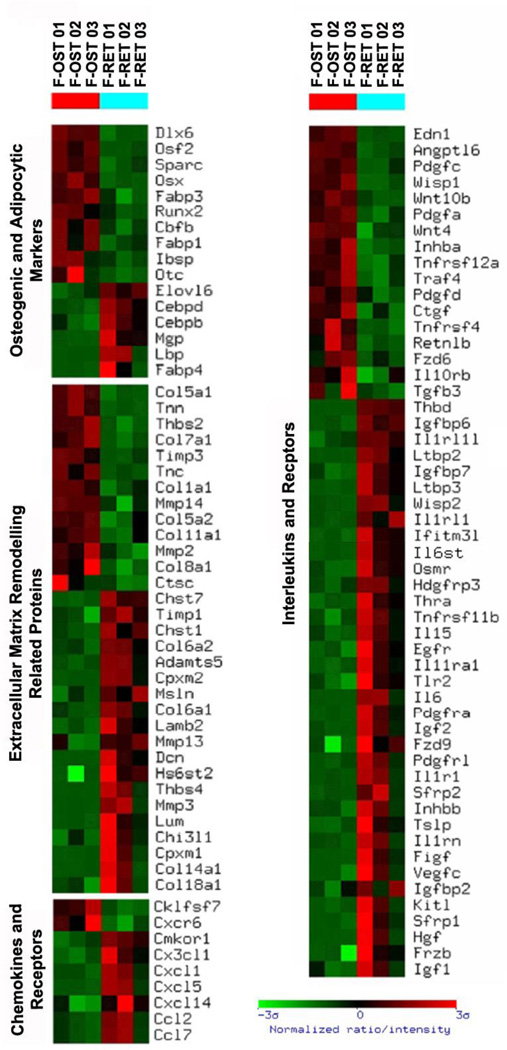
Figure 3. Gene expression profile based on gene function and physiology of subendosteal stromal cells (F-RET) compared to endosteal osteoblasts (F-OST).
Functional clustering was achieved and gene groups of particular interest are shown. The profile was created using the Ingenuity program. The brighter the red, the higher the gene expression level observed. The brighter the green, the lower the gene expression level observed. Only gene expression differences statistically significant are shown.
For adhesion molecules and cell surface membrane markers, osteoblasts expressed higher levels of contactins-3 and -6, NCAM, and, unexpectedly, CD146, all known to participate in cell-cell and cell-matrix interactions (Figure 4). Expression of CD29 and glypican-1 was also higher in the endosteal cells compared with subendosteal stroma. In contrast, subendosteal cells expressed higher levels of ICAM1, VCAM1, CD164, CD109, Thy1, and Cadherin-13.
Figure 4. Gene expression profile of membrane-associated molecules of subendosteal stromal cells (F-RET) compared to endosteal osteoblasts (F-OST).
Functional clustering was achieved and gene groups of particular interest are shown. The profile was created using the Ingenuity program. The brighter the red, the higher the gene expression level observed. The brighter the green, the lower the gene expression level observed. Only gene expression differences statistically significant are shown.
Among several cytokines and growth factors tested, osteoblasts express higher levels of endothelin-1 (End1) and angiopoietin-like 6 (Angptl6). Osteoblasts also expressed higher levels of Pdgfa, Pdgfc, and Pdgfd, members of the Platelet-Derived Growth Factor family, which signal through two receptors: Pdgfra and Pdgfrb. Subendosteal stroma expressed higher levels of Pdgfra (Figure 3, Table 2).
Members of the CCN family (Cystein-rich61 / Connective Tissue Growth Factor / Nephroblastoma Overexpressed), which belong to the TGFβ family, were also detected in the two stromal populations. Ctgf (CCN2) and Wisp1 (CCN4) expression were higher in osteoblasts than in subendosteal stroma. Conversely, the subendostal fraction expressed higher levels of Wisp2 (CCN5) and TGFβ3. Subendosteal stroma also expressed higher levels of Ltbp-2 and -3, which are thought to play a role in TGFβ deposition into the extracellular matrix (Table 2).
Members of Wnt family were also evaluated. No differences in the expression of Wnt3 and Wnt5a were noted between the two stromal cell populations were detected. However, expression of Wnt4 and Wnt10b was higher osteoblasts than subendosteal stroma. Fzd6 (Frizzled-6) was also expressed at higher levels by osteoblasts (Table 2). Interestingly, subendosteal reticular cells expressed higher levels of Fzd9, as well as critical Wnt modulators including Sfrp1, Sfrp2, and Frzb. Subendosteal stroma expressed higher levels of IGF–I and –II.
Also very interestingly, subendosteal reticular cells were observed to express higher levels of IL-6 (interleukin 6), HGF (Hepatocyte Growth Factor), and SCF (Stem Cell Factor), all shown to stimulate HSC high proliferation in vitro and pool expansion in vivo. Additionally, subendosteal reticular cells expressed higher levels of IL1r1, IL11ra1, Osmr (Oncostatin M Receptor), Egfr (Epidermal Growth Factor Receptor). In contrast, subendosteal reticular cells expressed higher levels of many chemokines (CXCL1, CXCL5, CXCL14, CCL2, and CCL7). Interestingly, subendosteal stroma also expressed higher levels of CXCL12 (SDF1a). This difference was not observed in the microarray data as statistical analyses indicated a 0.6 value for P. However, the difference in the expression of CXCL12 by endosteal and subendosteal stroma was confirmed using real time RT-PCR analysis (Figure 5).
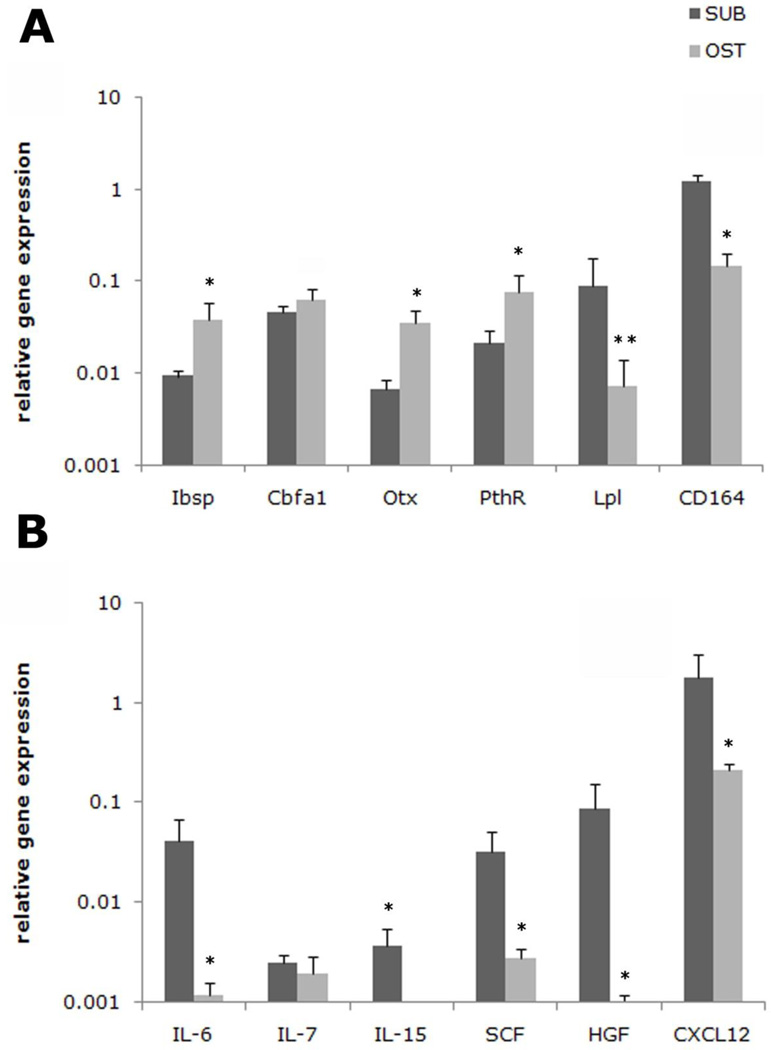
Figure 5. Differences in relevant HSC niche gene expression by subendosteal stromal cells (F-RET) compared to endosteal osteoblasts (F-OST).
Real time RT-PCR analysis was performed on the subendosteal cells (SUB) and osteoblasts (OST) for the following genes identified using the microarray profiles: (A) Bone sialoprotein (Ibsp), Cbfa-1, osterix (Otx), Pth receptor (PthR), lipoprotein lipase (LPL) and CD164 and (B) interleukin 6 (IL-6), interleukin 7 (IL-7), interleukin 15 (IL-15), stem cell factor (SCF), hepatocyte growth factor (HGF), and stromal derived factor-1α (CXCL12) are presented. The results were normalized to GAPDH, which was also used to determine relative gene expression (dCT). Primers used are listed on Table 1.
Discussion
In the present work, we establish a method to isolate separately the stromal populations from the endosteal and the subendosteal/perivascular niches, also known as HSC “quiescent” and “proliferative” niches. Even though these two niches are organized in a very close range, the isolation protocol herein presented allowed us to identify endosteal stroma cells as fully committed osteoblasts and subendosteal stroma cells as uncommitted mesenchymal cells. Microarray analyses further indicated a few possible mechanisms used by the two stromal compartments to create and coordinate the “quiescent” and “proliferative” niches in which hematopoietic stem cells and progenitors reside.
Early histological and functional analyses showed that hematopoietic stem cells and progenitors preferentially colonize the endosteal and subendosteal/perivascular niches (Lord et al., 1975; Lambertsen and Weiss, 1984; Eliasson and Jönsson, 2001; Nilsson and Simmons, 2004; Haylock et al., 2007; Lo Celso et al., 2009). Based on a stereological study, Lambertsen and Weiss (1984) demonstrated that the endosteal niche extended 0.02mm (20µm) from the bone surface, and the subendosteal niche extended 0.08mm (80µm) from the endosteal niche limit. A similar extension was observed in our previous work (Balduino et al., 2005). Importantly, the authors showed that HSCs were heterogeneously distributed in these two niches, in which long-term HSC would be mostly concentrated in the endosteal fraction. This observation has been corroborated by recent studies in which slow-cycling HSC are found in association with endosteal osteoblasts, and fast-cycling HSC mostly reside in the perivascular niches distributed in the subendosteal zone (Sugiyama et al., 2006; Kiel et al., 2007). The method used in the present work is exclusively based on a time-dependent enzymatic digestion separation procedure (Balduino et al., 2005), and is in agreement with the bone marrow compartment organization set by Lambertsen and Weiss (1984). We show that, regardless the close range distribution, as for the HSC, the endosteal and subendosteal zones, identified by Lambertsen and Weiss, are created by two very distinct stromal compartments.
As widely discussed, osteoblasts play crucial role on HSC self-renewal and maintenance in vivo (Calvi et al., 2003; Zhang et al., 2003; Visnjic et al., 2004). In addition to the endosteal niche, hematopoietic stem cells also reside in the abluminal side of bone marrow sinusoids, in association with perivascular reticular cells and endothelial cells, identified as the perivascular niche (Kiel et al., 2005; Sugiyama et al., 2006). On the same study mentioned before, Lambertsen and Weiss (1984) showed that most of the perivascular niches harboring HSC are distributed in the subendosteal zone. Sugiyama et al. (2006) demonstrated that HSCs in the perivascular niches are in close association with the perisinusoidal reticular cells, which express high levels of CXCL12, also known as SDF1α, a critically important chemokine which plays a significant role on HSC retention, homing, and lodging in the marrow cavity. Using the real time RT-PCR, it was demonstrated that subendosteal stromal cells expressed higher levels (almost 10 times more) of CXCL12 than osteoblasts. When compared to the stroma isolated from a regular flushed marrow, CXCL12 level was also high in subendosteal cells (Figure 5). In accordance with these observations, our data suggest that the subendosteal stromal cells isolated using the method described in the present study are likely comprised of perivascular CXCL12-positive reticular cells described by Sugiyama et al. (2006) Furthermore, both stromal cell populations described by both groups, are derived mostly from the trabecular zone (Figure 1). Thus, our data demonstrate the validity of a method to isolate and study two critical components of the HSC niche stromal populations. As such this approach provides an opportunity to further explore the mechanisms that regulate HSC control and behavior. Different markers, such as Osteopontin and Sca-1, have been evaluated for osteoblasts identification and isolation from endosteal/subendosteal niches (Mayack et al., 2008; Nakamura et al., 2010). However, these markers are not only expressed by endosteal cells, but also by osteoprogenitors and uncommitted mesenchymal cells found in the subendosteal niche.
As previously described, slow-cycling hematopoietic stem cells are found in close association with endosteal osteoblasts, as fast-cycling HSC reside in the perivascular niche (Calvi et al., 2003; Zhang et al., 2003; Stier et al., 2005; Sugiyama et al., 2006). This has been well demonstrated in in vivo long-term BrdU retaining assays and myeloablation models. Although it is clear that HSC behavior in either niche is very distinct, how much of this behavior is controlled by the niche stromal cells it is not known. Therefore, both stromal cells must be compared. A wide molecular comparison would provide clues on the mechanisms used by stromal cells to drive HSC quiescence and/or proliferation; on how these two niches communicate in order to maintain a balanced hematopoiesis; and how peripheral factors may influence HSC behavior. Yet both stromal cells herein evaluated express many overlapping and similar levels of HSC self-renewal inducing factors as well. In our previous work, we found that osteoblasts expressed higher levels of Jagged-1 as compared to the subendosteal stroma. However, in that study we analyzed the cells after several passages, thus the culture conditions may have influenced cell phenotype and, consequently, Jagged-1 expression. Among all cytokines tested, our major finding was that subendosteal reticular cells express higher levels of three of the most important cytokines known to increase HSC proliferation. Hepatocyte Growth Factor (HGF), Stem Cell Factor (SCF), and Interleukin-6 (IL-6) have been shown to induce HSC proliferation and to increase HSC pool in vivo and in vitro (Petzer et al., 1996; Miller and Eaves, 1997; Han et al., 1993; Miura et al., 1993; Taichman et al., 2001), thus providing an explanation for the fast-cycling behavior of HSCs and how perivascular reticular cells create a “proliferative” niche. In addition, the high concentration of different chemokines secreted by these stromal cells may indicate that the subendosteal zone is also responsible for harboring intermediate progenitors, which also present fast-cycling profile.
Two major MMPs were detected on endosteal and subendosteal stromal cells: MMP2 and MMP3, respectively. Both MMPs are able to activate MMP9, which plays a very crucial role on HSC mobilization from the quiescent to the proliferative niche (Heissig et al., 2002). Both endosteal and perivascular niches thus appear to participate on HSC mobilization. The mechanisms of HSC mobilization in G-CSF stimulated or myeloablated animals have been elucidated in the last few years (Heissig et al., 2002; Levesque et al., 2003; 2004; Lucas et al., 2008; Shirvaikar et al., 2010). Nonetheless, the physiological exit of HSC from the quiescent niche and its transition to the proliferative niche is not clear. Osteoblasts higher expression of Timp3, a potent inhibitor of MMP9 activity (Brew et al., 2000), suggests a “protective” activity played by endosteal niche over the slow-cycling HSC pool (Figure 3). Furthermore, HSC exit from quiescent niche does not seem to happen randomly, but in a niche controlled fashion. Complementarily, the fact that subendosteal cells express higher levels of Wnt modulators (Fzdb, Sfrp1, and Sfrp2) supports the hypothesis that subendosteal-perivascular niche play a significant role upon over HSC recruitment from a quiescent niche into a proliferative niche and/or, maybe, its engagement into the differentiation cascade (Figure 3, Table 2). Active egression and/or release of HSC from their protective and restrictive environment seems to be the result of a finely controlled balance between the endosteal and subendosteal/perivascular reticular cells, and may not represent a random process, just like mentioned before. Additionally, expression of receptors for cytokines known to increase or decrease production of different types of blood cells were highly expressed on subendosteal reticular cells as compared to osteoblasts, suggest that the perivascular niche may be best positioned to promptly respond to a peripheral demand. Based on our results, we propose that the hematopoietic stem cell niche in the marrow cavity is the result of a fine balance between endosteal and perivascular/subendosteal regions (Figure 6). To our knowledge, it is the first time that these two stromal cell types have been shown to play active and complementary roles on maintaining and/or recruiting hematopoietic stem cells under physiological conditions. Endosteal and subendosteal niches may be generated by two separate stroma compartments. Several quantitative and qualitative analyses in vivo are definitely needed to fully describe the HSC niche organization. Further molecular evaluations of the stromal components contributing to all HSC niches will be crucial in order to understand how both contribute to hematopoiesis.
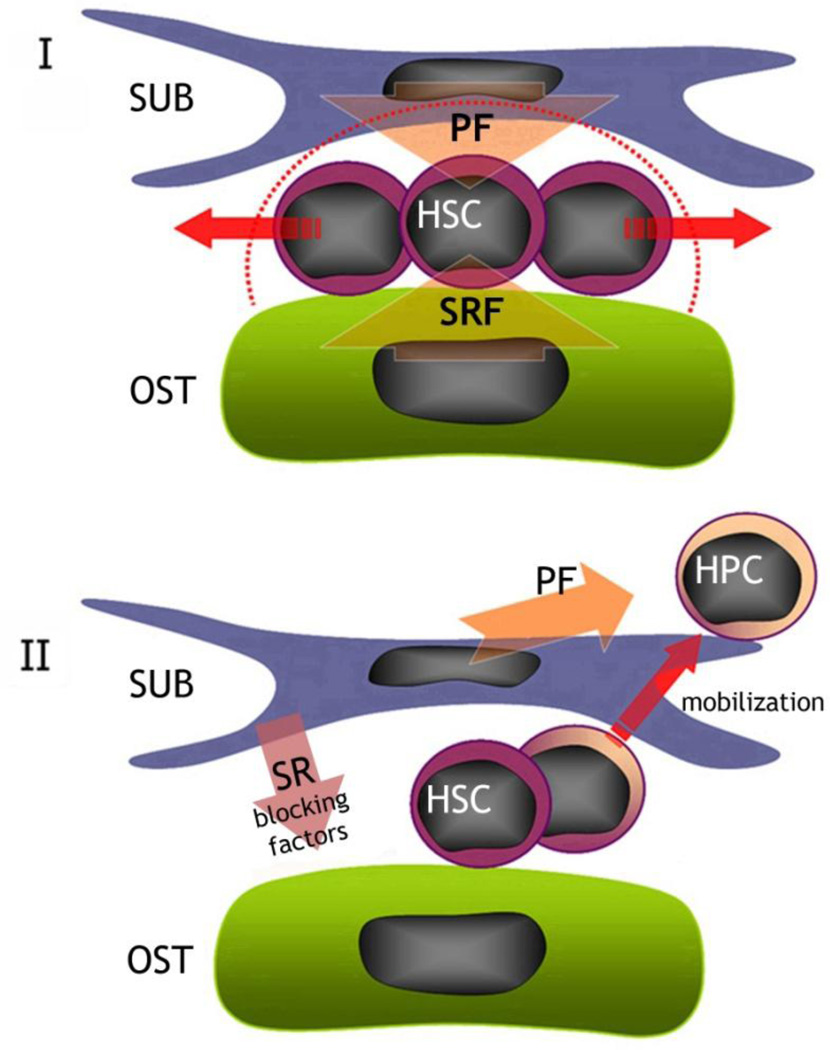
Figure 6. Potential differences and roles for endosteal osteoblasts (F-OST) and subendosteal stromal cells (F-RET) in maintaining the HSC niche in the bone marrow.
Our hypothesis says that, in the HSC niche, endosteal osteoblasts (F-OST) may supply the specific signals for self-renewal and quiescence (SRF). However, subendosteal stromal cells (F-RET) secrete growth factors and chemokines (PF) that, in a balance with stemness inducing factors secreted by osteoblasts, will promote HSC pool expansion (I). Conversely, subendosteal stromal cells (F-RET) secrete factors (SR blocking factors) which neutralize self-renewal factors presented by endosteal osteoblasts (F-OST), promoting HSC exit from the niche, proliferation and migration towards the central space.
Supplementary Material
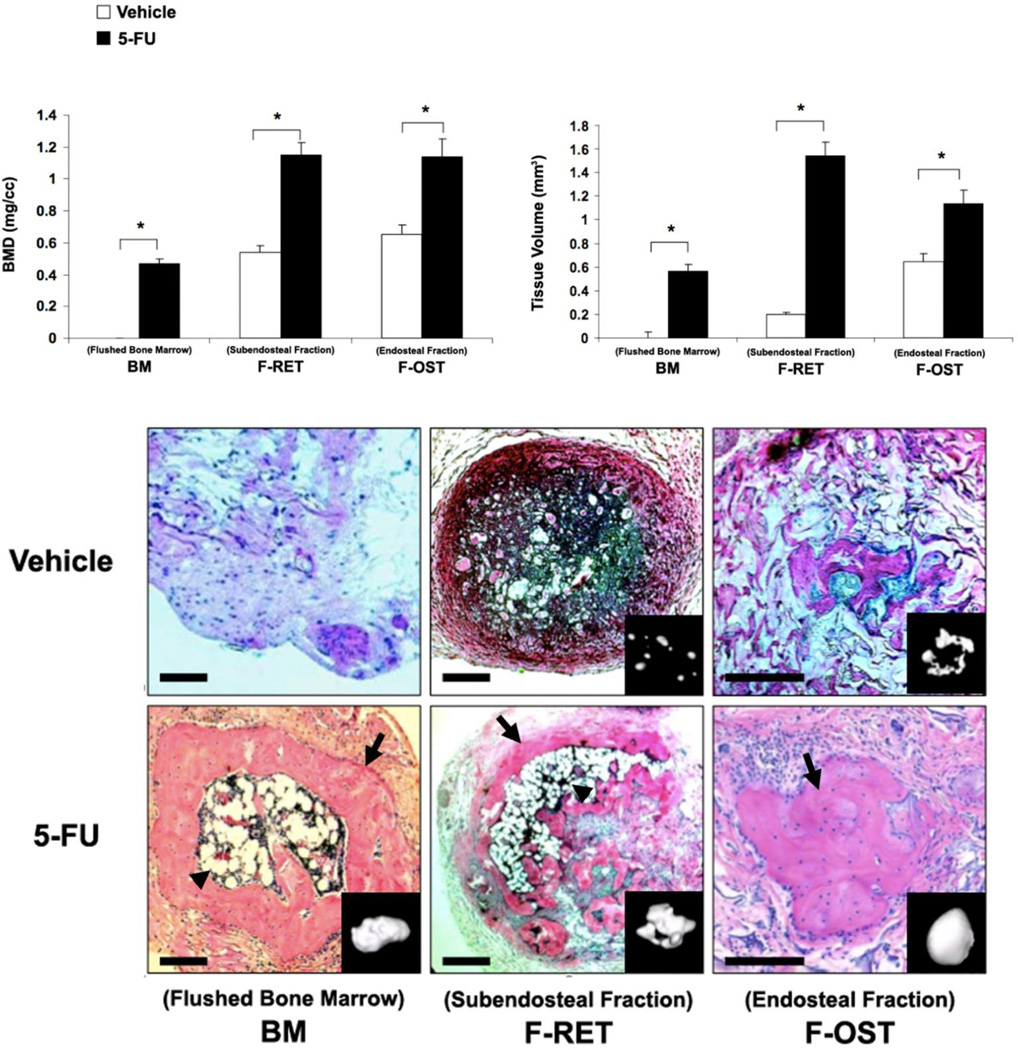
Figure 2. Histological and morphometric analyses of stromal cells fractions implants.
As described in the Materials and Methods, 2×106 of flushed bone marrow cells (BM), subendosteal stroma (F-RET), and endosteal osteoblasts (F-OST) isolated from normal and 5-FU treated mice were mixed up in gelatin sponge and transplanted into SCID mice subcutaneously. After 5 weeks, the implants were harvested for micro-CT scanning (inserts) and sectioned for histological staining using hematoxylin and eosin of the BM, F-FRET and F-OST tissues (lower panels). Original Magnification of 400×. Bone mass density (BMD) and tissue volume were measured (upper panels).
Acknowledgments
This research was, in part, supported by CNPq, FAPERJ, and the Intramural Research Program of the National Institute on Aging, National Institutes of Health, and by NIH awards CA-93900 and DK082481.
Funding: National Counsel of Technological and Scientific Development and NIH awards CA-93900 and DK082481. This work has also been supported, in part, by the National Institute on Aging, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship and Conflict of Interest Statements
Alex Balduino - designed research; performed research; analyzed data; wrote paper
Valeria Mello Coelho - designed research; analyzed data; wrote paper
Zhou Wang – performed research; analyzed data; wrote paper
Russell S. Taichman – designed research; analyzed data; wrote paper
Paul H. Krebsbach – analyzed data; wrote paper
Ashani T. Weeraratna - designed research; performed research; analyzed data
Kevin G. Becker - designed research; performed research; analyzed data
Wallace de Mello - performed research; analyzed
Dennis D. Taub - designed research; analyzed data; wrote paper
Radovan Borojevic - designed research; analyzed data; wrote paper
Conflict of interest statement
The authors declare no competing financial or potential conflict of interests.
References
- Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, Ito K, Koh GY, Suda T. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Balduino A, Hurtado SP, Frazão P, Takiya CM, Alves LM, Nasciutti LE, El-Cheikh MC, Borojevic R. Bone marrow subendosteal microenvironment harbours functionally distinct haemosupportive stromal cell populations. Cell Tissue Res. 2005;319:255–266. doi: 10.1007/s00441-004-1006-3. [DOI] [PubMed] [Google Scholar]
- Bourke VA, Watchman CJ, Reith JD, Jorgensen ML, Dieudonnè A, Bolch WE. Spatial gradients of blood vessels and hematopoietic stem and progenitor cells within the marrow cavities of the human skeleton. Blood. 2009;114:4077–4080. doi: 10.1182/blood-2008-12-192922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta. 2000;1477:267–283. doi: 10.1016/s0167-4838(99)00279-4. [DOI] [PubMed] [Google Scholar]
- Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, Seandel M, Shido K, White IA, Kobayashi M, Witte L, May C, Shawber C, Kimura Y, Kitajewski J, Rosenwaks Z, Bernstein ID, Rafii S. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6:251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson P, Jönsson JI. The hematopoietic stem cell niche: low in oxygen but a nice place to be. J Cell Physiol. 2010;222:17–22. doi: 10.1002/jcp.21908. [DOI] [PubMed] [Google Scholar]
- Haylock DN, Williams B, Johnston HM, Liu MC, Rutherford KE, Whitty GA, Simmons PJ, Bertoncello I, Nilsson SK. Hemopoietic stem cells with higher hemopoietic potential reside at the bone marrow endosteum. Stem Cells. 2007;25:1062–1069. doi: 10.1634/stemcells.2006-0528. [DOI] [PubMed] [Google Scholar]
- Han M, Kobayashi M, Imamura M, Hashino S, Kobayashi H, Maeda S, Iwasaki H, Fujii Y, Musashi M, Sakurada K. In vitro expansion of murine hematopoietic progenitor cells in liquid cultures for bone marrow transplantation: effects of stem cell factor. Int J Hematol. 1993;57:113–120. [PubMed] [Google Scholar]
- Heissig B, Hattori K, Dias S, Friedrich M, Ferris B, Hackett NR, Crystal RG, Besmer P, Lyden D, Moore MA, Werb Z, Rafii S. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of kit-ligand. Cell. 2002;109:625–637. doi: 10.1016/s0092-8674(02)00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Wang J, Schneider A, Sun YX, Koh-Paige AJ, Osman NI, McCauley LK, Taichman RS. Regulation of SDF-1 (CXCL12) production by osteoblasts; a possible mechanism for stem cell homing. Bone. 2006;38:497–508. doi: 10.1016/j.bone.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Iwashita T, Yilmaz OH, Morrison SJ. Spatial differences in hematopoiesis but not in stem cells indicate a lack of regional patterning in definitive hematopoietic stem cells. Dev Biol. 2005;283:29–39. doi: 10.1016/j.ydbio.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Radice GL, Morrison SJ. Lack of evidence that hematopoietic stem cells depend on N-cadherin-mediated adhesion to osteoblasts for their maintenance. Cell Stem Cell. 2007;1:204–217. doi: 10.1016/j.stem.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, He S, Ashkenazi R, Gentry SN, Teta M, Kushner JA, Jackson TL, Morrison SJ. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollet O, Dar A, Shivtiel S, Kalinkovich A, Lapid K, Sztainberg Y, Tesio M, Samstein RM, Goichberg P, Spiegel A, Elson A, Lapidot T. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat Med. 2006;12:657–664. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SA, Krebsbach PH, Satomura K, Kerr J, Riminucci M, Benayahu D, Robey PG. Single-colony derived strains of human marrow stromal fibroblasts form bone after transplantation in vivo. J Bone Miner Res. 1997;12(9):1335–1347. doi: 10.1359/jbmr.1997.12.9.1335. [DOI] [PubMed] [Google Scholar]
- Lambertsen RH, Weiss L. A model of intramedullary hematopoietic microenvironments based on stereologic study of the distribution of endocloned marrow colonies. Blood. 1984;63:287–297. [PubMed] [Google Scholar]
- Lévesque JP, Hendy J, Winkler IG, Takamatsu Y, Simmons PJ. Granulocyte colony-stimulating factor induces the release in the bone marrow of proteases that cleave c-KIT receptor (CD117) from the surface of hematopoietic progenitor cells. Exp Hematol. 2003;31:109–117. doi: 10.1016/s0301-472x(02)01028-7. [DOI] [PubMed] [Google Scholar]
- Levesque JP, Liu F, Simmons PJ, Betsuyaku T, Senior RM, Pham C, Link DC. Characterization of hematopoietic progenitor mobilization in protease-deficient mice. Blood. 2004;104:65–72. doi: 10.1182/blood-2003-05-1589. [DOI] [PubMed] [Google Scholar]
- Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Côté D, Rowe DW, Lin CP, Scadden DT. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord BI, Testa NG, Hendry JH. The relative spatial distributions of CFUs and CFUc in the normal mouse femur. Blood. 1975;46:65–72. [PubMed] [Google Scholar]
- Lucas D, Battista M, Shi PA, Isola L, Frenette PS. Mobilized hematopoietic stem cell yield depends on species-specific circadian timing. Cell Stem Cell. 2008;3:364–366. doi: 10.1016/j.stem.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig A, Carter A, Bertak D, Enika D, Vandanmagsar B, Wood W, Becker KG, Weeraratna AT, Taub DD. Transcriptome analysis of murine thymocytes reveals age-associated changes in thymic gene expression. Int J Med Sci. 2009;6:51–64. doi: 10.7150/ijms.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayack SR, Wagers AJ. Osteolineage niche cells initiate hematopoietic stem cell mobilization. Blood. 2008;112:519–531. doi: 10.1182/blood-2008-01-133710. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Miller CL, Eaves CJ. Expansion in vitro of adult murine hematopoietic stem cells with transplantable lympho-myeloid reconstituting ability. Proc Natl Acad Sci U S A. 1997;94:13648–13653. doi: 10.1073/pnas.94.25.13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura N, Okada S, Zsebo KM, Miura Y, Suda T. Rat stem cell factor and IL-6 preferentially support the proliferation of c-kit-positive murine hemopoietic cells rather than their differentiation. Exp Hematol. 1993;21:143–149. [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Arai F, Iwasaki H, Hosokawa K, Kobayashi I, Gomei Y, Matsumoto Y, Yoshihara H, Suda T. Isolation and characterization of endosteal niche cell populations that regulate hematopoietic stem cells. Blood. 2010;116:1422–1432. doi: 10.1182/blood-2009-08-239194. [DOI] [PubMed] [Google Scholar]
- Nilsson SK, Johnston HM, Coverdale JA. Spatial localization of transplanted hemopoietic stem cells: inferences for the localization of stem cell niches. Blood. 2001;97:2293–2299. doi: 10.1182/blood.v97.8.2293. [DOI] [PubMed] [Google Scholar]
- Nilsson SK, Simmons PJ. Transplantable stem cells: home to specific niches. Curr Opin Hematol. 2004;11:102–106. doi: 10.1097/01.moh.0000133651.06863.9c. [DOI] [PubMed] [Google Scholar]
- Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc Natl Acad Sci USA. 2007;104:5431–5436. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzer AL, Hogge DE, Landsdorp PM, Reid DS, Eaves CJ. Self-renewal of primitive human hematopoietic cells (long-term-culture-initiating cells) in vitro and their expansion in defined medium. Proc Natl Acad Sci USA. 1996;93:1470–1474. doi: 10.1073/pnas.93.4.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey PG. Cell sources for bone regeneration: the good, the bad, and the ugly (but promising) Tissue Eng Part B Rev. 2011;17(6):423–430. doi: 10.1089/ten.teb.2011.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirvaikar N, Marquez-Curtis LA, Shaw AR, Turner AR, Janowska-Wieczorek A. MT1-MMP Association with membrane lipid rafts facilitates G-CSF-induced hematopoietic stem/progenitor cell mobilization. Exp Hematol. 2010;38:823–835. doi: 10.1016/j.exphem.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Stier S, Ko Y, Forkert R, Lutz C, Neuhaus T, Grünewald E, Cheng T, Dombkowski D, Calvi LM, Rittling SR, Scadden DT. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med. 2005;201:1781–1791. doi: 10.1084/jem.20041992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Taichman RS, Emerson SG. Human osteoblasts support hematopoiesis through the production of granulocyte colony-stimulating factor. J Exp Med. 1994;179:1677–1682. doi: 10.1084/jem.179.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taichman RS, Reilly MJ, Emerson SG. Human osteoblasts support human hematopoietic progenitor cells in vitro bone marrow cultures. Blood. 1996;87:518–524. [PubMed] [Google Scholar]
- Taichman RS, Emerson SG. The role of osteoblasts in the hematopoietic microenvironment. Stem Cells. 1998;16:7–15. doi: 10.1002/stem.160007. [DOI] [PubMed] [Google Scholar]
- Taichman R, Reilly M, Verma R, Ehrenman K, Emerson S. Hepatocyte growth factoris secreted by osteoblasts and cooperatively permits the survival of haematopoietic progenitors. Br J Haematol. 2001;112:438–448. doi: 10.1046/j.1365-2141.2001.02568.x. [DOI] [PubMed] [Google Scholar]
- Taichman RS, Wang Z, Shiozawa Y, Jung Y, Song J, Balduino A, Wang J, Patel LR, Havens AM, Kucia M, Ratajczak MZ, Krebsbach PH. Prospective Identification and Skeletal Localization of Cells Capable of Multi-Lineage Differentiation In Vivo. Stem Cells Dev. 2010;19:1557–1570. doi: 10.1089/scd.2009.0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visnjic D, Kalajzic Z, Rowe DW, Katavic V, Lorenzo J, Aguila HL. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood. 2004;103:3258–3264. doi: 10.1182/blood-2003-11-4011. [DOI] [PubMed] [Google Scholar]
- Wang Z, Song J, Taichman RS, Krebsbach PH. Ablation of proliferating marrow with 5-fluorouracil allows partial purification of mesenchymal stem cells. Stem Cells. 2006;24(6):1573–1582. doi: 10.1634/stemcells.2005-0399. [DOI] [PubMed] [Google Scholar]
- Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol. 2006;6:93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- Winkler IG, Barbier V, Wadley R, Zannettino A, Williams S, Lévesque JP. Positioning of bone marrow hematopoietic and stromal cells relative to blood flow in vivo: Serially reconstituting hematopoietic stem cells reside in distinct non-perfused niches. Blood. 2010;116:375–385. doi: 10.1182/blood-2009-07-233437. [DOI] [PubMed] [Google Scholar]
- Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, Feng JQ, Harris S, Wiedemann LM, Mishina Y, Li L. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.