Abstract
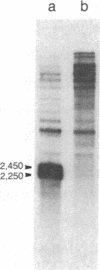
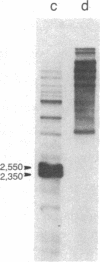
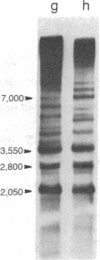
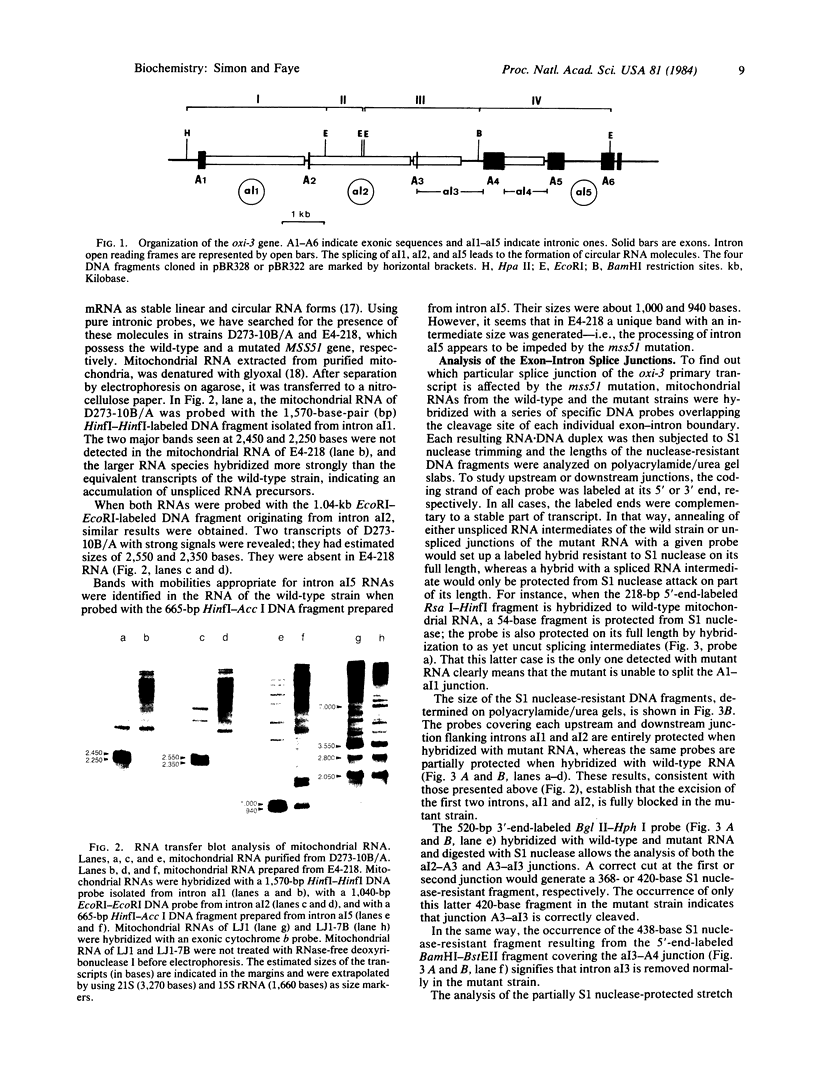
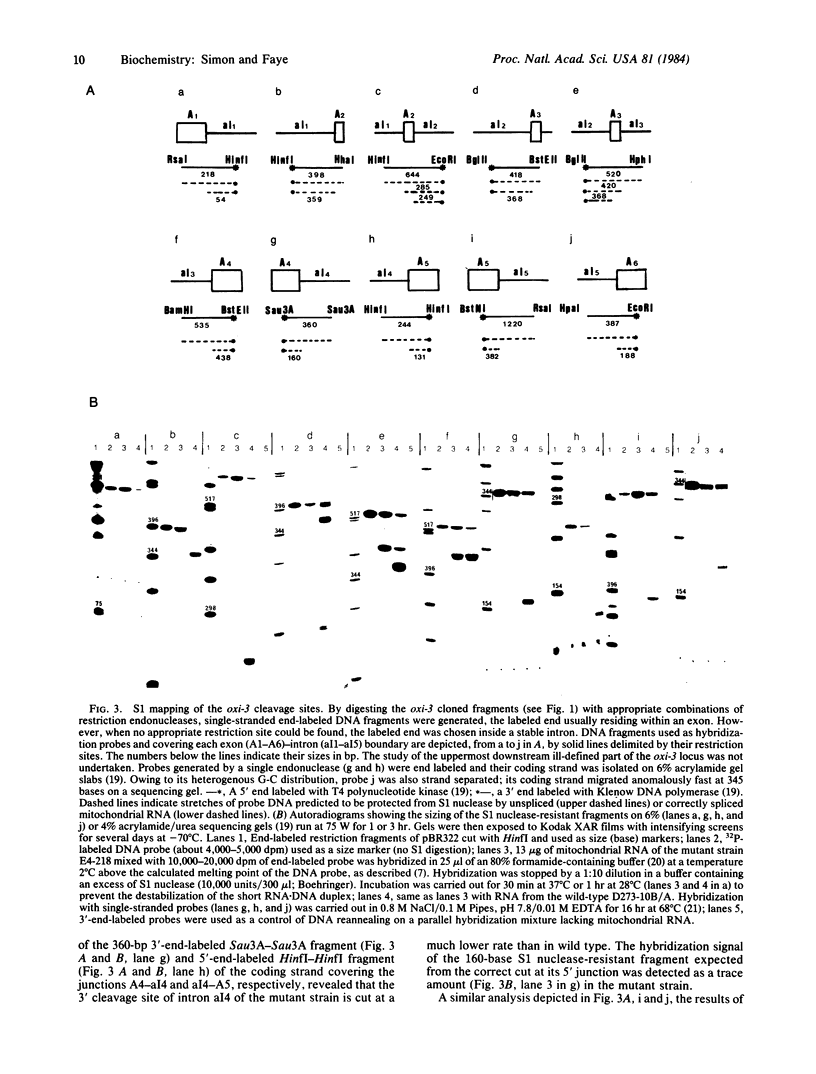
In Saccharomyces cerevisiae, the mitochondrial gene encoding the subunit I of cytochrome c oxidase (oxi-3 gene) is interrupted by intervening sequences. In this report, a nuclear mutation [referred to as mss51 in Faye, G. & Simon, M. (1983) Cell 32, 77-87] that specifically affects the processing of oxi-3 pre-mRNA was further characterized. DNA probes covering each oxi-3 exon-intron boundary were individually hybridized to wild-type and mutant mitochondrial RNA. By a technique relying on the S1 nuclease resistance or sensitivity of the RNA X DNA hybrids thereof, we have shown which site needs the MSS51 gene product to be cleaved. The mutation in the MSS51 gene gave rise to a complex pattern of splicing: the third intron was excised efficiently but the first two introns remained bracketed by their flanking exons. Further, the fourth and fifth introns were only partially split from their common exon and remained fused to their upstream and downstream flanking exon, respectively. Several plausible roles for the MSS51 gene product are discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnberg A. C., Van Ommen G. J., Grivell L. A., Van Bruggen E. F., Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980 Feb;19(2):313–319. doi: 10.1016/0092-8674(80)90505-x. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bonitz S. G., Coruzzi G., Thalenfeld B. E., Tzagoloff A., Macino G. Assembly of the mitochondrial membrane system. Structure and nucleotide sequence of the gene coding for subunit 1 of yeast cytochrme oxidase. J Biol Chem. 1980 Dec 25;255(24):11927–11941. [PubMed] [Google Scholar]
- Carmichael G. G., McMaster G. K. The analysis of nucleic acids in gels using glyoxal and acridine orange. Methods Enzymol. 1980;65(1):380–391. doi: 10.1016/s0076-6879(80)65049-6. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- De La Salle H., Jacq C., Slonimski P. P. Critical sequences within mitochondrial introns: pleiotropic mRNA maturase and cis-dominant signals of the box intron controlling reductase and oxidase. Cell. 1982 Apr;28(4):721–732. doi: 10.1016/0092-8674(82)90051-4. [DOI] [PubMed] [Google Scholar]
- Faye G., Simon M. Analysis of a yeast nuclear gene involved in the maturation of mitochondrial pre-messenger RNA of the cytochrome oxidase subunit I. Cell. 1983 Jan;32(1):77–87. doi: 10.1016/0092-8674(83)90498-1. [DOI] [PubMed] [Google Scholar]
- Groudinsky O., Dujardin G., Slonimski P. P. Long range control circuits within mitochondria and between nucleus and mitochondria. II. Genetic and biochemical analyses of suppressors which selectively alleviate the mitochondrial intron mutations. Mol Gen Genet. 1981;184(3):493–503. doi: 10.1007/BF00352529. [DOI] [PubMed] [Google Scholar]
- Hudspeth M. E., Shumard D. S., Tatti K. M., Grossman L. I. Rapid purification of yeast mitochondrial DNA in high yield. Biochim Biophys Acta. 1980 Dec 11;610(2):221–228. doi: 10.1016/0005-2787(80)90003-9. [DOI] [PubMed] [Google Scholar]
- Jacq C., Lazowska J., Slonimski P. P. Sur un nouveau mécanisme de la régulation de l'expression génétique. C R Seances Acad Sci D. 1980 Jan 14;290(2):89–92. [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Hall B. D., Gillam S., Smith M. Identification and isolation of the yeast cytochrome c gene. Cell. 1978 Jul;14(3):673–680. doi: 10.1016/0092-8674(78)90250-7. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K., Hall B. D., Astell C., Smith M. A position effect in the control of transcription at yeast mating type loci. Nature. 1981 Jan 22;289(5795):244–250. doi: 10.1038/289244a0. [DOI] [PubMed] [Google Scholar]
- Schmelzer C., Schweyen R. J. Evidence for ribosomes involved in splicing of yeast mitochondrial transcripts. Nucleic Acids Res. 1982 Jan 22;10(2):513–524. doi: 10.1093/nar/10.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R., Breitenbach M., Schweyen R. J. Mitochondrial circular RNAs are absent in sporulating cells of Saccharomyces cerevisiae. Nucleic Acids Res. 1983 Mar 25;11(6):1735–1746. doi: 10.1093/nar/11.6.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Berk A. J., Berget S. M. Transcription maps of adenovirus. Methods Enzymol. 1980;65(1):750–768. doi: 10.1016/s0076-6879(80)65071-x. [DOI] [PubMed] [Google Scholar]
- Soberon X., Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. IV. Deletion derivatives of pBR322 and pBR325. Gene. 1980 May;9(3-4):287–305. doi: 10.1016/0378-1119(90)90328-o. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Needleman R. B. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J Biol Chem. 1975 Oct 25;250(20):8228–8235. [PubMed] [Google Scholar]
- Wolf K., Dujon B., Slonimski P. P. Mitochondrial genetics. V. Multifactorial mitochondrial crosses involving a mutation conferring paromomycin-resistance in Saccharomyces cerevisiae. Mol Gen Genet. 1973 Sep 5;125(1):53–90. doi: 10.1007/BF00292983. [DOI] [PubMed] [Google Scholar]