Abstract
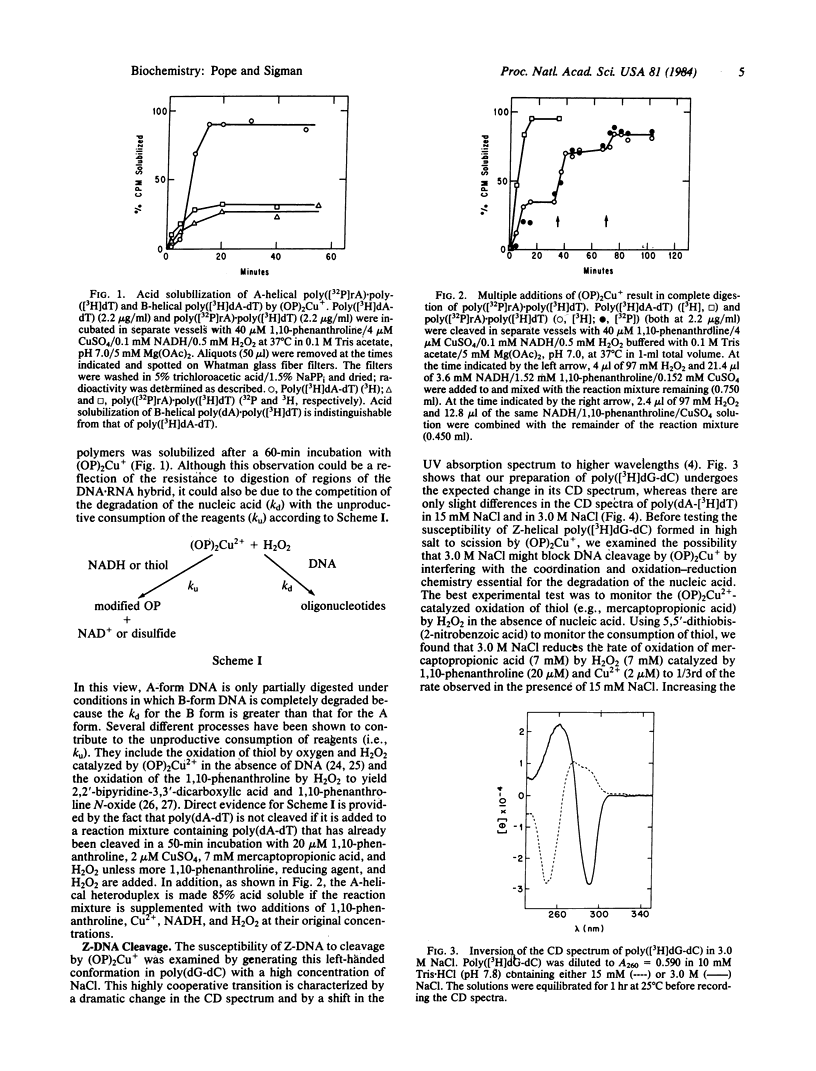
The artificial DNase activity of the 1,10-phenanthroline-cuprous ion complex [(OP)2Cu+] and H2O2 cleaves the A, B, and Z forms of DNA at different rates. The B structure, formed by most DNAs including poly(dA-dT) and poly(dA) X poly(dT), is the most susceptible to cleavage. It is completely degraded within 1 min by 40 microM 1,10-phenanthroline/4 microM Cu2+/7 mM H2O2/7 mM 3-mercaptopropionic acid. The A structure, formed by RNA X DNA hybrids such as poly(rA) X poly(dT), is cleaved in both strands roughly 10-20% as rapidly as poly(dA-dT) under comparable conditions. In contrast, the left-handed Z structure, formed by poly(dG-dC) in 3.0 M NaCl, is completely resistant to cleavage even though the same copolymer in the B structure at 15 mM NaCl is readily degraded. Poly(dA-dT) is rendered acid soluble at both salt concentrations at similar rates. The basis for the secondary structure specificity of the DNA cleavage reaction most likely resides in the requisite formation of a productive complex between (OP)2Cu+ and DNA during the course of this reaction. Previous studies have suggested that strand scission is due to oxidative destruction of the deoxyribose by hydroxyl radicals produced by the oxidation of DNA-bound Cu+ by H2O2. Apparently, the Z and A structures are unable to form a stable noncovalent complex with the same optimal geometry for cleavage as the B structure and are less susceptible to degradation. This artificial DNase activity may provide an approach to assess the formation of non-B-DNA structures in solution.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S. The geometry of nucleic acids. Prog Biophys Mol Biol. 1970;21:265–319. doi: 10.1016/0079-6107(70)90027-1. [DOI] [PubMed] [Google Scholar]
- Cooper P. J., Hamilton L. D. The A-B conformational change in the sodium salt of DNA. J Mol Biol. 1966 Apr;16(2):562–563. doi: 10.1016/s0022-2836(66)80193-6. [DOI] [PubMed] [Google Scholar]
- D'Aurora V., Stern A. M., Sigman D. S. 1,10-Phenanthroline-cuprous ion complex, a potent inhibitor of DNA and RNA polymerases. Biochem Biophys Res Commun. 1978 Feb 28;80(4):1025–1032. doi: 10.1016/0006-291x(78)91348-7. [DOI] [PubMed] [Google Scholar]
- D'Aurora V., Stern A. M., Sigman D. S. Inhibition of E. coli DNA polymerase I by 1,10-phenanthroline. Biochem Biophys Res Commun. 1977 Sep 9;78(1):170–176. doi: 10.1016/0006-291x(77)91236-0. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R., Conner B. N., Wing R. M., Fratini A. V., Kopka M. L. The anatomy of A-, B-, and Z-DNA. Science. 1982 Apr 30;216(4545):475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovin T. M., Englund P. T., Bertsch L. L. Enzymatic synthesis of deoxyribonucleic acid. XXVI. Physical and chemical studies of a homogeneous deoxyribonucleic acid polymerase. J Biol Chem. 1969 Jun 10;244(11):2996–3008. [PubMed] [Google Scholar]
- Lafer E. M., Möller A., Nordheim A., Stollar B. D., Rich A. Antibodies specific for left-handed Z-DNA. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3546–3550. doi: 10.1073/pnas.78.6.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps H. J., Nordheim A., Lafer E. M., Ammermann D., Stollar B. D., Rich A. Antibodies against Z DNA react with the macronucleus but not the micronucleus of the hypotrichous ciliate stylonychia mytilus. Cell. 1983 Feb;32(2):435–441. doi: 10.1016/0092-8674(83)90463-4. [DOI] [PubMed] [Google Scholar]
- MARVIN D. A., SPENCER M., WILKINS M. H., HAMILTON L. D. The molecular configuration of deoxyribonucleic acid. III. X-ray diffraction study of the C form of the lithium salt. J Mol Biol. 1961 Oct;3:547–565. doi: 10.1016/s0022-2836(61)80021-1. [DOI] [PubMed] [Google Scholar]
- Marshall L. E., Graham D. R., Reich K. A., Sigman D. S. Cleavage of deoxyribonucleic acid by the 1,10-phenanthroline-cuprous complex. Hydrogen peroxide requirement and primary and secondary structure specificity. Biochemistry. 1981 Jan 20;20(2):244–250. doi: 10.1021/bi00505a003. [DOI] [PubMed] [Google Scholar]
- Milman G., Langridge R., Chamberlin M. J. The structure of a DNA-RNA hybrid. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1804–1810. doi: 10.1073/pnas.57.6.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Hao W. M., Wogan G. N., Rich A. Salt-induced conversion of B-DNA to Z-DNA inhibited by aflatoxin B1. Science. 1983 Mar 25;219(4591):1434–1436. doi: 10.1126/science.6402818. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Pardue M. L., Lafer E. M., Möller A., Stollar B. D., Rich A. Antibodies to left-handed Z-DNA bind to interband regions of Drosophila polytene chromosomes. Nature. 1981 Dec 3;294(5840):417–422. doi: 10.1038/294417a0. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien E. J., MacEwan A. W. Molecular and crystal structure of the polynucleotide complex: polyinosinic acid plus polydeoxycytidylic acid. J Mol Biol. 1970 Mar 14;48(2):243–261. doi: 10.1016/0022-2836(70)90159-2. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M., Baehr W., Holbrook J. J. Ethidium bromide as a cooperative effector of a DNA structure. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3805–3809. doi: 10.1073/pnas.69.12.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Pohl F. M. Polymorphism of a synthetic DNA in solution. Nature. 1976 Mar 25;260(5549):365–366. doi: 10.1038/260365a0. [DOI] [PubMed] [Google Scholar]
- Pope L. M., Reich K. A., Graham D. R., Sigman D. S. Products of DNA cleavage by the 1,10-phenanthroline-copper complex. Inhibitors of Escherichia coli DNA polymerase I. J Biol Chem. 1982 Oct 25;257(20):12121–12128. [PubMed] [Google Scholar]
- Riley M., Maling B. Physical and chemical characterization of two- and three-stranded adenine-thymine and adenine-uracil homopolymer complexes. J Mol Biol. 1966 Sep;20(2):359–389. doi: 10.1016/0022-2836(66)90069-6. [DOI] [PubMed] [Google Scholar]
- Sage E., Leng M. Conformation of poly(dG-dC) . poly(dG-dC) modified by the carcinogens N-acetoxy-N-acetyl-2-aminofluorene and N-hydroxy-N-2-aminofluorene. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4597–4601. doi: 10.1073/pnas.77.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santella R. M., Grunberger D., Weinstein I. B., Rich A. Induction of the Z conformation in poly(dG-dC).poly(dG-dC) by binding of N-2-acetylaminofluorene to guanine residues. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1451–1455. doi: 10.1073/pnas.78.3.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selsing E., Wells R. D., Alden C. J., Arnott S. Bent DNA: visualization of a base-paired and stacked A-B conformational junction. J Biol Chem. 1979 Jun 25;254(12):5417–5422. [PubMed] [Google Scholar]
- Selsing E., Wells R. D. Polynucleotide block polymers consisting of a DNA.RNA hybrid joined to a DNA.DNA duplex. Synthesis and characterization of dGn.rCidCk duplexes. J Biol Chem. 1979 Jun 25;254(12):5410–5416. [PubMed] [Google Scholar]
- Sigman D. S., Graham D. R., D'Aurora V., Stern A. M. Oxygen-dependent cleavage of DNA by the 1,10-phenanthroline . cuprous complex. Inhibition of Escherichia coli DNA polymerase I. J Biol Chem. 1979 Dec 25;254(24):12269–12272. [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Blakesley R. W., Hardies S. C., Horn G. T., Larson J. E., Selsing E., Burd J. F., Chan H. W., Dodgson J. B., Jensen K. F. The role of DNA structure in genetic regulation. CRC Crit Rev Biochem. 1977;4(3):305–340. doi: 10.3109/10409237709102561. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Goodman T. C., Hillen W., Horn G. T., Klein R. D., Larson J. E., Müller U. R., Neuendorf S. K., Panayotatos N., Stirdivant S. M. DNA structure and gene regulation. Prog Nucleic Acid Res Mol Biol. 1980;24:167–267. doi: 10.1016/s0079-6603(08)60674-1. [DOI] [PubMed] [Google Scholar]
- van de Sande J. H., Jovin T. M. Z* DNA, the left-handed helical form of poly[d(G-C)] in MgCl2-ethanol, is biologically active. EMBO J. 1982;1(1):115–120. doi: 10.1002/j.1460-2075.1982.tb01133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


