Abstract
Telomeres protect the ends of linear chromosomes, which if eroded to a critical length can become uncapped and lead to replicative senescence. Telomerase maintains telomere length in some cells, but inappropriate expression facilitates the immortality of cancer cells. Recently, proteins involved in RNA processing and ribosome assembly, such as hnRNP (heterogeneous nuclear ribonucleoprotein) A1, have been found to participate in telomere maintenance in mammals. The S. cerevisiae protein Npl3 shares significant amino acid sequence similarities with hnRNP A1. We found that deleting NPL3 accelerated the senescence of telomerase null cells. The highly conserved RNA recognition motifs (RRM) in Npl3 appear to be important for preventing faster senescence. Npl3 preferentially binds telomere sequences in vitro, suggesting that Npl3 may affect telomeres directly. Despite similarities between the two proteins, human hnRNP A1 was unable to complement the lack of Npl3 to rescue accelerated senescence in tlc1 npl3 cells. Deletion of CBC2, which encodes another hnRNP-related protein that associates with Npl3, also accelerates senescence. Potential mechanisms by which hnRNP-related proteins maintain telomeres are discussed.
Keywords: Npl3, Cbc2, hnRNP A1, Senescence, Telomere Capping
1. Introduction
Telomeres contain highly repeated DNA sequences and are typically characterized by G-rich single stranded 3′-overhangs. Telomeres shorten when chromosomes are replicated, but the enzyme telomerase is able to restore telomere lengths. Removing components of the telomerase complex in S. cerevisiae (the catalytic component, Est2 or the RNA template, TLC1) results in progressively shortened telomeres and cell senescence. In addition to being lengthened by telomerase, telomere integrity is maintained by a large set of proteins that organize the chromosome terminus into a protective “capped” structure. Capping the telomeres resists typical responses to DNA ends, including exonucleotypic degradation, end-joining, recombination, and activation of checkpoint-mediated arrest.
We are particularly interested in understanding how proteins with well-known roles for RNA processing contribute to maintaining telomere integrity. Mammalian heterogeneous ribonucleoprotein (hnRNP) A1 regulates splicing and exports mRNA (reviewed in[1]), and several studies have shown its role in telomere maintenance. Deletion in hnRNP A1 causes significantly short telomeres [2]. hnRNP A1 also interacts with telomeric DNA [3, 4] and telomerase [5, 6] as well as stimulates telomerase-dependent telomere maintenance in vivo [7]. hnRNP A1 likely has protective roles at the telomeres.
The RNA processing protein in yeast, Npl3 is very similar to hnRNP A1 (Fig. S1). Both contain two highly conserved RNA recognition motifs (RRMs) [8] and RGG boxes, which are closely spaced arginine-glycine-glycine repeats. Like hnRNP A1, Npl3 is involved in mRNA transport [9–11]. While the C-terminus of hnRNP A1 has nuclear localization sequences, Npl3 lacks them but uses methylation of the arginine residues within the RGG for protein localization [12–14]. Npl3 also appears to be a multi-faceted protein that binds to the 3′ end of mRNA to regulate transcription termination [15, 16], is involved in spliceosome assembly [17], and promotes translation termination [18–20]. With mRNA cap-binding proteins, Cbc1/Cbc2, Npl3 co-transcriptionally packages mRNA for nuclear export [21].
Telomere maintenance roles have been attributed to some yeast proteins involved in RNA metabolism[22], but no such role has yet been reported for yeast hnRNPs. Based on similarities to RNA processing protein hnRNP A1, which has telomere maintenance roles in mammalian cells, we asked if Npl3 has similar functions in yeast. Here, we show that Npl3 participates in the maintenance of telomeres by preventing the faster senescence of telomerase mutants and demonstrate that the conserved RRMs are involved. Our results support a possible telomere-capping role for Npl3.
2. Materials and methods
2.1. Yeast Strains and Senescence Assays
All cells were derived from the S288C strain and cultured in standard yeast media at 30°C. The TLC1/tlc1::LEU2, NPL3/npl3::kanR diploid (yJL325) was generated by mating tlc1 and npl3 haploids. Diploid cells were propagated through several re-streaks to normalize the telomere lengths from the telomerase mutations. The CBC2 gene was removed by complete PCR-mediated ORF deletion in yJL325 cells to generate TLC1/tlc1::LEU2, NPL3/npl3::kanR, CBC2/cbc2::HygroR diploids. Cells were sporulated and dissected to compare segregants that have lost telomerase at the same time. Liquid senescence assays were performed as previously described [23]. To determine colony-forming efficiency, 1000 cells were spread on YPAD plates, and the numbers of colonies were counted 2–3 days later. yJL325 cells transformed with plasmids were sporulated and dissected on SC-URA plates and maintained in SC-URA liquid medium for senescence assays.
2.2. Plasmids
Wild type and mutant proteins were expressed from subcloned plasmids (Table SI). A 461-bp fragment of the NPL3 promoter region was amplified by primer pair 1 & 2 (Table SII) and inserted into SacI/EagI-digested pRS316 (low copy expression plasmid) to generate pJL215. WT and mutant NPL3 sequences with 6xHis tag at the 3′ end were amplified with primer pair 3 & 4 and inserted into MluI/BamHI-digested pJL215. Wild type NPL3 sequence was amplified from plasmid pPS1152 and the F160L mutant of NPL3 (npl3-F160L) from pPS879. The NPL3 sequence containing the RK1-15 mutation where all 15 RGG triplets were converted to KGG (npl3-RK-15) was amplified from pAM420. Mutant NPL3 with F160L, L225S, G241N, and E244K mutations (herein referred to as npl3-LSNK) and mutant NPL3 with L225S, G241N, and E244K mutations (or npl3-SNK) were generated by site-directed mutagenesis at GENEWIZ, Inc (South Plainfield, NJ). Plasmid expressing hnRNP A1 was generated by inserting amplified hnRNP A1 sequences (with primer pair 5 & 6 from plasmid pBS0A1) into pJL215.
2.3. Southern Blot Analysis
DNA was extracted from liquid cultures of senescing cells by the standard phenol chloroform protocol. After XhoI digestion, DNA was separated in a 1% agarose gel and transferred onto Hybond XL membrane (Amersham). Blots were hybridized to 32P-labeled 784bp PCR-generated Y’ fragment [24] or TG repeats amplified from plasmid containing yeast chromosome I-L telomeres [23]. Telomere fragment lengths (TFL) were measured using the NIH Image J software. Slopes representing rates of telomere shortening were analyzed and compared using Student’s t-test.
2.4. Western Blot Analysis
Whole cell lysates from transformed cells were prepared using the Yeast Protein Extraction Buffer Kit (GE Healthcare) following the manufacturer’s instructions. Yeast suspension and protein extraction buffers were supplemented with EDTA-free protease inhibitor cocktail (Sigma). Hexahistidine-tagged WT and Npl3 proteins from senescent survivors were isolated using the Y-PER 6xHis Fusion Protein Purification Kit with chelated nickel columns, following the manufacturer’s instructions (Thermo Scientific). The Y-PER cell lysis reagent was supplemented with EDTA-free protease inhibitor cocktail, 1mM PMSF, 5μg/ml leupeptin, 100μg/ml bacitracin, 5μg/ml chymostatin, 1mM benzamidine, and 10μg/ml pepstatin. Eluted proteins were concentrated and dialyzed with Amicon Ultra-2 centrifugal filter units (Millipore). Protein concentrations were estimated by the bicinchoninic acid (BCA) assay. Cell lysates or purified proteins were separated on 4–15% Tris-Glycine SDS-PAGE gels, transferred to Hybond C membranes (Amersham), and immunoblotted with anti-6xHis antibody (HIS.H8, Thermo Scientific Pierce) or anti-hnRNP A1 (4B10, Abcam).
2.5. Gel Shift Assay
Purified 6xHis-tagged WT or mutant Npl3 were mixed with oligonucleotides synthesized and purified by RNase-free HPLC (Integrated DNA Technologies). Double stranded G-rich telomere sequences (dsTelo, 3137 DS) were generated by annealing 3137 For (ssTelo) and 3137 Rev oligos in dH2O (Table SII). All telomere sequences, including the G-rich RNA sequences (TeloRNA, oligo 3137 RNA), were derived from chromosome I-L ([23]). Non-telomeric dsDNA was generated from annealing oligos 10 & 11 (dsDNA). Oligos were denatured (except the annealed double strands), labeled with γ– 32p-ATP by T4 polynucleotide kinase, and purified by ethanol precipitation. Proteins and radiolabeled nucleic acid (0.5 – 1.5 pmol per reaction) were incubated for 30 minutes at RT in binding buffer (1mM magnesium acetate, 50mM potassium acetate, 10% glycerol, 20mM HEPES, pH 7.2, 1mM DTT, and 10ng/ml Poly dI dC) [25]. Glycerol was added to 25% before loading in 5% 30:0.8 acrylamide:bis-acrylamide gels in 0.5X TBE. All buffers were prepared in DEPC-treated dH2O. Gels were dried and exposed to X-ray film.
3. Results
3.1. Npl3 prevents rapid senescence in cells lacking telomerase
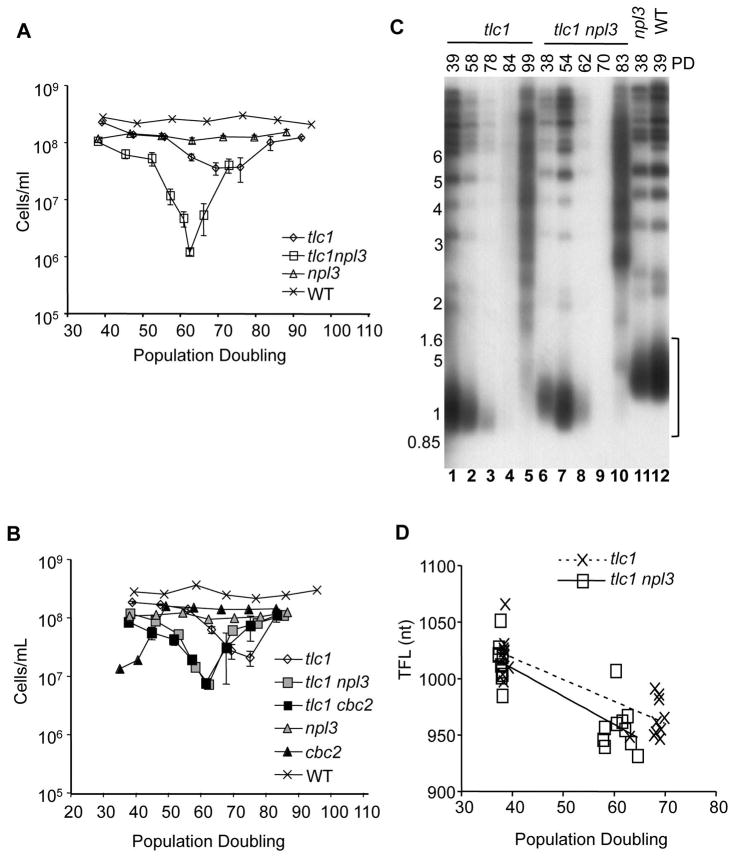
To examine if Npl3 has a role at the telomeres in yeast, we measured steady state telomere lengths of npl3 mutants and found them to be comparable to those in WT cells with the analyses of 12 pairs of npl3 and NPL3 cells (examples are shown in Fig. 1C, lanes 11 and 12). Since Npl3 has no apparent role in the control of steady-state telomere length when telomerase is present, we wanted to determine if Npl3 might affect telomere maintenance in the absence of telomerase. When the rates of senescence were compared, we found that the tlc1 npl3 double mutants senesced much faster than the tlc1 single mutants. The tlc1 npl3 cells reached the lowest point in senescence much sooner (around 62 population doublings [PDs] after the loss of telomerase) than the tlc1 single mutants (74 PDs). As expected, WT and npl3 single mutant with intact telomerase did not senesce (Fig. 1A). Both the tlc1 and tlc1 npl3 mutants eventually escaped senescence by recombining telomeres, but the double mutants reached a point of greater reduced growth before survivor formation. Telomeres relengthened by recombination are seen in lanes 5 and 10 in tlc1 and tlc1 npl3 mutant survivors, respectively (Fig. 1C). NPL3 deletion in est2 mutants, lacking the catalytic function of telomerase, also caused accelerated senescence (Fig. S2). The npl3 single mutants were able to form colonies as efficiently as the WT haploids (nearly 100% of cells spread on YPAD plates grew and formed colonies; data not shown); this indicated that the accelerated senescence is not due to a higher baseline probability of cell cycle arrest due to the npl3 mutation alone. The rates of telomere shortening were similar in tlc1 versus tlc1 npl3 senescing cells (examples are shown in lanes 1–3 and 6–8 in Fig. 1C) with mean rates and standard deviations of 1.9 ± 0.5 and 2.2 ± 1.7 bp per population doubling for tlc1 and tlc1 npl3 mutants, respectively (N = 10 segregant pairs, P = 0.66) (Fig. 1D). The faster senescence caused by the npl3 mutation thus may not be linked to faster telomere shortening. Taken together, Npl3 may be involved in telomere maintenance but is working in a pathway separate from the role telomerase has in extending telomeres.
Figure 1. Deletion of NPL3 or CBC2 accelerates the senescence of tlc1 mutants, but telomere fragment lengths are comparable between senescing tlc1 and tlc1 npl3 cells.
Population doublings in senescence assays were determined daily for 2x106 cells after 22 hours of growth for sporulated and dissected TLC1/tlc1 Npl3/npl3 (A) or TLC1/tlc1 Npl3/npl3 CBC2/cbc2 cells (B). Error bars represent standard error of the mean, N = 2 – 5. (C) Telomere fragments (indicated by a bracket) from DNA of senescing cultures at specified population doublings (PD) were hybridized with 32P-labeled TG repeats. Ladder marker (in kb) is indicated. (D) Telomere fragment lengths (TFL) from autoradiographs were extrapolated using the Image J software and analyzed. Trend lines indicate average rates of telomere shortening for 10 segregant pairs of tlc1 and tlc1 npl3 mutants.
Because Npl3 interacts with the nuclear mRNA cap binding protein complex, Cbc1/Cbc2, to shuttle mRNA to the cytoplasm, and Cbc2 is an hnRNP-like protein containing an RRM and one RGG box (Fig. S1), we asked if Cbc2 also has a role at the telomeres and if Npl3 and Cbc2 work in the same pathway. Steady state telomeres in cbc2 mutants are elongated compared to CBC2 cells ([26] and Fig. S3), suggesting that Cbc2 may cap telomeres and regulate telomere length. To examine if Npl3 and Cbc2 may work together in the absence of telomerase, we sporulated TLC1/tlc1, NPL3/npl3, CBC2/cbc2 diploids. In liquid senescence assay, the deletion in CBC2 accelerated the senescence of tlc1 mutants, similar to the deletion in NPL3 (Fig. 1B). Therefore, like Npl3, Cbc2 is also involved in telomere maintenance. In 40 sets of dissected tetrads, no cbc2 npl3 double or tlc1 cbc2 npl3 triple mutants were recovered. The synthetic lethality of npl3 and cbc2 could be due to the two proteins contributing important roles at the telomeres and/or in other critical RNA processes. Given the similar accelerated senescence by the loss of either protein, Npl3 and Cbc2 may be working similarly as capping proteins at the telomere.
3.2. The RRMs in Npl3 are required for preventing accelerated senescence in tlc1 mutant cells
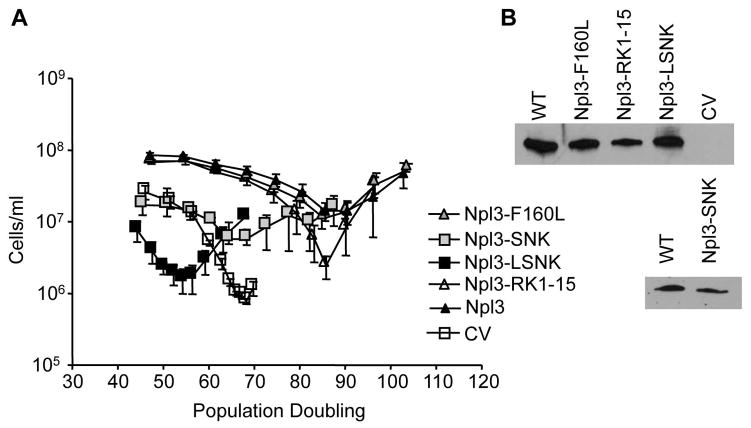
Three specific protein domains in Npl3 are important for its RNA processing functions: two RRMs and the RGG-rich C-terminus. We asked whether these three conserved domains might be involved in preventing faster senescence. We generated plasmids that expressed the WT NPL3 genes, no NPL3 (CV for control vector), the mutant npl3 genes expressing mutant Npl3 with all the RGG’s replaced with KGG (npl3-RK1-15), mutant Npl3 with no functional RRM1 (npl3-F160L), mutant Npl3 with no functional RRM2 (npl3-SNK), or mutant Npl3 with disrupted RRM1 and RRM2 (npl3-LSNK) (Table SI). The RRM mutations were chosen based on previously published reports [15, 25]. Each plasmid was transformed into TLC1/tlc1, NPL3/npl3 diploids then sporulated and dissected. In liquid senescence assays, the exogenously expressed WT, Npl3-RK1-15, and Npl3-F160L proteins were all able to rescue the accelerated senescence of the tlc1 npl3 double mutants. These cells reached their lowest point of senescence after approximately 85 PDs after the loss of telomerase and senesced similar to the tlc1 single mutants containing the same expression plasmids (Fig. 2A, and data not shown). Interestingly, the tlc1 npl3 double mutants expressing Npl3-SNK protein with mutated RRM2 also senesced quickly and reached a growth nadir at approximately 68 PDs, and these cells, interestingly, formed survivors three days earlier than the tlc1 npl3 cells with control vector. All cultures eventually formed survivors indicating their ability to re-lengthen telomeres using telomerase-independent mechanisms. The expression of Npl3-LSNK proteins with non-functional RRM1 and RRM2 caused the tlc1 npl3 mutants to senesce the fastest; senescence reached the lowest point as early as 54 PDs. The mutant Npl3-LSNK protein may have dominant negative activity since its expression also caused tlc1 cells (with intact NPL3) to be very sick. In npl3 cells with intact telomerase, however, the expression of WT or any of the mutant Npl3 had no effect on colony forming efficiency (Data not shown). Expressions of 6XHis-tagged WT and mutant proteins were confirmed by western blots (Fig. 2B). These results imply that the RRM2 region but not RRM1 or the RGGs of Npl3 is required in the protein’s telomere maintenance role.
Figure 2. RRM2 in Npl3 is required for preventing accelerated senescence in tlc1 mutant cells.
(A) Before sporulation and dissection, TLC1/tlc1::LEU2, NPL3/npl3::KanR diploid cells (yJL325) were transformed with plasmids. Only the tlc1 npl3 strains are shown. Plasmids expressed no Npl3 (CV for control vector), 6xHis-tagged WT or mutant Npl3 (Npl3-F160L has mutated RRM1, Npl3-SNK has mutated RRM2, Npl3-LSNK has mutated RRM1 and RRM2, and Npl3-RK1-15 has KGGs in place of RGGs). Senescence assay was performed as described but in SC-URA medium. Error bars represent standard error of the mean, N = 3 – 5. (B) Cells from survivors were expanded; 6xHis-tagged WT and mutant Npl3 were purified by nickel column. Proteins were analyzed by SDS-PAGE and immunoblotted with anti-6xHis antibody.
3.3. Npl3 binds double stranded and single stranded telomere sequences
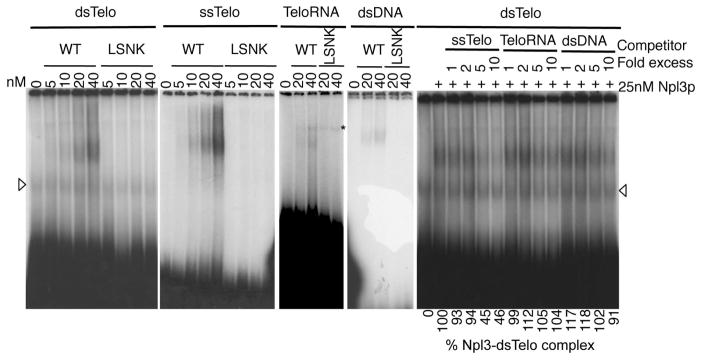
Npl3 binds to RNA in vitro with a preference for G/U-rich sequences [25]. We wanted to ask if this binding is specific for telomere sequences, both DNA and RNA. Purified 6xHis-tagged Npl3 was mixed with double stranded TG-rich telomere sequences (dsTelo), TG-rich single strands (ssTelo), or UG-rich RNA (TeloRNA) (see Table SII for sequences). We observed shifted complexes with all three types of nucleic acid, although the binding to TeloRNA was weak (Fig. 3). Npl3 also bound non-telomeric dsDNA; however, binding to the G-rich dsTelo could be readily displaced by ssTelo but not as much by the non-telomeric dsDNA or TeloRNA. This indicated that Npl3 prefers to bind telomeric DNA sequences. Binding to all types of nucleic acids was significantly diminished with mutant Npl3-LSNK indicating the requirement of the RRMs.
Figure 3. Npl3 binds telomeric sequences.
Purified 6xHis-tagged WT Npl3 or mutant Npl3-LSNK were incubated with double stranded telomeric DNA (dsTelo), single stranded telomeric DNA (ssTelo), G-rich RNA (TeloRNA), all derived from chromosome I-L, or non-telomeric double stranded DNA (dsDNA). Oligonucleotide sequences are listed in Table SII. For the right most panel, WT Npl3 (25nM) was mixed with unlabeled ssTelo, TeloRNA, or dsDNA (1-, 2-, 5-, or 10- fold excess) prior to the addition of radiolabeled dsTelo oligo. Percent Npl3-dsTelo complex is the average density of each protein complex (minus background) relative to the Npl3-dsTelo binding with no unlabelled competitor. Non-specific bands are indicated by asterisks or open triangles (secondary structures formed by G-rich dsTelo oligos). Figure is representative of two to three independent experiments.
3.4. Human hnRNP A1 is unable to substitute for the lack of Npl3 to rescue accelerated senescence in tlc1 npl3 cells
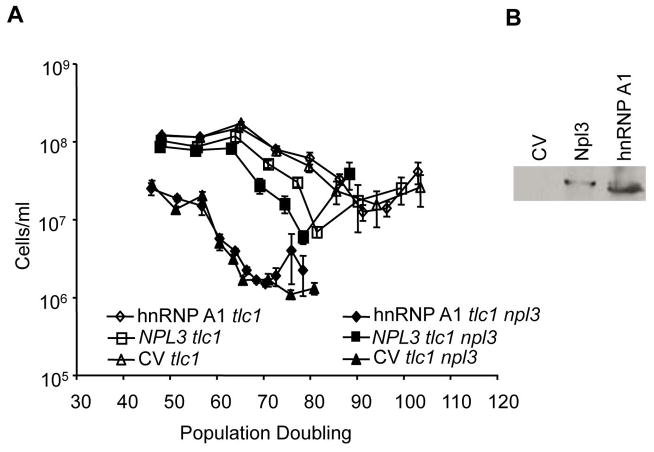
Because Npl3 is similar to human hnRNP A1, we asked if hnRNP A1 is able to substitute for Npl3 during the senescence of yeast lacking telomerase. Diploid TLC1/tlc1, NPL3/npl3 cells were transformed with control vector or plasmids that expressed 6xHis-tagged- human hnRNP A1 or Npl3. While the exogenously expressed WT Npl3 was able to rescue the faster senescence of tlc1 npl3 double mutants, hnRNP A1 was not. The rate of senescence for exogenously expressed hnRNP A1 in tlc1 npl3 mutants was accelerated, comparable to the control tlc1 npl3 + CV cells with the lowest point of senescence at around 70 PDs for both strains (Fig. 4A). In tlc1 NPL3 strains expressing endogenous Npl3, the control or hnRNP A1-expression vectors had no effects on tlc1 senescence; lowest points of senescence in these cells were at 94 PDs for tlc1 + CV cells and 92 PDs for tlc1 + hnRNP A1 cells. The plasmid expressing NPL3 caused the tlc1 and tlc1 npl3 mutants to senesce faster (bottoming out at 78 and 81 PDs, respectively) compared to the tlc1 + hnRNP A1 cells (92 PDs) but slower compared to tlc1 npl3 + CV cells (~70 PDs). It is unclear whether the levels of Npl3 were too high and caused some toxicity or too low to fully rescue the faster senescence. By western blot analysis, hnRNP A1 protein was detected in cell lysates of tlc1 npl3 survivors (Fig. 4B). These results show that hnRNP A1 was expressed but was unable to rescue the faster senescence of tlc1 npl3 mutants.
Figure 4. Human hnRNP A1 is unable to rescue accelerated senescence in tlc1 npl3 cells.
(A) yJL325 diploid cells were transformed with plasmids that expressed 6xHis tagged- human hnRNP A1 or Npl3 or control vector. Transformed cells were sporulated, dissected, and maintained in SC-URA medium for the senescence assay as described. Error bars indicate standard error of the mean, N = 5. (B) Whole cell lysates from survivors were prepared, analyzed by SDS-PAGE, and immunoblotted with anti-hnRNP A1 antibody.
4. Discussion
Npl3 is involved in multiple RNA processes including splicing, 3′ end formation, export, and stability. This report now attributes telomere maintenance as another function for Npl3. The second RNA recognition motif (RRM2) appears to be critical for this maintenance. Both RRM1 and RRM2 are required for Npl3 to bind preferentially to telomeric DNA. Based on our data and other published reports, we speculate that Npl3, perhaps with Cbc2, may help maintain telomeres by working as a telomere capping protein.
Npl3 and Cbc2 may protect chromosome ends as telomere capping proteins in several ways. They may prevent cells from detecting the telomere ends as double stranded breaks thus becoming substrates for homologous recombination or chromosome end fusion. Longer telomeres in mutants such as cbc2 compared to WT suggest that capping proteins may also regulate the access of telomerase. Without caps and without functional telomerase to elongate the ends, telomeres may be perceived as damaged DNA thereby causing senescence. Steady state telomere lengths being similar in npl3 and WT cells support the idea that Npl3 may be dispensable as a telomere capping protein and have additional roles in maintaining telomeres. Telomere binding proteins may interact with telomeres by binding directly to the single stranded overhang or the double stranded regions. Npl3 is interesting in that it has preference for single stranded or double stranded telomeric DNA. It is possible that Npl3 facilitates the formation or stabilizes secondary structures at the telomere thus helping to protect chromosome ends.
Npl3 may also work at the subtelomeric region. It physically or genetically interacts with RNA polymerase components as well as histones and histone modifying proteins in direct binding assays [27, 28] and genome-wide studies [29, 30]. Perhaps Npl3 helps maintain the heterochromatin and prevents the activation of subtelomeric transcription. It will be interesting to measure the levels of non-coding RNA telomeric repeat containing RNA (TERRA) in npl3 mutants.
Instead of Npl3 working directly at the telomeres to prevent faster senescence of telomerase null cells, it may modulate the levels of other telomere binding proteins. Npl3 targets a large number of mRNAs [31]. In the absence of Npl3, mRNAs that encode essential telomere maintenance proteins (Ten1 and Stn1, for example) may not be properly processed causing telomere instability and faster senescence. However, since Npl3 association with these and the transcripts of other telomere-binding genes (i.e. RAP1, CDC13, SGS1) is absent or relatively low [31], Npl3 is not likely to cause faster senescence by regulating the expression of these genes.
Collectively, we have shown that RNA processing proteins, Npl3 and Cbc2, may function in maintaining telomere stability perhaps as a telomere capping proteins. Further understanding of how Npl3 works with or without Cbc2 to prevent fast senescence will shed light on how proteins involved in RNA metabolism may participate in protecting the stability of chromosome ends.
Supplementary Material
Yeast hnRNP-related proteins are able to prevent faster senescence in telomerase-null cells.
The conserved RRMs in Npl3 are important for telomere maintenance.
Human hnRNP A1 is unable to complement the lack of NPL3 in yeast.
Npl3 and Cbc2 may work as telomere capping proteins.
Acknowledgments
Authors gratefully acknowledge funding from NIH/NIA (R15 AG035257). We thank members of the Lee-Soety lab for technical assistance, Jim Watrous for help with statistical analysis, and Brad Johnson for yeast strains and comments on the manuscript. We also thank Karen Hubbard, Anne McBride, and Pamela Silver for sharing their plasmids.
Abbreviations
- hnRNP
heterogeneous nuclear ribonucleoprotein
- RRM
RNA recognition motif
- PD
Population Doubling
- TFL
Telomere Fragment Length
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weighardt F, Biamonti G, Riva S. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 2.LaBranche H, Dupuis S, Ben-David Y, et al. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 3.Ding J, Hayashi MK, Zhang Y, et al. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dallaire F, Dupuis S, Fiset S, et al. Heterogeneous nuclear ribonucleoprotein A1 and UP1 protect mammalian telomeric repeats and modulate telomere replication in vitro. J Biol Chem. 2000;275:14509–14516. doi: 10.1074/jbc.275.19.14509. [DOI] [PubMed] [Google Scholar]
- 5.Fiset S, Chabot B. hnRNP A1 may interact simultaneously with telomeric DNA and the human telomerase RNA in vitro. Nucleic Acids Res. 2001;29:2268–2275. doi: 10.1093/nar/29.11.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohyama T, Furukawa A, Miyoshi T, et al. Interactions with RNA/DNA of proteins involved in the regulation of transcription, translation and telomere elongation. Nucleic Acids Symp Ser (Oxf) 2007:77–78. doi: 10.1093/nass/nrm039. [DOI] [PubMed] [Google Scholar]
- 7.Zhang QS, Manche L, Xu RM, et al. hnRNP A1 associates with telomere ends and stimulates telomerase activity. Rna. 2006;12:1116–1128. doi: 10.1261/rna.58806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flach J, Bossie M, Vogel J, et al. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 11.Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen EC, Henry MF, Weiss VH, et al. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBride AE, Cook JT, Stemmler EA, et al. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J Biol Chem. 2005;280:30888–30898. doi: 10.1074/jbc.M505831200. [DOI] [PubMed] [Google Scholar]
- 14.Xu C, Henry MF. Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p. Mol Cell Biol. 2004;24:10742–10756. doi: 10.1128/MCB.24.24.10742-10756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bucheli ME, Buratowski S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J. 2005;24:2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dermody JL, Dreyfuss JM, Villen J, et al. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE. 2008;3:e3273. doi: 10.1371/journal.pone.0003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell. 2008;32:727–734. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estrella LA, Wilkinson MF, Gonzalez CI. The shuttling protein Npl3 promotes translation termination accuracy in Saccharomyces cerevisiae. J Mol Biol. 2009;394:410–422. doi: 10.1016/j.jmb.2009.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Windgassen M, Sturm D, Cajigas IJ, et al. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol. 2004;24:10479–10491. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong CM, Qiu H, Hu C, et al. Yeast cap binding complex impedes recruitment of cleavage factor IA to weak termination sites. Mol Cell Biol. 2007;27:6520–6531. doi: 10.1128/MCB.00733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen EC, Stage-Zimmermann T, Chui P, et al. The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J Biol Chem. 2000;275:23718–23724. doi: 10.1074/jbc.M002312200. [DOI] [PubMed] [Google Scholar]
- 22.Askree SH, Yehuda T, Smolikov S, et al. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci U S A. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JY, Kozak M, Martin JD, et al. Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol. 2007;5:e160. doi: 10.1371/journal.pbio.0050160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson FB, Marciniak RA, McVey M, et al. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001;20:905–913. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deka P, Bucheli ME, Moore C, et al. Structure of the yeast SR protein Npl3 and Interaction with mRNA 3′-end processing signals. J Mol Biol. 2008;375:136–150. doi: 10.1016/j.jmb.2007.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatbonton T, Imbesi M, Nelson M, et al. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2:e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurt E, Luo MJ, Rother S, et al. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci U S A. 2004;101:1858–1862. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tardiff DF, Abruzzi KC, Rosbash M. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc Natl Acad Sci U S A. 2007;104:19948–19953. doi: 10.1073/pnas.0710179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan X, Ye P, Yuan DS, et al. A DNA Integrity Network in the Yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 30.Wilmes GM, Bergkessel M, Bandyopadhyay S, et al. A Genetic Interaction Map of RNA-Processing Factors Reveals Links between Sem1/Dss1-Containing Complexes and mRNA Export and Splicing. Molecular Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Guisbert K, Duncan K, Li H, et al. Functional specificity of shuttling hnRNPs revealed by genome-wide analysis of their RNA binding profiles. Rna. 2005;11:383–393. doi: 10.1261/rna.7234205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.