Abstract
Activation of the fibroblast growth factor receptor pathway is a common event in many cancer types. Here we review the role of fibroblast growth factor receptor signalling in breast cancer, from SNPs in FGFR2 that influence breast cancer risk and SNPs in FGFR4 that associate with breast cancer prognosis, and potential therapeutic targets such as receptor amplification and aberrant autocrine and paracrine ligand expression. We discuss the multiple therapeutic strategies in preclinical and clinical development and the current and future challenges to successfully targeting this pathway in cancer.
Introduction
Activation of tyrosine kinase growth factor receptors presents one of the most common oncogenic events in cancer. Targeting these receptors is a proven therapeutic strategy, as exemplified by the efficacy of trastuzumab in HER2 amplified breast cancer. However, in the ~85% of breast cancers that do not have HER2 amplification there has been limited progress with targeting other growth factor receptors. Studies have found potential evidence of efficacy targeting epidermal growth factor receptor (EGFR) in combination with endocrine therapy [1], and insulin-like growth factor 1 receptor in combination with mammalian target of rapamycin inhibitors [2], although none of these approaches have as yet proceeded beyond phase II trials.
Preclinical evidence suggests that activation of fibroblast growth factor receptor (FGFR) signalling is a common event in cancer [3]. Yet the clinical development of therapies targeting the FGFR signalling pathway presents multiple challenges, with diverse mechanisms of pathway activation combined with multiple inhibitors of differing potency and with antibodies in preclinical development. In the present review we discuss the multiple mechanisms through which FGFR signalling contributes to the pathogenesis of breast cancer, and also review the challenges of translating this evidence into clinical trials of therapies targeting the FGFRs.
The fibroblast growth factor signalling system
The fibroblast growth factors (FGFs) and their receptors (FGFRs) play an important role in a wide range of biological functions, controlling developmental events such as brain patterning, morphogenesis and limb development [4,5] with multiple physiological functions in the adult including angiogenesis, wound repair and endocrine functions [6].
The FGF family consists of 18 ligands; FGF ligand nomenclature extends to FGF23 although only 18 FGFs function as ligands, which signal through four high-affinity FGFRs (FGFR1 to FGFR4) [3,6,7]. The majority of FGFs bind to heparan sulphate glycosaminoglycans on the cell surface or in the extracellular matrix, and consequently do not diffuse far from the site of production acting as paracrine or autocrine growth factors - although one FGF ligand family (FGF19, FGF21, FGF23) function as hormones and bind to FGFRs in complex with Klotho proteins [6]. As well as this spatial regulation of ligand-receptor interaction, alternative splicing of the third immunoglobulin domain in the receptor generates two different receptors with highly different ligand specificity (reviewed in [6]).
The majority of FGFs bind receptor in a trimeric complex with heparins, triggering a conformational change in the receptor that leads to activation of the FGFR that results in phosphorylation of multiple sites on the intracellular domain, adapter protein binding and intracellular signalling (reviewed in detail elsewhere [8]). Under physiological conditions, the highly complex FGF signalling pathway is tightly regulated [3]. The deregulation of FGF signalling in cancer results in activation of the pathway without appropriate regulation leading to/contributing to development of cancer, promoting cancer cell proliferation, survival and migration [9-13].
FGFR signalling in breast cancer pathogenesis
The mouse mammary tumour virus was a major cause of mammary tumours in multiple laboratory mouse strains, vertically transmitted from mother to pup. Mouse mammary tumour virus is a retrovirus that is oncogenic through integration into the genome activating the expression of nearby genes, with FGF3 and FGF8 being, along with WNT genes, the commonest site of integration [14,15]. The link between FGF activation and mammary carcinoma was subsequently confirmed by experiments with transgenic mice, with both epithelial FGF3 overexpression [16] and FGFR1 activation [17] leading to epithelial proliferation and invasive lesions [17].
Genome-wide association studies have subsequently identified SNPs within the second intron of the FGFR2 gene that are associated with increased risk of developing breast cancer [18,19]. The minor, predisposing, allele is present in approximately 40% of western populations, although the associated increased risk is relatively small: 1.26-fold for heterozygotes and 1.63-fold for homozygotes [18]. The minor allele increases the risk of developing oestrogen receptor (ER)-positive breast cancer, with only a minor effect on ER-negative breast cancer [20]. Multiple SNPs in the second intron are in very high linkage disequilibrium, and from genetic data it is not possible to pinpoint the causative SNP(s) - although strong biochemical evidence suggests that rs2981578 may be causative through creation of an OCT1/RUNX2 binding site [21], potentially resulting in increased FGFR2 expression in breast cancers with the minor allele variant [21]. Whether this reflects increased epithelial or stromal expression is less clear [22]. FGFR2 IIIb knockout mice have a gross failure of branching morphogenesis in the breast [23], raising the possibility that increased FGFR2 expression may simply result in nonspecific changes in breast epithelium that predispose to breast cancer. Further research with transgenic models is required to establish how increased FGFR2 expression results in breast cancer predisposition.
A SNP in FGFR4 (G388R, Gly338-Arg338) has been shown to confer a more aggressive behaviour and poor prognosis in multiple cancer types, including breast cancer [24-28]. This SNP may increase invasion and motility through altering receptor internalisation, potentially leading to abnormally sustained signalling [29-31]. Recent data have suggested in addition that the FGFR4 Arg388 allele may be associated with a pathological complete response to chemotherapy [32], although potentially conflicting data have also been reported [27]. Although the SNPs in both FGFR2 and FGFR4 illustrate the potential importance of FGF signalling in breast cancer pathogenesis, there is no current evidence that either SNP presents a therapeutic target in established breast cancer.
Potential therapeutic targets in breast cancer
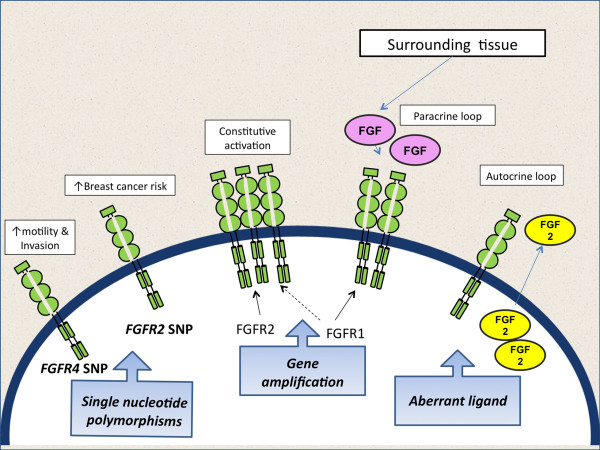
There are multiple mechanisms through which FGFR signalling may be activated in breast cancer, that may present potential therapeutic targets (Figure 1).
Figure 1.
Alterations in the FGFR signalling pathway that influence breast cancer. Single nucleotide polymorphisms (SNPs) influence breast cancer risk (FGFR2), and prognosis in established cancers likely through effects on motility and invasive capacity (FGFR4). Somatic alterations presenting potential therapeutic targets include amplification of FGFR1 and FGFR2, and aberrant FGF2 ligand expressed in a paracrine or autocrine fashion.
FGFR2 gene amplification
Amplification of the FGFR2 gene occurs in a small subset of breast cancer, although in these cancers preclinical evidence suggests this gene is potentially an excellent therapeutic target. Breast cancer cell lines with FGFR2 amplification show high sensitivity to FGFR inhibitors in vitro [33,34], and the FGFR2-amplified MFM223 cell line is sensitive in vivo to an FGFR2 targeting antibody [35]. FGFR2 is highly overexpressed in amplified cell lines, along with expression of a C-terminal truncated form that results in impaired receptor internalisation [36], and FGFR2 is constitutively active and ligand independent in the amplified cell lines. FGFR2 amplification is rare in breast cancer, however, present in only 1 to 2% of breast cancer overall [37], although this is enriched to an estimate of ~4% of breast cancers with the aggressive triple-negative breast cancers [33]. FGFR2 amplifications have also been described in approximately 10% of gastric cancers usually associated with the poor-prognosis diffuse-type histology [38].
FGFR1 gene amplification
The FGFR1 gene is one of the most commonly amplified genes in cancer [39]. Amplification of the chromosomal region 8p11-12, the genomic location of FGFR1, is seen in approximately 10% of the breast cancers, predominantly in the ER-positive breast cancers [40-44]. The oncogenic driver of 8p11-12 amplifications has been a source of substantial discord in the scientific literature for the last 15 years, although in the last few years clarity has finally emerged.
Prior misunderstandings have arisen in part from attempts to find a single oncogenic driver within the region, a view that follows the paradigm of HER2 and 17q21 amplification. Evidence that this simplified model is incorrect emerged from high-resolution comparative genomic hybridisation analysis of breast cancer suggesting two major cores, or peaks, of amplification (core A1 distal at 36.5 to 37.8 Mb, and core A2 proximal at 38.1 to 38.9 Mb; Genome Build 35) [41]. Although the most common pattern was for amplification of both cores, amplification of either core alone occurred in a minority of cancers. Further evidence supporting the existence of two separate cores, and therefore at least two driver oncogenes, has subsequently come from cross-cancer comparisons. Amplification of 8p11-12 is also found in ~10% of squamous lung cancers but with a different genomic structure, with, at least in the published data-sets, a frequent pattern of amplification of the proximal A2 core without amplification of the distal A1 core [13].
Following on from clarity on the genomic structure, pointing to at least two oncogenic drivers, has come further clarity on the likely oncogenic drivers for each amplification core. ZNF703 has been demonstrated, with high likelihood, to be the principle oncogenic driver of the distal A1 core [45,46], resulting in induction of stem-cell-like phenotypes, potentially also suppressing ER and promoting E2F1 transcriptional activity [47]. In contrast, FGFR1 is the likely target of proximal A2 amplifications - although other genes have been implicated, such as the phosphatase PPAPDC1B [47]. FGFR1 promotes the growth of both breast cancer and lung cancer cell lines with FGFR1 amplification [13,43,48], with FGFR1 mRNA overexpression tightly linked to FGFR1 amplification [12,13], although cases of FGFR1 amplification without receptor overexpression have been demonstrated [12].
Amplification of FGFR1 is associated with a marked poor prognosis in breast cancer, specifically in ER-positive breast cancer [11]. We have recently provided evidence that FGFR1 amplification promotes resistance to endocrine therapy [12], potentially through enhanced ligand-dependent signalling in FGFR1 amplified cell lines. FGFR1 signalling promoted cyclin D1 expression and suppressed progesterone receptor expression, and similarly FGFR1 overexpressed cancers were more likely to be progesterone receptor negative and high in proliferation. Up to 25% of luminal-B-type breast cancers potentially have amplification of FGFR1 [12], and in these cancers FGFR1 may present an alternative growth/survival signal to escape the effects of endocrine therapy. An association has been reported between increased FGFR1 expression [49], FGFR1 amplification [43], and lobular breast cancer, although the enrichment for FGFR1 amplification in lobular cancers is relatively weak [11].
Some important questions remain, however, regarding the role of FGFR1 as an oncogene and therapeutic target. In contrast to FGFR2, where an aberrant form of the receptor is expressed, all data currently suggest that wild-type FGFR1 is overexpressed in amplified cancers. Ligand-independent signalling can be seen at very high levels of wild-type FGFR1 expression, presumably from local crowding of the receptors at the cell surface promoting transient receptor dimerisation [12]. There is little evidence of ligand-independent signalling in amplified breast cell lines or tumours, however, with the limited evidence suggesting enhanced ligand-dependent signalling [12]. This raises important, and unanswered, questions regarding which extracellular splice variants are expressed, and which of the multiple potential ligands activate the receptor.
Cooperative effects of FGFR gene amplification
There is substantial evidence that FGFR signalling cooperates with other oncogenic drivers to drive tumorigenesis. FGFR1 activation substantially accelerated the development of mammary carcinomas in a murine Wnt1 model of mammary carcinoma, and in this model FGFR signalling potentially accelerated tumour development through the promotion of cap-dependent translation [50]. FGFR signalling has also been shown to upregulate the EGFR ligands amphiregulin and epiregulin in mouse mammary cells and MCF7 breast cancer cells [51], and FGFR2 activates EGFR family receptors in FGFR2 amplified gastric cell lines [38], suggesting cooperation of FGFR and EGFR signalling in oncogenesis. Whether EGFR family signalling is important in the pathogenesis of FGFR amplified breast cancers is unknown. In certain contexts, FGFR1 transformed cells have been shown to be dependent on ribosomal S6 kinase signalling [49] - potentially because FGFR may directly phosphorylate RSK2 and possibly other ribosomal S6 kinase isoforms [52].
FGFR1 is frequently co-amplified with CCND1 on 11q, and in vitro evidence suggests substantial functional interaction between the genes on 8p11-12 and 11q [53]. An uncertain area around FGFR1 as a potential therapeutic target, however, is the relationship between FGFR1 and ZNF703. Whether co-amplification of ZNF703 affects sensitivity to FGFR inhibition in breast cancer will be an important question for future research.
FGFR mutations
Although FGFR activating mutations are found in multiple other cancer types, including FGFR2 in endometrial cancer [54] and FGFR3 in bladder cancer [55], there is no evidence for common mutational activation of the FGFRs in breast cancer.
Aberrant autocrine and paracrine signalling
Extending the evidence that FGFR2 amplifications are enriched in triple-negative breast cancer cell lines, we recently demonstrated that a number of triple-negative breast cancer cell lines are sensitive to FGFR inhibitors in vitro [33]. Sensitive cells lines were of the claudin-low subtype, and expressed autocrine FGF2 ligand. Sensitivity was found predominantly in anchorage-independent conditions in vitro, and CAL51 cell line xenografts were also sensitive in vivo [33]. Expression of cytoplasmic FGF2 ligand was also found to be specific to basal-like breast cancers by immunohistochemistry [33]. This raises the possibility that autocrine FGF2 ligand may be a therapeutic target in basal-like breast cancer, although there is uncertainty as to whether this is specific to the subset of basal-like breast cancers with a claudin-low-type expression pattern.
Assessment of the tumour stromal ligand concentration has shown FGF2 ligand to be expressed at high levels in tumour stroma [56]. Indeed, assessment of elevated FGF2 content in nipple aspirates has been suggested to be a potential diagnostic test for breast cancer [57]. Presumably FGF2 is secreted by activated stromal fibroblasts, but there is no direct evidence for the cell of origin and how this relates to cancer biology is unclear. Elevated FGF2 ligand may potentially be a source for signalling by amplified and overexpressed FGFR1. FGF2 is an angio-genic signalling peptide that is also released in an autocrine/paracrine fashion from activated endothelial cells [58]. There is, however, no association between FGF2 ligand concentrations and microvessel density [56], which has been interpreted as evidence that FGF2 does not promote de novo angiogenesis in breast cancer [56]. FGF2 has been shown to cause resistance to VEFGR targeting in vitro [59], although it is unknown whether promotes resistance to bevacizumab in breast cancer.
A potential role for paracrine FGF9/FGFR signalling has also been identified in the oestrogen-mediated expansion of a breast cancer stem-cell-like subpopulation in vitro, potentially through promoting expression of the Tbx3 transcription factor [60]. The full potential role of FGF autocrine and paracrine signalling in breast cancer is therefore yet to be fully elucidated.
Targeting FGFR signalling
The past decade has seen a marked increase in our understanding of the FGF signalling pathway. Given its role in the pathogenesis of various cancers, several pharmaceutical companies have developed agents targeting FGFs or FGFRs, the most common being small-molecule receptor tyrosine kinase inhibitors targeting the FGFR (Table 1).
Table 1.
Fibroblast growth factor targeting therapies in clinical development
| Drug class | Drug name | Target | Stage of clinical development |
|---|---|---|---|
| First-generation TKIs | TKI258 (dovitinib) [64] | FGFR, PDGFR and VEGFR | Phase III |
| BMS540215 (brivanib) [71] | FGFR and VEGFR | Phase II | |
| BIBF 1120 [72] | FGFR, PDGFR and VEGFR | Phase III | |
| Ponatinib [73] | ABL, FGFR, VEGFR2, PDGFRα, FLT3 | Phase II | |
| E7080 [74] | VEFGR, PDGFR, FGFR, KIT and RET | Phase I | |
| E3810 [75] | VEGFR1 to VEGFR3 and FGFR1 inhibitor | Phase I | |
| Sulfatinib [76] | VEGFR and FGFR inhibitor | Phase I | |
| Second-generation TKIs | AZD 4547 [77,78] | Selective FGFR1, FGFR2 and FGFR3 inhibitor | Phase II |
| BGJ398 [79] | Selective pan-FGFR inhibitor | Phase I | |
| FGFR antibodies | IMC-A1 [62] | FGFR1-IIIc-specific antibody | Preclinical |
| GP369 [35] | FGFR2 blocking antibody | Preclinical | |
| PRO-001 [80] | FGFR3-specific blocking antibody | Preclinical | |
| R3Mab [61] | FGFR3-specific antibody | Preclinical | |
| FGFR ligand traps | FP-1039 [81] | FGF ligand trap (blocks multiple FGFs) | Phase I |
ABL, c-abl oncogene 1, non-receptor tyrosine kinase; FGF, fibroblast growth factor; FGFR, fibroblast growth factor receptor; FLT3, fms-related tyrosine kinase 3; KIT, Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog; PDGFR, platelet-derived growth factor receptor; RET, ret proto-oncogene; TKI, tyrosine kinase inhibitor; VEGFR, vascular endothelial growth factor receptor.
Tyrosine kinase inhibitors
Multiple FGFR tyrosine kinase inhibitors are currently in early clinical development, although the inhibitors vary substantially in potency (Table 1). The first generation of inhibitors are multi-targeting ATP competitive inhibitors, with most originally developed as VEGFR inhibitors that also inhibit the FGFRs due to similarity in the ATP binding pocket structure. These inhibitors have varying potency against the FGFRs, and in cellular assays, in particular, have relatively low potency. Consequently, a number of pharmaceutical companies have developed second-generation inhibitors, developing inhibitors that specifically target FGFRs with selectivity over VEGFR and other kinases, with substantially increased potency (Table 1). A number of additional selective FGFR inhibitors are in preclinical development. The kinase domains of FGFR1 to FGFR3 are highly similar and the kinase inhibitors in development inhibit all three members, to a lesser or greater extent. FGFR4 has diverged from the other kinases, and consequently many inhibitors are less potent against FGFR4.
Antibodies
Multiple FGFR antibodies are in preclinical development, with evidence of efficacy for FGFR2 targeting antibodies in FGFR2 amplified breast cancer models [35] and FGFR3 targeting antibodies in FGFR3-driven models [61]. FGFR1 inhibitory antibodies are in preclinical development, but have not proceeded beyond preclinical toxicity testing due to appetite suppression and weight loss, potentially due to FGFR1 targeting in the hypothalamus [62]. A second potential approach is to develop anti-bodies against specific FGFs, such as FGF2, although none of these antibodies have yet emerged from the early preclinical development. The potential disadvantage of targeting a single FGF is the potential for rescue of any effect by alternative ligands.
Ligand traps
Another approach to targeting ligand-dependent signalling has been to develop ligand traps - such as FP-1039 based on a modified extracellular domain of FGFR1 fused to Fc, which has the potential to sequester multiple ligands including FGF2 [63]. Whether such approaches can work on autocrine ligand production is yet to be fully addressed.
Early clinical trial evidence
The first clinical trial evidence to support FGFR1 as a potential therapeutic target was presented at the 2011 American Society of Clinical Oncology annual meeting. Andre and colleagues presented the results of the phase II (n = 81) multicentre trial of dovitinib, a multi-tyrosine kinase inhibitor that targets FGFR, VEGFR and platelet-derived growth factor receptor in patients with metastatic breast cancer prescreened for FGFR1 amplification [64]. An unconfirmed response was observed in 15% of women with FGFR1 amplified ER-positive breast cancer, with no responses in nonamplified ER-positive breast cancer, although this level of response failed to meet the predefined criteria for a positive study [64]. Many patients withdrew from the study for reasons other than disease progression, with the drug less well tolerated than expected in a very heavily pretreated population [64]. Interestingly this study suggested that co-amplification of the 11q genomic region, encompassing CCND1, FGF3, FGF4 and FGF19, possibly identified sensitive tumours, potentially supporting in vitro evidence of cooperation between CCND1 and FGFR1 in oncogenesis [53,65].
Recently a second multi-targeting inhibitor has reported very preliminary evidence of activity, with responses reported in FGFR1 amplified cancers in the dose escalation study of E3810 [66]
Roadmap for clinical development
The multiple different mechanisms through which FGF signalling can be activated necessitate a complex approach to clinical development. Only a subset of breast cancers are likely to be sensitive to FGFR inhibitors, and screening will be required to specifically identify cancers with amplification, or potentially with FGF2 ligand expression.
Yet this complex approach presents substantial challenges for rare targets such as FGFR2 amplification. One approach is to screen a very large number of patients, as has been done for ELM4-ALK translocations in nonsmall-cell lung cancer leading to the licence of crizotinib [67]. Another approach is to potentially combine different cancer types with the same genetic aberration into a single trial - but this requires the target to be the same in different cancer subtypes. FGFR2 amplification occurs in both breast cancer and gastric cancer, and based on current evidence appears to be a similarly good potential target in both cancers. In contrast, it is not clear that FGFR1 amplification found in breast cancers, squamous lung cancers [13] as well as oral squamous cell carcinomas [68] is similar in the different cancers, as we have discussed previously.
Matching therapeutic approaches to targets
Multiple different therapeutics are in clinical development, so it is important to consider whether different therapeutic approaches lend themselves to specific oncogenic aberrations. Different FGFR tyrosine kinase inhibitors vary substantially in potency against FGFRs. Kinases with constitutive ligand-independent activation, through mutation or amplification, are generally more sensitive to tyrosine kinase inhibitors than wild-type receptors. Consequently, for targeting oncogenic aberrations such as FGFR2 amplification, which results in constitutive activation, it is likely that multi-targeted first-generation inhibitors will be of sufficient potency to induce tumour shrinkage. For most of the multi-targeted inhibitors, however, the maximum tolerated dose is not defined by the side effects of FGFR inhibition, and consequently may be administered at a dose below that required to achieve full wild-type FGFR inhibition. Targets such as FGF2 ligand autocrine expression, and potentially FGFR1 amplification, which signal through a wild- type receptor, may therefore be best approached through antibodies or more potent second-generation inhibitors.
The only first-generation inhibitor that has been shown, at the time of writing, to have inhibitory properties in clinical trials against wild-type FGFR signalling is dovitinib/TKI258, which results in a moderate increase in FGF23 ligand. FGF23 is secreted in bone, and hormonally regulates phosphate excretion from the kidney [64], and inhibition of FGFR in the kidney is expected to increase FGF23 levels. Recent data, however, have suggested that FGFR signalling also promotes FGF23 expression in bone, making interpretation of FGF23 levels complex [69]. This observation emphasises the importance of assessing further biomarkers in inhibitor development, although at present there are no biomarkers that can be used on clinical tumour material to assess FGFR directly, and this is an area that requires urgent attention to direct future development.
The second-generation inhibitors have potentially different challenges around high potency inhibition of multiple FGFRs, which have important physiological roles such as phosphate excretion (bone-derived FGF23 hormonally acting on renal FGFR1) [6]. The potential toxicity of pan-FGFR inhibition could therefore be avoided by use of FGFR inhibitory antibodies whose side effects would be limited to those of a single FGFR member, although FGFR1/FGFR2 antibodies have yet to progress beyond preclinical development.
Challenges to study design
Conducting clinical trials in small subsets presents challenges of recruitment in a study that only enrols a small proportion of potentially eligible patients. For example, considering the 10% rate of FGFR1 amplification in breast cancer, nearly 1,000 patients would need to be screened for a 100-patient phase II trial; and an even larger number would be needed for a phase III trial. The complexity of targets such as FGFR1 amplification potentially also requires even larger trials to identify within amplified cancers those cancers that are sensitive to FGFR inhibition. This factor potentially argues for a different approach to clinical development, focused on biomarker analysis - ideally with biopsy at study entry, as biomarkers may alter through prior therapy, paired with biopsy on study completion to confirm target inhibition and to identify potential determinants of sensitivity.
Conclusion
Substantial progress is being made in understanding how FGF signalling may impact breast cancer pathogenesis and progression, but we are only at the beginning of understanding how, and in which cancers, FGF signalling might be targeted for therapeutic benefit. Should FGFR inhibitors be developed in combination with conventional therapies? How does FGFR signalling effect respond to chemotherapy? With everolimus heading towards licensing in metastatic breast cancer [70], how will mammalian target of rapamycin inhibition impact on FGFR signalling? We look forward to further scientific and clinical research to clarify the potential role of FGFR targeting in breast cancer treatment.
Abbreviations
FGF: fibroblast growth factor; FGFR: fibroblast growth factor receptor; EGFR: epidermal growth factor receptor; ER: oestrogen receptor; HER2: human epidermal growth factor receptor 2; SNP: single nucleotide polymorphism.
Competing interests
NCT has consulted for and received honoraria from Novartis, and has received research funding from AstraZeneca. VKJ declares that he has no competing interests.
Contributor Information
Vikram K Jain, Email: vikram.jain@rmh.nhs.uk.
Nicholas C Turner, Email: nicholas.turner@icr.ac.uk.
Acknowledgements
NCT is funded by a Cancer Research UK Clinician Scientist fellowship. The authors acknowledge National Health Service funding from the National Institute for Health Research Biomedical Research Centre.
References
- Osborne CK, Neven P, Dirix LY, Mackey JR, Robert J, Underhill C, Schiff R, Gutierrez C, Migliaccio I, Anagnostou VK, Rimm DL, Magill P, Sellers M. Gefitinib or placebo in combination with tamoxifen in patients with hormone receptor-positive metastatic breast cancer: a randomized phase II study. Clin Cancer Res. 2011;17:1147–1159. doi: 10.1158/1078-0432.CCR-10-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cosimo S, Bendell JC, Cervantes-Ruiperez A, Roda D, Prudkin L, Stein MN, Leighton-Swayze A, Song Y, Ebbinghaus S, Baselga J. A phase I study of the oral mTOR inhibitor ridaforolimus (RIDA) in combination with the IGF-1R antibody dalotozumab (DALO) in patients (pts) with advanced solid tumors [abstract 3008] J Clin Oncol. 2010;28(Suppl):15s.. [Google Scholar]
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M. Synergistic induction of mesoderm by FGF and TGF-beta and the identification of an mRNA coding for FGF in the early Xenopus embryo. Cell. 1987;51:869–877. doi: 10.1016/0092-8674(87)90110-3. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DE, Williams LT. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D. et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JGt, Cheuk AT, Tsang PS, Chung JY, Song YK, Desai K, Yu Y, Chen QR, Shah K, Youngblood V, Fang J, Kim SY, Yeung C, Helman LJ, Mendoza A, Ngo V, Staudt LM, Wei JS, Khanna C, Catchpoole D, Qualman SJ, Hewitt SM, Merlino G, Chanock SJ, Khan J. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119:3395–3407. doi: 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, Ellis IO, Reis-Filho JS. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9:R23. doi: 10.1186/bcr1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A, Gillett C, Grigoriadis A, Tutt A, Reis-Filho JS, Ashworth A. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, Ullrich RT, Menon R, Maier S, Soltermann A, Moch H, Wagener P, Fischer F, Heynck S, Koker M, Schottle J, Leenders F, Gabler F, Dabow I, Querings S, Heukamp LC, Balke-Want H, Ansen S, Rauh D, Baessmann I, Altmuller J, Wainer Z, Conron M, Wright G, Russell P. et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleford GM, MacArthur CA, Kwan HC, Varmus HE. Mouse mammary tumor virus infection accelerates mammary carcinogenesis in Wnt-1 transgenic mice by insertional activation of int-2/Fgf-3 and hst/Fgf-4. Proc Natl Acad Sci USA. 1993;90:740–744. doi: 10.1073/pnas.90.2.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou V, Kimm MA, Boer M, Wessels L, Theelen W, Jonkers J, Hilkens J. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet. 2007;39:759–769. doi: 10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- Muller WJ, Lee FS, Dickson C, Peters G, Pattengale P, Leder P. The int-2 gene product acts as an epithelial growth factor in transgenic mice. EMBO J. 1990;9:907–913. doi: 10.1002/j.1460-2075.1990.tb08188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welm BE, Freeman KW, Chen M, Contreras A, Spencer DM, Rosen JM. Inducible dimerization of FGFR1: development of a mouse model to analyze progressive transformation of the mammary gland. J Cell Biol. 2002;157:703–714. doi: 10.1083/jcb.200107119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF Jr, Hoover RN, Thomas G, Chanock SJ. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, Bojesen SE, Nordestgaard BG, Axelsson CK, Arias JI, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Zamora P, Brauch H, Justenhoven C, Hamann U, Ko YD, Bruening T, Haas S, Dork T, Schurmann P, Hillemanns P, Bogdanova N, Bremer M, Karstens JH, Fagerholm R, Aaltonen K, Aittomaki K. et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KB, Maia AT, O'Reilly M, Teschendorff AE, Chin SF, Caldas C, Ponder BA. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008;6:e108. doi: 10.1371/journal.pbio.0060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijts PE, van Dongen M, de Goeij MC, van Moolenbroek AJ, Blanken F, Vreeswijk MP, de Kruijf EM, Mesker WE, van Zwet EW, Tollenaar RA, Smit VT, van Asperen CJ, Devilee P. Allele-specific regulation of FGFR2 expression is cell type-dependent and may increase breast cancer risk through a paracrine stimulus involving FGF10. Breast Cancer Res. 2011;13:R72. doi: 10.1186/bcr2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Bresnick J, Rosewell I, Crafton T, Poulsom R, Stamp G, Dickson C. Fibroblast growth-factor receptor signaling has a role in lobuloalveolar development of the mammary-gland. J Cell Sci. 1997;110:1261–1268. doi: 10.1242/jcs.110.11.1261. [DOI] [PubMed] [Google Scholar]
- Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, Knyazeva T, Muller S, Gartner S, Sures I, Wang H, Imyanitov E, Haring HU, Knayzev P, Iacobelli S, Hofler H, Ullrich A. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 2002;62:840–847. [PubMed] [Google Scholar]
- Spinola M, Leoni V, Pignatiello C, Conti B, Ravagnani F, Pastorino U, Dragani TA. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J Clin Oncol. 2005;23:7307–7311. doi: 10.1200/JCO.2005.17.350. [DOI] [PubMed] [Google Scholar]
- Spinola M, Leoni VP, Tanuma J, Pettinicchio A, Frattini M, Signoroni S, Agresti R, Giovanazzi R, Pilotti S, Bertario L, Ravagnani F, Dragani TA. FGFR4 Gly388Arg polymorphism and prognosis of breast and colorectal cancer. Oncol Rep. 2005;14:415–419. [PubMed] [Google Scholar]
- Thussbas C, Nahrig J, Streit S, Bange J, Kriner M, Kates R, Ulm K, Kiechle M, Hoefler H, Ullrich A, Harbeck N. FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J Clin Oncol. 2006;24:3747–3755. doi: 10.1200/JCO.2005.04.8587. [DOI] [PubMed] [Google Scholar]
- Sugiyama N, Varjosalo M, Meller P, Lohi J, Hyytiainen M, Kilpinen S, Kallioniemi O, Ingvarsen S, Engelholm LH, Taipale J, Alitalo K, Keski-Oja J, Lehti K. Fibroblast growth factor receptor 4 regulates tumor invasion by coupling fibroblast growth factor signaling to extracellular matrix degradation. Cancer Res. 2010;70:7851–7861. doi: 10.1158/0008-5472.CAN-10-1223. [DOI] [PubMed] [Google Scholar]
- Wang J, Yu W, Cai Y, Ren C, Ittmann MM. Altered fibroblast growth factor receptor 4 stability promotes prostate cancer progression. Neoplasia. 2008;10:847–856. doi: 10.1593/neo.08450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Varjosalo M, Meller P, Lohi J, Chan KM, Zhou Z, Alitalo K, Taipale J, Keski-Oja J, Lehti K. FGF receptor-4 (FGFR4) polymorphism acts as an activity switch of a membrane type 1 matrix metalloproteinase-FGFR4 complex. Proc Natl Acad Sci USA. 2010;107:15786–15791. doi: 10.1073/pnas.0914459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitzer N, Mayr T, Streit S, Ullrich A. A single nucleotide change in the mouse genome accelerates breast cancer progression. Cancer Res. 2010;70:802–812. doi: 10.1158/0008-5472.CAN-09-3239. [DOI] [PubMed] [Google Scholar]
- Marme F, Werft W, Benner A, Burwinkel B, Sinn P, Sohn C, Lichter P, Hahn M, Schneeweiss A. FGFR4 Arg388 genotype is associated with pathological complete response to neoadjuvant chemotherapy for primary breast cancer. Ann Oncol. 2010;21:1636–1642. doi: 10.1093/annonc/mdq017. [DOI] [PubMed] [Google Scholar]
- Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, Geyer FC, van Kouwenhove M, Kreike B, Mackay A, Ashworth A, van de Vijver MJ, Reis-Filho JS. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–2023. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannheimer SL, Rehemtulla A, Ethier SP. Characterization of fibroblast growth factor receptor 2 overexpression in the human breast cancer cell line SUM-52PE. Breast Cancer Res. 2000;2:311–320. doi: 10.1186/bcr73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai A, Meetze K, Vo NY, Kollipara S, Mazsa EK, Winston WM, Weiler S, Poling LL, Chen T, Ismail NS, Jiang J, Lerner L, Gyuris J, Weng Z. GP369, an FGFR2-IIIb-specific antibody, exhibits potent antitumor activity against human cancers driven by activated FGFR2 signaling. Cancer Res. 2010;70:7630–7639. doi: 10.1158/0008-5472.CAN-10-1489. [DOI] [PubMed] [Google Scholar]
- Cha JY, Maddileti S, Mitin N, Harden TK, Der CJ. Aberrant receptor internalization and enhanced FRS2-dependent signaling contribute to the transforming activity of the fibroblast growth factor receptor 2 IIIb C3 isoform. J Biol Chem. 2009;284:6227–6240. doi: 10.1074/jbc.M803998200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiskanen M, Kononen J, Barlund M, Torhorst J, Sauter G, Kallioniemi A, Kallioniemi O. CGH, cDNA and tissue microarray analyses implicate FGFR2 amplification in a small subset of breast tumors. Anal Cell Pathol. 2001;22:229–234. doi: 10.1155/2001/981218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunii K, Davis L, Gorenstein J, Hatch H, Yashiro M, Di Bacco A, Elbi C, Lutterbach B. FGFR2-amplified gastric cancer cell lines require FGFR2 and Erbb3 signaling for growth and survival. Cancer Res. 2008;68:2340–2348. doi: 10.1158/0008-5472.CAN-07-5229. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, McHenry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D. et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, Rodriguez C, Theillet C. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 1997;57:4360–4367. [PubMed] [Google Scholar]
- Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rouge C, Lasorsa L, Letessier A, Ginestier C, Monville F, Esteyries S, Adelaide J, Esterni B, Henry C, Ethier SP, Bibeau F, Mozziconacci MJ, Charafe-Jauffret E, Jacquemier J, Bertucci F, Birnbaum D, Theillet C, Chaffanet M. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res. 2005;3:655–667. doi: 10.1158/1541-7786.MCR-05-0128. [DOI] [PubMed] [Google Scholar]
- Letessier A, Sircoulomb F, Ginestier C, Cervera N, Monville F, Gelsi-Boyer V, Esterni B, Geneix J, Finetti P, Zemmour C, Viens P, Charafe-Jauffret E, Jacquemier J, Birnbaum D, Chaffanet M. Frequency, prognostic impact, and subtype association of 8p12, 8q24, 11q13, 12p13, 17q12, and 20q13 amplifications in breast cancers. BMC Cancer. 2006;6:245. doi: 10.1186/1471-2407-6-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, Grigoriadis A, Sarrio D, Savage K, Dexter T, Iravani M, Fenwick K, Weber B, Hardisson D, Schmitt FC, Palacios J, Lakhani SR, Ashworth A. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–6662. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- Jacquemier J, Adelaide J, Parc P, Penault-Llorca F, Planche J, deLapeyriere O, Birnbaum D. Expression of the FGFR1 gene in human breast-carcinoma cells. Int J Cancer. 1994;59:373–378. doi: 10.1002/ijc.2910590314. [DOI] [PubMed] [Google Scholar]
- Holland DG, Burleigh A, Git A, Goldgraben MA, Perez-Mancera PA, Chin SF, Hurtado A, Bruna A, Ali HR, Greenwood W, Dunning MJ, Samarajiwa S, Menon S, Rueda OM, Lynch AG, McKinney S, Ellis IO, Eaves CJ, Carroll JS, Curtis C, Aparicio S, Caldas C. ZNF703 is a common luminal B breast cancer oncogene that differentially regulates luminal and basal progenitors in human mammary epithelium. EMBO Mol Med. 2011;3:167–180. doi: 10.1002/emmm.201100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sircoulomb F, Nicolas N, Ferrari A, Finetti P, Bekhouche I, Rousselet E, Lonigro A, Adelaide J, Baudelet E, Esteyries S, Wicinski J, Audebert S, Charafe-Jauffret E, Jacquemier J, Lopez M, Borg JP, Sotiriou C, Popovici C, Bertucci F, Birnbaum D, Chaffanet M, Ginestier C. ZNF703 gene amplification at 8p12 specifies luminal B breast cancer. EMBO Mol Med. 2011;3:153–166. doi: 10.1002/emmm.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard-Pierrot I, Gruel N, Stransky N, Vincent-Salomon A, Reyal F, Raynal V, Vallot C, Pierron G, Radvanyi F, Delattre O. Characterization of the recurrent 8p11-12 amplicon identifies PPAPDC1B, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer Res. 2008;68:7165–7175. doi: 10.1158/0008-5472.CAN-08-1360. [DOI] [PubMed] [Google Scholar]
- Sharpe R, Pearson A, Herrera-Abreu MT, Johnson D, Mackay A, Welti JC, Natrajan R, Reynolds AR, Reis-Filho JS, Ashworth A, Turner NC. FGFR signaling promotes the growth of triple-negative and basal-like breast cancer cell lines both in vitro and in vivo. Clin Cancer Res. 2011;17:5275–5286. doi: 10.1158/1078-0432.CCR-10-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian W, Pappas L, Pandya D, Selfors LM, Derksen PW, de Bruin M, Gray NS, Jonkers J, Rosen JM, Brugge JS. Fibroblast growth factor receptor 1-transformed mammary epithelial cells are dependent on RSK activity for growth and survival. Cancer Res. 2009;69:2244–2251. doi: 10.1158/0008-5472.CAN-08-3398. [DOI] [PubMed] [Google Scholar]
- Pond AC, Herschkowitz JI, Schwertfeger KL, Welm B, Zhang Y, York B, Cardiff RD, Hilsenbeck S, Perou CM, Creighton CJ, Lloyd RE, Rosen JM. Fibroblast growth factor receptor signaling dramatically accelerates tumorigenesis and enhances oncoprotein translation in the mouse mammary tumor virus-Wnt-1 mouse model of breast cancer. Cancer Res. 2010;70:4868–4879. doi: 10.1158/0008-5472.CAN-09-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bade LK, Goldberg JE, Dehut HA, Hall MK, Schwertfeger KL. Mammary tumorigenesis induced by fibroblast growth factor receptor 1 requires activation of the epidermal growth factor receptor. J Cell Sci. 2011;124(Pt 18):3106–3117. doi: 10.1242/jcs.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Dong S, Gu TL, Guo A, Cohen MS, Lonial S, Khoury HJ, Fabbro D, Gilliland DG, Bergsagel PL, Taunton J, Polakiewicz RD, Chen J. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell. 2007;12:201–214. doi: 10.1016/j.ccr.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwek SS, Roy R, Zhou H, Climent J, Martinez-Climent JA, Fridlyand J, Albertson DG. Co-amplified genes at 8p12 and 11q13 in breast tumors cooperate with two major pathways in oncogenesis. Oncogene. 2009;28:1892–1903. doi: 10.1038/onc.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C, Nicoletti R, Winckler W, Grewal R, Hanna M, Wyhs N, Ziaugra L, Richter DJ, Trovik J, Engelsen IB, Stefansson IM, Fennell T, Cibulskis K, Zody MC, Akslen LA, Gabriel S, Wong KK, Sellers WR, Meyerson M, Greulich H. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci USA. 2008;105:8713–8717. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, Chopin D, Thiery JP, Radvanyi F. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- Relf M, LeJeune S, Scott PA, Fox S, Smith K, Leek R, Moghaddam A, Whitehouse R, Bicknell R, Harris AL. Expression of the angiogenic factors vascular endothelial cell growth factor, acidic and basic fibroblast growth factor, tumor growth factor beta-1, platelet-derived endothelial cell growth factor, placenta growth factor, and pleiotrophin in human primary breast cancer and its relation to angiogenesis. Cancer Res. 1997;57:963–969. [PubMed] [Google Scholar]
- Liu Y, Wang JL, Chang H, Barsky SH, Nguyen M. Breast-cancer diagnosis with nipple fluid bFGF. Lancet. 2000;356:567. doi: 10.1016/S0140-6736(00)02587-3. [DOI] [PubMed] [Google Scholar]
- Schweigerer L, Neufeld G, Friedman J, Abraham JA, Fiddes JC, Gospodarowicz D. Capillary endothelial cells express basic fibroblast growth factor, a mitogen that promotes their own growth. Nature. 1987;325:257–259. doi: 10.1038/325257a0. [DOI] [PubMed] [Google Scholar]
- Welti JC, Gourlaouen M, Powles T, Kudahetti SC, Wilson P, Berney DM, Reynolds AR. Fibroblast growth factor 2 regulates endothelial cell sensitivity to sunitinib. Oncogene. 2011;30:1183–1193. doi: 10.1038/onc.2010.503. [DOI] [PubMed] [Google Scholar]
- Fillmore CM, Gupta PB, Rudnick JA, Caballero S, Keller PJ, Lander ES, Kuperwasser C. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci USA. 2010;107:21737–21742. doi: 10.1073/pnas.1007863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing J, Du X, Chen Y, Chan P, Li H, Wu P, Marsters S, Stawicki S, Tien J, Totpal K, Ross S, Stinson S, Dornan D, French D, Wang QR, Stephan JP, Wu Y, Wiesmann C, Ashkenazi A. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. J Clin Invest. 2009;119:1216–1229. doi: 10.1172/JCI38017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HD, Malabunga M, Tonra JR, DiRenzo R, Carrick FE, Zheng H, Berthoud HR, McGuinness OP, Shen J, Bohlen P, Leibel RL, Kussie P. Monoclonal antibody antagonists of hypothalamic FGFR1 cause potent but reversible hypophagia and weight loss in rodents and monkeys. Am J Physiol Endocrinol Metab. 2007;292:E964–E976. doi: 10.1152/ajpendo.00089.2006. [DOI] [PubMed] [Google Scholar]
- Marshall ME, Hinz TK, Kono SA, Singleton KR, Bichon B, Ware KE, Marek L, Frederick BA, Raben D, Heasley LE. Fibroblast growth factor receptors are components of autocrine signaling networks in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2011;17:5016–5025. doi: 10.1158/1078-0432.CCR-11-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre F, Bachelot TD, Campone M, Dalenc F, Perez-Garcia JM, Hurvitz SA, Turner NC, Rugo HS, Shi MM, Zhang Y, Kay ACM, Yovine AJ, Baselga J. A multicenter, open-label phase II trial of dovitinib, an FGFR1 inhibitor, in FGFR1 amplified and non-amplified metastatic breast cancer [abstract 508] J Clin Oncol. 2011;29(Suppl) [Google Scholar]
- Tashiro E, Maruki H, Minato Y, Doki Y, Weinstein IB, Imoto M. Overexpression of cyclin D1 contributes to malignancy by up-regulation of fibroblast growth factor receptor 1 via the pRB/E2F pathway. Cancer Res. 2003;63:424–431. [PubMed] [Google Scholar]
- Soria JC, Dienstmann R, de Braud F, Cereda R, Bahleda R, Hollebecque A, Tabernero J, Delmonte A, Dromain C, Lassau N , Farace F, Zucchetti M, Marsoni S, Camboni MG. First-In-Man study of E-3810, a novel VEGFR and FGFR inhibitor, in patients with advanced solid tumors. Annals Oncol. 2012;23(Suppl 1):i15–i25. [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier K, Schwaenen C, Sticht C, Flechtenmacher C, Muhling J, Hofele C, Radlwimmer B, Lichter P, Joos S. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC) Oral Oncol. 2007;43:60–66. doi: 10.1016/j.oraloncology.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Wohrle S, Bonny O, Beluch N, Gaulis S, Stamm C, Scheibler M, Muller M, Kinzel B, Thuery A, Brueggen J, Hynes NE, Sellers WR, Hofmann F, Graus-Porta D. FGF receptors control vitamin D and phosphate homeostasis by mediating renal FGF-23 signaling and regulating FGF-23 expression in bone. J Bone Miner Res. 2011;26:2486–2497. doi: 10.1002/jbmr.478. [DOI] [PubMed] [Google Scholar]
- Baselga J, Campone M, Piccart M, Burris HA, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, Beck JT, Ito Y, Yardley D, Deleu I, Perez A, Bachelot T, Vittori L, Xu Z, Mukhopadhyay P, Lebwohl D, Hortobagyi GN. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratain MJ, Schwartz GK, Oza AM, Rudin CM, Kaye SB, De Jonge MJ, Khayat D, Awada A, Sawyer MB, Obel JC, Medioni J, Evans TRJ, De Greve J, Soetekouw PM, Baurain J, O'Dwyer PJ, Hartman C, Poulart V, Walters IB. Brivanib (BMS-582664) in advanced solid tumors (AST): results of a phase II randomized discontinuation trial (RDT) [abstract 3079] J Clin Oncol. 2011;29(Suppl) [Google Scholar]
- Bouché O, Ducreux M, Lledo G, André T, Maindrault-Goebel F, Stopfer P, Oum'Hamed Z, Chadjaa M, de Gramont A. A phase II trial of weekly alternating sequential administration of BIBF1120 and BIBW2992 in patients with advanced colorectal cancer [abstract 15001] J Clin Oncol. 2008;26(Suppl) [Google Scholar]
- Talpaz M, Shah NP, Deininger MW, Mauro MJ, Flinn IW, Lustgarten S, Lindmark W, Gozgit JM, Clackson T, Turner CD, Haluska FG, Kantarjian H, Cortes JE. Ponatinib in patients with acute myeloid leukemia (AML): preliminary findings from a phase I study in hematologic malignancies [abstract 6518] J Clin Oncol. 2011;29(Suppl) [Google Scholar]
- An Open Label Phase I Dose Escalation Study of E7080 Administered to Patients with Solid Tumors. http://clinicaltrials.gov/ct2/show/NCT00280397?term=e7080&rank=4
- Study of Oral E-3810, a Dual VEGFR-FGFR Tyrosine Kinase Inhibitor, in Patients with Solid Tumors. http://clinicaltrials.gov/ct2/show/NCT01283945?term=e3810&rank=5
- Xu J, Wang Y, Ye CY, Huang C, Zhang XH, Sai Y, Shen L, MU H. Phase I study on safety and pharmacokinetics of sulfatinib, a selective VEGFR/FGFR dual inhibitor, in Chinese patients with advanced solid tumors [abstract e13558] J Clin Oncol. 2011;29(Suppl) [Google Scholar]
- Xie L, Su X, Zhang D, Tang L, Xu J, Wang M, Yin L, Zhang J, Ye K, Wang Z, Kilgour E, Qunsheng J. Proceedings of the 102nd Annual Meeting of the American Association for Cancer Research; 2-6 April 2011; Orlando, FL. Philadelphia, PA: AACR; 2011. AZD4547, a potent and selective inhibitor of FGF-receptor tyrosine kinases 1, 2 and 3, inhibits the growth of FGF-receptor 2 driven gastric cancer models in vitro and in vivo [abstract 1643] [Google Scholar]
- A Randomised Open-Label Phase IIa Study to Assess the Efficacy and Safety of AZD4547 Monotherapy versus Paclitaxel in Patients with Advanced Gastric or Gastro-oesophageal Junction Cancer with FGFR2 Polysomy or Gene Amplification (SHINE Study) http://clinicaltrials.gov/ct2/show/NCT01457846?term=AZD4547&rank = 2
- A Phase I, Open-label, Multi-center, Dose Escalation Study of Oral BGJ398, a Pan FGF-R Kinase Inhibitor, in Adult Patients with Advanced Solid Malignancies. http://clinicaltrials.gov/ct2/show/NCT01004224
- Trudel S, Stewart AK, Rom E, Wei E, Li ZH, Kotzer S, Chumakov I, Singer Y, Chang H, Liang SB, Yayon A. The inhibitory anti-FGFR3 antibody, PRO-001, is cytotoxic to t(4;14) multiple myeloma cells. Blood. 2006;107:4039–4046. doi: 10.1182/blood-2005-10-4179. [DOI] [PubMed] [Google Scholar]
- A Phase 1, Open-label, Dose-finding Study Evaluating the Safety and Pharmacokinetics of FP-1039 in Subjects with Metastatic or Locally Advanced Unresectable Solid Tumors. http://clinicaltrials.gov/ct2/show/NCT00687505?term=fp-1039&rank=2