Abstract
We describe the synthesis and properties of new fluorescence quenchers containing aldehyde, hydrazine and aminooxy groups, allowing convenient bioconjugation as oximes or hydrazones. Conjugation to oligonucleotides proceeded in high yield with aniline as catalyst. Kinetics studies of conjugation show that, under optimal conditions, a hydrazine or aminooxy quencher can react with aldehyde-modified DNA to form a stable hydrazone or oxime adduct in as little as five minutes. The resulting quencher-containing DNAs were assessed for their ability to quench the emission of fluorescein in labeled complements and compared to the commercially available dabcyl and Black Hole Quencher 2 (BHQ2), which were conjugated as phosphoramidites. Results show that the new quenchers possess slightly different absorbance properties compared to dabcyl and are as efficient as the commercial quenchers in quenching fluorescein emission. Hydrazone-based quenchers were further successfully incorporated into molecular beacons and shown to give high signal:background in single nucleotide polymorphism detection in vitro . Finally, aminooxy and hydrazine quenchers were applied to quenching of an aldehyde-containing fluorophore associated with living cells, demonstrating cellular quenching within one hour.
Introduction
Fluorescence quenchers have garnered a wide variety of uses in assays for biomolecular detection and analysis.1–11 Potent fluorescence quenching is often attained by placing the quencher molecule in close spatial proximity to the fluorophore to allow for contact quenching, where the fluorophore undergoes direct electronic interactions with the quencher moiety.11–16 The resulting emission can be greatly reduced compared to the “on” state of the fluorophore separated from the quencher, which is important for lowering background signals. Many different fluorescence quenchers have been designed, including the widely used and commercially available Dabcyl (4-((4-(dimethylamino)phenyl)azo)benzoic acid), Black Hole Quenchers (BHQ)17, the QSY series18 and BlackBerry Quenchers.19 Other quenchers with broadened absorbance properties have been designed, based on such scaffolds as azaphthalocyanine,20,21 azulene,22 cyanine,8 or napthalimide.23
Despite their broad use, however, these quenchers are limited by their chemistry of conjugation. Chemical attachment is nearly always done by one of two irreversible approaches: as a phosphodiester linkage via automated DNA synthesis, or as an amide link via reaction of carboxylate and amine. Development of new bioorthogonal means of attaching quencher molecules could allow for increased ease and speed of conjugation, could enable conjugation of complex molecules with multiple functional groups, and could provide greater freedom for design of multiplex assays.
Hydrazone and oxime chemistry has been advanced in recent years as a bioorthogonal means of attaching a variety of labels to molecules of interest, even in cellular conditions. Although the uncatalyzed condensations possess slow reaction kinetics, the development of aniline catalysis to greatly enhance reaction rate has allowed for a variety of useful applications.24–26 Surprisingly, although these bond formations have been used to attach fluorophores to many molecules of interest,25,27 we are aware of no reports of the use of oxime or hydrazone chemistry to attach a quencher to a fluorescently labeled molecule. Such an approach may be desirable for a variety of reasons. First, the mild chemistry allows conjugations in molecules that may have otherwise sensitively reactive groups. Second, this synthetic approach can allow for temporal control of when a quencher will be introduced into a system, yielding quenching after some fluorescence analysis has taken place. Further, if the reactive groups are placed in direct conjugation, formation of either on oxime or hydrazone can result in direct electronic changes of the quencher itself (such as redshifting), which has been observed with hydrazone formation involving dyes.28,29 Finally, since hydrazone/oxime chemistry can proceed intracellularly,27,29 this offers the possibility of performing quenching experiments directly in cells.
Here we report the preparation and properties of multiple new hydrazine, aminooxy, and aldehyde-containing fluorescence quenchers (Figure 1) that can easily be conjugated to aldehyde, aminooxy or hydrazide modified molecules (such as DNA) (Figure 2) via the stable formation of oximes or hydrazones. The quenching efficiencies of these molecules are comparable to or slightly better than a commercially available dabcyl quencher, but the new compounds offer a convenient bioorthogonal means of attaching the quencher moiety. We find that with aniline catalysis a quencher can form a stable DNA hydrazone adduct in high yield in as little as five minutes. Finally, we present preliminary experiments showing evidence of fluorescence quenching within living cells by cell-permeable quencher groups.
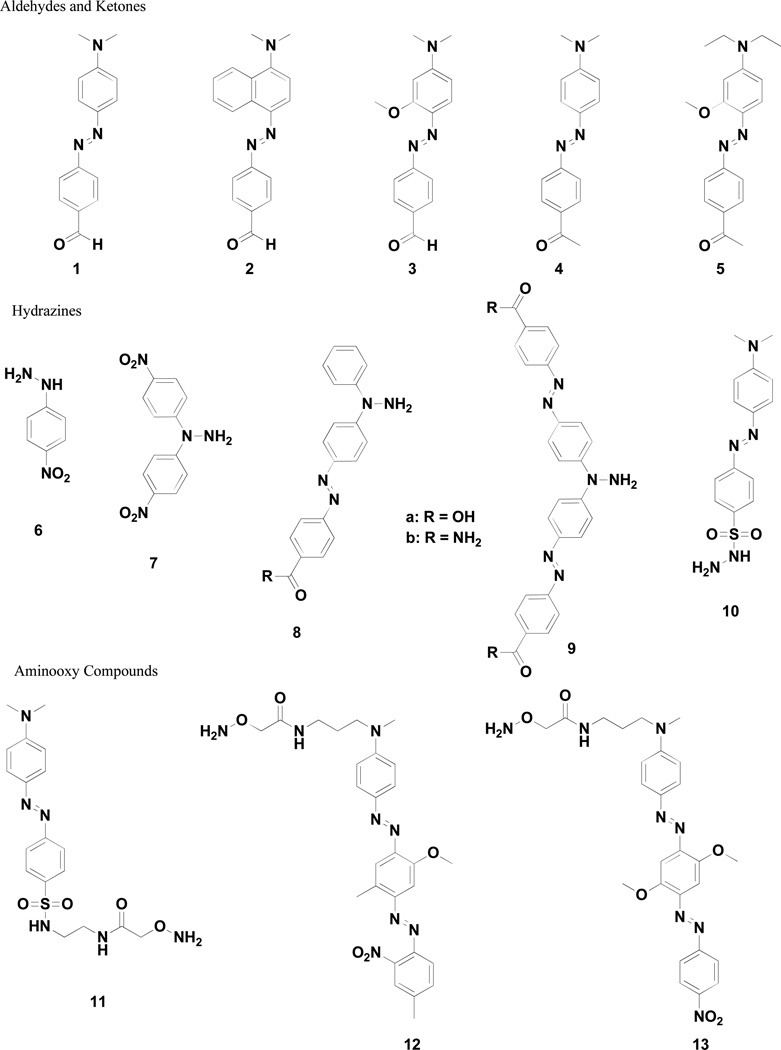
Figure 1.
Aldehyde-, hydrazine- and aminooxy- modified quenchers prepared for this study
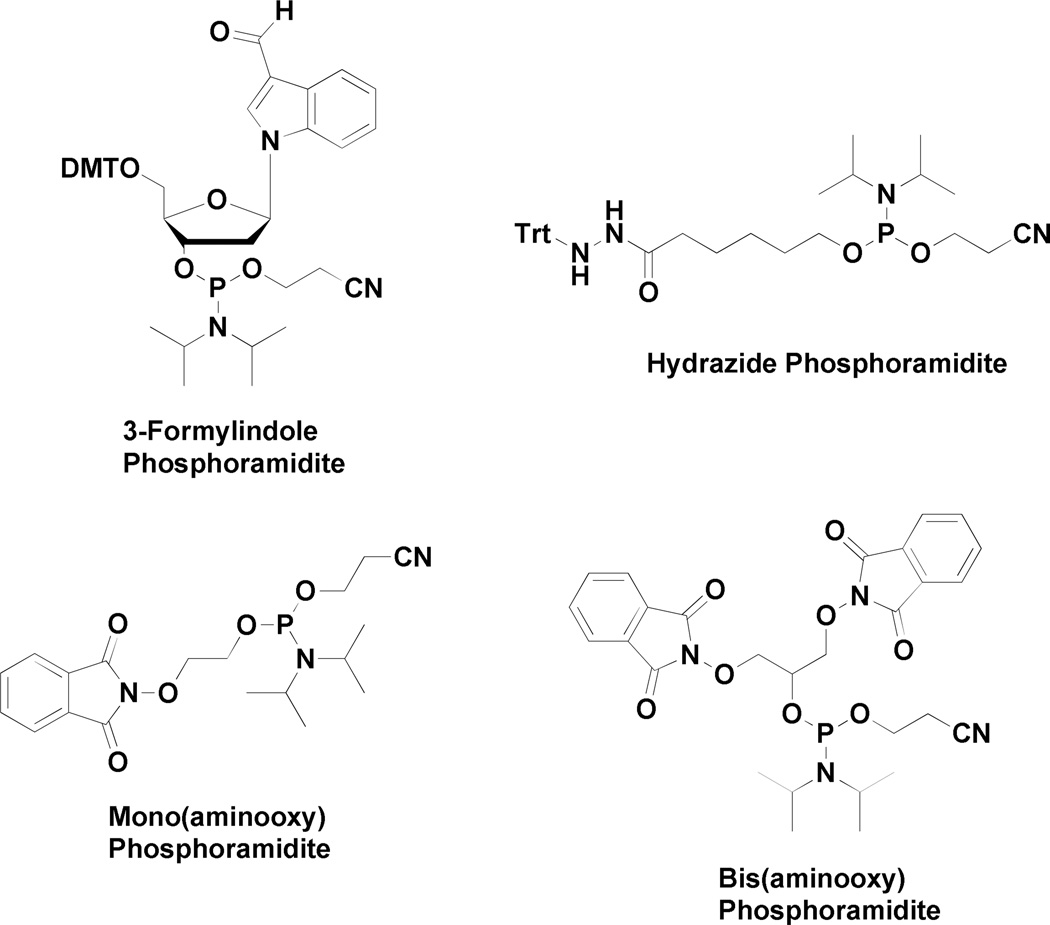
Figure 2.
Aldehyde, hydrazide and aminooxy phosphoramidites employed for oxime and hydrazone formation on DNA.
Experimental
Chemicals and Reagents
Chemicals were purchased from Sigma-Aldrich or TCI America and used without further purification. Solvents were purchased from Acros Organics and used as received. DNA phosphoramidites and solid supports were purchased from Glen Research. 5’-Dabcyl phosphoramidite, 3-formylindole phosphoramidite and 3’-Fluorescein CPG were purchased from Glen Research. 5’-BHQ2 phosphoramidite was purchased from Biosearch Technologies.
Instrumentation
1H- and 13C-NMR spectra were recorded on a Varian 500 MHz NMR spectrometer and internally referenced to the residual solvent signal; J values are reported in Hz. Oligonucleotides sequences were synthesized on an ABI 394 DNA synthesizer using standard reagents and phosphoramidites. Analytical and semi-preparative high performance liquid chromatography was performed on a LC-CAD Shimadzu liquid chromatograph, equipped with a SPD-M10A VD diode array detector and a SCL 10A VP system controller using reverse phase C18 columns (Grace ProSphere C18-300 10µ for semi-preparative scale HPLC, Alltech Platinum C18-100A 5µ for analytical HPLC). DNA concentrations were determined on a Cary 100 Bio UV-Visible Spectrophotometer at 90 °C by measuring absorbance at 260 nm. Fluorescence measurements were performed on a Fluorolog 3 Jobin Yvon fluorescence spectrometer. ESI MS were determined by the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry. Oligonucleotide masses were determined by the Stanford University Protein and Nucleic Acid Facility using a Perspective Voyager-DE RP Biospectrometry MALDI-TOF mass-spectrometry instrument using a 3-hydroxypicolinic acid/diammonium hydrogen citrate matrix.
Synthetic Procedures
Compounds 1 and 10 were prepared according to published procedures.30,31 Compound 6 is commercially available. Compounds 2 – 5 were prepared analogously to 1, substituting the appropriate aniline for N-N-dimethylaniline in 2, 3, and 5 or using 4-aminoacetophenone in place of 4-aminobenzaldehyde for 4 and 5. Representative reaction conditions and purification procedures are provided below. Hydrazide and aminooxy phosphoramidites were prepared according to published procedures.32–34
Representative conditions for synthesis of 2–5
0.454 g 4-aminobenzaldehyde or 0.507 g 4’-aminoacetophenone (3.75 mmol) was dissolved in 12 mL 1 M HCl and stirred at 0 °C. Separately, 0.272 g sodium nitrite (3.94 mmol, 1.05 eq) was dissolved in 2 mL water and added dropwise. The resulting solution was stirred at 0 °C for 30 minutes. A solution of the corresponding aniline (3.94 mmol, 1.05 eq) dissolved in 9 mL 1 M HCl was added followed by a solution of 2.15 g sodium acetate in 15 mL of water. The resulting red-orange suspension was stirred and allowed to warm to room temperature over a period of 1 hour. The suspension was filtered, washed with water, and purified as described below to provide the desired aldehyde or ketone dye.
4-((4-(dimethylamino)naphthalen-1-yl)diazenyl)benzaldehyde (2)
purified by column chromatography over silica gel eluting with chloroform to provide a red solid.
1H NMR (500 MHz, CDCl3): 3.05 (s, 6H), 7.07 (d, 1H, J = 8 Hz), 7.57 (dd, 1H, J = 7Hz, 1.5 Hz), 7.65 (dd, 1H, J = 7Hz, 1.5 Hz), 7.94 (d, 1H, J = 8.5 Hz), 8.02 – 8.04 (m, 2H), 8.08 – 8.11 (m, 2H), 8.22 (d, 1H, J = 8 Hz), 9.02 (d, 1H, J = 9 Hz), 10.09 (s, 1H)
13C NMR (125 MHz, CDCl3): 44.9, 113.0, 113.2, 123.3, 123.8, 124.8, 125.5, 127.2, 128.0, 130.8, 133.6, 136.8, 142.7, 155.6, 156.8, 191.8
ESI MS (Calc M+H = 304.14): 304.33
4-((4-(dimethylamino)-2-methoxyphenyl)diazenyl)benzaldehyde (3)
purified by column chromatography over silica gel eluting with dichloromethane to provide a dark red solid.
1H NMR (500 MHz, CDCl3): 3.13 (s, 6H), 4.04 (s, 3H), 6.19 (d, 1H, J = 2 Hz), 6.35 (dd, 1H, J = 9.5 Hz, 2 Hz), 7.84 (d, 1H, J = 9.5 Hz), 7.91 – 7.96 (m, 4H), 10.03 (s, 1H)
13C NMR (125 MHz, CDCl3): 40.4, 56.2, 94.3, 105.1, 118.4, 122.7, 130.8, 134.0, 135.8, 155.0, 157.6, 160.2, 191.9
ESI MS (Calc M+H = 284.14): 284.33
1-(4-((4-(dimethylamino)phenyl)diazenyl)phenyl)ethanone (4)
recrystallized from ethanol to provide a red solid.
1H NMR (500 MHz, CDCl3): 2.64 (s, 3H), 3.10 (s, 6H), 6.75 (d, 2H, J = 9 Hz), 7.87 – 7.91 (m, 4H), 8.06 (d, 2H, J = 8.5 Hz)
13C NMR (125 MHz, CDCl3): 26.9, 40.3, 111.5, 122.2, 125.6, 129.4, 136.9, 143.7, 153.0, 156.0, 197.7
ESI MS (Calc M+H = 268.14): 268.33
1-(4-((4-(diethylamino)-2-methoxyphenyl)diazenyl)phenyl)ethanone (5)
recrystallized from ethanol to provide a dark red solid.
1H NMR (500 MHz, CDCl3): 1.24 (t, 6H, J = 7 Hz), 2.62 (s, 3H), 3.45 (q, 4H, J = 7Hz), 4.02 (s, 3H), 6.18 (d, 1H, J = 3 Hz), 6.31 (dd, 1H, J = 9.5 Hz, 3 Hz), 7.81 – 7.85 (m, 3H), 8.03 (d, 2H, J = 6.5 Hz)
13C NMR (125 MHz, CDCl3): 12.8, 26.8, 45.0, 56.2, 93.9, 104.9, 118.6, 122.2, 129.4, 133.5, 136.4, 152.7, 156.7, 160.3, 197.7
ESI MS (Calc M+H = 326.19): 326.33
tert-butyl 2-(4-((4-(ethoxycarbonyl)phenyl)diazenyl)phenyl)-2-phenylhydrazinecarboxylate (14)
315 mg of tert-butyl 2-(4-nitrophenyl)-2-phenylhydrazinecarboxylate35 was dissolved in 10 mL MeOH. A catalytic amount of 10% palladium on carbon was added and the solution stirred at room temperature overnight under a hydrogen atmosphere, after which TLC showed consumption of starting materials. The resulting solution was filtered through Celite and washed with methanol and the filtrate concentrated to afford the crude amino product. The crude amino product was dissolved in 7 mL toluene and treated with 256 mg ethyl 4-nitrosobenzoate36 and 282 µL acetic acid. The resulting solution was stirred at 90 °C overnight under an argon atmosphere. TLC indicated the formation of a bright orange product. Volatiles were removed in vacuo and the crude material purified by silica column chromatography eluting with 5:2 Hexanes:EtOAc to afford 440 mg of a bright orange solid (quantitative). An analytically pure sample was obtained by recrystallization from methanol.
1H NMR (500 MHz, CDCl3): 1.33 (br s, 3H), 1.42 (t, 3H, J = 7 Hz), 1.51 (br s, 6H), 4.40 (q, 2H, J = 7 Hz), 7.11 – 7.19 (m, 3H), 7.31 – 7.40 (m, 4H), 7.88 (m, 4H), 8.16 (d, 2H, J = 8.5 Hz)
13C NMR (125 MHz, CDCl3): 14.4, 28.1, 28.3, 61.2, 81.8. 115.6, 115.9, 122.3, 122.5, 123.2, 124.9, 125.5, 129.5, 130.6, 131.3, 144.6, 147.0, 149.9, 154.7, 155.5, 166.3
ESI MS (Calc M+H = 461.22): 461.33
tert-butyl 2,2-bis(4-nitrophenyl)hydrazinecarboxylate (15)
300 mg of tert-butyl 2-(4-nitrophenyl)diazenecarboxylate35 was dissolved in 10 mL methanol. To this solution, 240 mg 4-nitrophenylboronic acid (1.2 eq) and 18 mg (0.075 eq) copper acetate monohydrate were added. The resulting solution was stirred overnight at 45 °C. Volatiles were removed in vacuo and the resulting solid purified by column chromatography eluting with 3:1 hexanes:EtOAc → 2:1 hexanes:EtOAc to afford 390 mg (87%) of the desired product as a bright yellow solid.
1H NMR (500 MHz, CDCl3): 1.30 (br s, 3H), 1.51 (br s, 6H), 7.31 (d, 4H, J = 9 Hz), 8.22 (d, 4H, J = 9 Hz)
13C NMR (125 MHz, CDCl3): 28.0, 28.3, 83.3, 119.1, 125.7, 143.4, 149.5
ESI MS (Calc M+H = 375.13): 373.42
tert-butyl 2-(4-((4-(ethoxycarbonyl)phenyl)diazenyl)phenyl)-2-phenylhydrazinecarboxylate (16)
300 mg of 15 was dissolved in 10 mL MeOH. A catalytic amount of 10% palladium on carbon was added and the solution stirred at room temperature overnight under a hydrogen atmosphere (balloon), after which TLC showed consumption of starting materials. The resulting solution was filtered through Celite and washed with methanol and the filtrate concentrated to afford the crude diamino product. The crude diamino product was dissolved in 7 mL toluene and treated with 515 mg ethyl 4-nitrosobenzoate36 and 270 µL acetic acid. The resulting solution was stirred at 90 °C overnight under an argon atmosphere. TLC indicated the formation of a bright orange product. Volatiles were removed in vacuo and the crude material purified by column chromatography eluting with 3:1 hexanes:EtOAc → 2:1 hexanes:EtOAc to afford 297 mg (58%) of a bright red foam. An analytically pure sample was obtained by recrystallization from methanol.
1H NMR (500 MHz, CDCl3): 1.33 (br s, 3H), 1.43 (t, 6H, J = 7.5 Hz), 1.53 (br s, 6H), 4.41 (q, 4H, J = 7.5 Hz), 7.36 (d, 4H, J = 9 Hz), 7.91 – 7.96 (m, 8H), 8.18 (d, 4H, J = 8.5 Hz)
13C NMR (125 MHz, CDCl3): 14.4, 28.1, 28.4, 61.3, 82.4, 119.5, 122.5, 124.8, 130.6, 131.8, 148.1, 148.5, 155.3, 166.2
ESI MS (Calc M+H = 637.28): 637.42
tert-butyl 2-((2-(4-((4-(dimethylamino)phenyl)diazenyl)phenylsulfonamido)ethyl)amino)-2- oxoethoxycarbamate (17)
113 mg of Boc(aminooxy)acetic acid was dissolved in 6 mL CH2Cl2. To this solution, 125 mg EDCI hydrochloride and 88 mg HOAt were added followed by 230 µL triethylamine. 188 mg of N-(2-aminoethyl)-4-((4-(dimethylamino)phenyl)diazenyl) benzenesulfonamide37 was added and the resulting suspension stirred overnight at room temperature. Solvents were eliminated and the crude solid was purified by column chromatography eluting with 3% MeOH in CH2Cl2 to afford 116 mg (38%) of a bright orange solid.
1H NMR (500 MHz, CDCl3): 1.48 (s, 9H), 3.12 (s, 6H), 3.19 (t, 2H, J = 5 Hz), 3.40 (t, 2H, J = 5 Hz), 4.28 (s, 2H), 5.71 (br s, 1H), 6.75 (d, 2H, J = 9.5 Hz), 7.74 (s, 1H), 7.88 – 7.90 (m, 4H), 7.95 (d, 2H, J = 7 Hz), 8.25 (br s, 1H)
13C NMR (125 MHz, CDCl3): 28.2, 38.5, 40.5, 43.6, 76.5, 83.8, 111.5, 122.7, 125.8, 128.0, 139.7, 143.6, 153.1, 155.6, 158.1, 169.8
ESI MS (Calc M-H = 519.20): 519.19
tert-butyl 2-((3-(methyl(phenyl)amino)propyl)amino)-2-oxoethoxycarbamate (18)
300 mg of Boc(aminooxy)acetic acid was dissolved in 10 mL CH2Cl2. To this solution, 451 mg of EDCI hydrochloride, 0.657 mL Et3N and 0.515 mL N-methyl-N-phenylpropane-1,3-diamine were added and the resulting solution stirred overnight at room temperature. The reaction was diluted with CH2Cl2, washed twice with water and once with brine. The organic layer was dried over MgSO4, filtered and concentrated, then purified by silica column chromatography eluting with 3:2 EtOAc:hexanes to afford 230 mg (43%) of a yellow oil.
1H NMR (500 MHz, CDCl3): 1.46 (s, 9H), 1.82 – 1.88 (m, 2H), 2.94 (s, 3H), 3.34 – 3.42 (m, 4H), 4.32 (s, 2H), 6.67 – 6.70 (m, 3H), 7.20 – 7.24 (m, 2H), 7.53 (s, 1H), 8.25 (br s, 1H)
13C NMR (125 MHz, CDCl3): 26.6, 28.1, 37.1, 38.5, 50.5, 76.5, 83.4, 112.4, 116.3, 129.3, 149.2, 158.0, 168.9
ESI MS (Calc M+H = 338.21): 338.27
Boc(aminooxy) BHQ1 (19)
183 mg of 18 was dissolved in 10 mL tetrahydrofuran. 271 mg of Fast Corinth V salt was added followed by 2 mL MeOH to aid in solubility. The red-brown suspension was stirred at room temperature overnight and slowly became a dark red solution. Volatiles were removed and the crude material was purified by column chromatography eluting with 1:1 Hexanes:EtOAc → 1:3 Hexanes:EtOAc to afford 47 mg (13%) of a dark red solid.
1H NMR (500 MHz, CDCl3): 1.47 (s, 9H), 1.92 (m, 2H), 2.51 (s, 3H), 2.70 (s, 3H), 3.10 (s, 3H), 3.39 (m, 2H), 3.54 (t, 3H, J = 7.5 Hz), 4.01 (s, 3H), 4.35 (s, 2H), 6.75 (d, 2H, J = 9 Hz), 7.39 (s, 1H), 7.47 (d, 1 H, J = 6.5 Hz), 7.57 (d, 2H, J = 8 Hz), 7.66 (d, 1H, J = 8 Hz), 7.70 (s, 1H), 7.91 (d, 2H, J = 9 Hz), 8.41 (br s, 1H)
13C NMR (125 MHz, CDCl3): 16.9, 21.4, 27.0, 28.2, 36.8, 38.8, 50.3, 56.4, 76.6, 83.6, 99.3, 111.4, 119.0, 119.1, 124.4, 126.0, 133.6, 133.6, 141.6, 149.5, 151.1, 169.0
ESI MS (Calc M+H = 649.31): 649.31
Boc(aminooxy) BHQ2 (20)
230 mg of 18 was dissolved in 15 mL tetrahydrofuran. 854 mg of Fast Black K salt was added followed by 3 mL MeOH to aid in solubility. The red-brown suspension was stirred at room temperature for two hours and slowly became a dark purple solution. Volatiles were removed and the crude material was purified by column chromatography eluting with 1:1 hexanes:EtOAc → EtOAc to afford 237 mg (53%) of a dark purple solid.
1H NMR (500 MHz, CDCl3): 1.47 (s, 9H), 1.93 (m, 2H), 3.11 (s, 3H), 3.40 (m, 2H), 3.55 (t, 3H, J = 7.5 Hz), 4.05 (s, 3H), 4.09 (s, 3H), 4.34 (s, 2H), 6.76 (dd, 2H, J = 7 Hz, 2 Hz), 7.46 (s, 1H), 7.50 (s, 1H), 7.64 (s, 1H), 7.93 (dd, 2H, J = 7 Hz, 2Hz), 8.04 (dd, 2H, J = 7 Hz, 2Hz), 8.37 (dd, 2H, J = 7 Hz, 2 Hz), 8.43 (br s, 1H)
13C NMR (125 MHz, CDCl3): 27.0, 28.2, 36.8, 38.9, 50.2, 56.8, 76.6, 83.5, 99.9, 101.0, 111.5, 123.8, 124.8, 126.3, 142.1, 144.5, 146.9, 148.4, 150.9, 152.0, 153.7, 156.5, 158.2, 169.1
ESI MS (Calc M+H = 651.29): 651.25
DNA Conjugation
Removal of Protecting Groups
For aminooxy modified DNA, the phthalimide group was removed selectively with hydrazine as recommended in the literature.34 For hydrazide modified DNA, the trityl group was removed on the DNA synthesizer using alternating 10 second cycles of deprotection reagent (3 % trichloroacetic acid in dichloromethane) and dichloromethane washes for 3 minutes. Commercially available 3-formylindole utilizes no protection of the aldehyde. For compounds 7 – 9 and 11 – 13, 20 mg of boc-protected precursor 14 – 17 or 19 – 20 was dissolved in 1 mL of 1:1 dichloromethane:trifluoroacetic acid and shaken at room temperature for two hours to provide the free hydrazine or aminooxy compound for conjugation. Volatiles were removed in vacuo and the crude product was used for DNA labeling without further purification.
DNA Labeling
A solution of approximately 25 mM of desired label in 750 µL 9:1 dimethylformamide:water was added to the DNA CPG. 7.5 µL of aniline was added and the resulting solution was shaken at room temperature overnight. The CPG was rinsed 4 × 500 µL dimethylformamide and 4 × 500 µL acetonitrile. Labeled DNA was cleaved using standard deprotection conditions (concentrated aqueous ammonia, 55 °C, overnight) and purified by HPLC. Purity of the collected product was assessed by analytical HPLC.
Fluorescence Experiments
Fluorescence of the hybridization buffer (10 mM MgCl2, 70 mM Tris•Borate, pH 7.55) alone was first determined and subtracted from all subsequent fluorescence data. Fluorescein-labeled 20mer was added to the buffer solution to a final concentration of 30 nM. The fluorescence of the solution was measured in triplicate (excitation 494 nm and emission 517 nm, slits both 2 nm) and averaged, subtracting background buffer emission. A concentrated solution of quencher labeled DNA was added to bring the final volume of quencher strand to 150 nM. The oligonucleotides were allowed to hybridize until the fluorescence emission remained constant, then the fluorescence spectrum was taken in triplicate and averaged, subtracting background buffer emission. Quenching efficiency was calculated at the emission maxima by dividing the quenched fluorescence by the initial fluorescence, multiplying by 100, then subtracting from 100. All experiments were performed in triplicate and averaged.
Molecular beacon experiments were performed with the same hybridization buffer and a 100 nM concentration of molecular beacon. 200 nM of either the match, mismatch or deletion sequence were added and the change in fluorescence monitored. The beacon sequence was 5’-Quencher-(3-formylindole)-GCG-AGA-AGT-TAA-GAC-CTA-TGC-TCG-C-FAM-3’, where the underlined regions represent the stem region.4 Match sequence (5’-CAT-AGT-TCT-TAA-CTT-3’), mismatch sequence (5’-CAT-AGT-TGT-TAA-CTT-3’) and deletion sequence (5’-CAT-AGT-T_T-TAA-CTT-3’) were purified by PolyPakII cartridges (Glen Research).4
Kinetics Experiments
A concentrated solution of 4-nitrophenylhydrazine in dimethylformamide (final concentration 200 µM) was added to a buffered solution (100 mM NH4OAc, pH 4.5) containing 100 µM aldehyde modified DNA and either 0 mM or 100 mM aniline to a total volume of 200 µL. 25 µL samples were taken and immediately injected into an HPLC at 5 minutes, 80 minutes, 150 minutes, and 210 minutes. Yield was determined by dividing the integrated area of the product by the total integrated area of both the product and starting material. The yield at time = 0 was assumed to be zero.
Cellular Experiments
CCL-2 HeLa cells (ATCC, Mannasas, VA) were grown at 37 °C (5% CO2) in Dulbecco’s modified Eagles’ medium (DMEM, GIBCO) – supplemented with 10% fetal bovine serum (FBS, GIBCO), 100 Sg/mL streptomycin (GIBCO) and 100 U/mL penicillin (GIBCO) – until 60–80% confluency was reached. In preparation for in vivo experiments, cells were trypsinized, suspended in DMEM + 10% FBS, transferred to 8-chambered microscope slides (Lab-Tek II, 500 µL cell suspension containing ca. 10,000 cells per chamber), and incubated for 24 hours. The following stock solution was prepared in DMEM (without phenol red indicator, GIBCO) supplemented with 100 mM sodium pyruvate: 10 µM 7-diethylamino-3-formylcoumarin. The following four stock solutions were prepared in Hanks’ buffered saline solution (HBSS, GIBCO): 100 µM dabcyl, 100 µM dabcyl + 10 mM aniline, 100 µM 8 or 11, and 100 µM 8 or 11 + 10 mM aniline.
To each 8-chambered microscope slide, media was removed, cells were washed with phosphate buffered saline (PBS, GIBCO) solution (2×), and fluorophore-containing media was added (500 µL). Each slide contained a control chamber with DMEM (without phenol indicator) only. After 1 hour of incubation, media was removed, cells were washed with PBS (2×) buffer, and quencher-containing media was added (500 µL). The control chamber and an additional chamber in each slide contained HBSS only. Slides were incubated at 1, 2, or 3 hours. After incubation period, media was removed, cells were washed with PBS (2×) buffer, removable chamber walls were detached, and the slide was prepared for fluorescence microscopy. Fluorescence microscopy was performed on a Nikon Eclipse E800 epifluorescence microscope equipped with a Spot RT Slider camera with an excitation filter of 400 – 440 nm and emission cutoff filter of greater than 470 nm. Camera and image settings were the same for all images.
Results
Synthesis of aldehyde, hydrazine, and aminooxy quencher derivatives
Both compounds 1 and 10 have been reported in the literature, but surprisingly have not been applied as fluorescence quenchers despite the obvious structural similarity to the commonly used quencher dabcyl.30,31 Compound 10 has only sparsely been employed in detecting reducing sugars via hydrazone formation.31 Synthetically, 2 – 5 were easily prepared using procedures analogous to that for the synthesis of 1 (Scheme S1).30 Absorption properties of the free compounds in methanol are provided in Table 1.
Table 1.
Absorption properties of free aldehyde or boc protected hydrazine dyes in methanol.
| Dye | Absorbance Maxima | εmax (L mol−1 cm−1) | ε260 (L mol−1 cm−1) |
|---|---|---|---|
| 1 | 449 nm | 29,300 | 9,000 |
| 2 | 451 nm | 22,300 | 12,400 |
| 3 | 464 nm | 42,100 | 11,300 |
| 4 | 443 nm | 40,800 | 12,200 |
| 5 | 476 nm | 41,400 | 9,000 |
| 10 | 439 nm | 40,200 | 14,000 |
| 14 | 412 nm | 35,900 | 19,000 |
| 15 | 383 nm | 28,800 | 7,300 |
| 17 | 437 nm | 55,600 | 24,500 |
Preparation of compounds 7 – 9 required a different approach (Scheme S2), particularly in the case of compounds 8 and 9 as no compounds containing both an aryl hydrazine and diazo bonds were found in a literature survey. However, the reported ability to cross-couple aryl boronic acids to asymmetric Boc-protected diazo compounds afforded a simple preparation of nitroarenes.35 While the nitroarene precursor for preparation of 14 is known, preparation of dinitro compound 15 was realized by the analogous reaction of tert-butyl 2-(4-nitrophenyl)diazenecarboxylate with 4-nitrophenylboronic acid in the presence of catalytic copper (II).35 Reduction of the nitro group followed by reaction with ethyl 4-nitrosobenzoate36 in the presence of acetic acid easily afforded the desired azo containing hydrazine precursors 14 and 16. Absorption properties of the Boc-protected dyes in methanol can be found in Table 1.
Preparation of quenchers 11 – 13 required a different approach, particularly as no dye containing an aminooxy group attached directly to the aryl ring could be prepared. Consequently, Boc(aminooxy)acetic acid was attached via simple linkers to dabcyl and Black Hole Quencher 1 and 2 scaffolds, as shown in Scheme S3. As no changes in the chromophore were made by this synthetic approach, the photophysical properties of these molecules were assumed to be the same as the parent dyes.17
Based upon literature reports regarding possible problems with the stability of free hydrazines and, to a lesser extent, free aminooxy compounds,25,38,39 we chose to store 7 – 9 and 11 – 13 as the Boc protected forms. The Boc group provides for a bench stable derivative and is easily cleaved with trifluoroacetic acid in dichloromethane to provide the free hydrazine or aminooxy compound for subsequent conjugation. MALDI-TOF data and spectral properties are consistent with the proper deprotection and labeling of DNA as the desired hydrazones or oximes.
Labeling of DNA
Conjugation experiments were performed by reaction of the dye with the modified DNA attached to the solid support in a DMF/water mixture in the presence of aniline at room temperature. Subsequent deprotection of the DNA was performed under standard aqueous ammonia conditions at 55 °C. Attachment of dyes to DNA via oxime formation (by reaction of aldehyde dyes with aminooxy DNA) was, qualitatively, higher yielding and provided more stable dyes than formation of hydrazone linkages, consistent with the greater stability of oximes versus hydrazones.40 However, use of diphenylhydrazine derivatives 7 – 9 provided a higher yield of labeled DNA when compared to simple labeling with dabsyl hydrazide 10, indicating that the diphenylhydrazine derivatives may be preferable to dabsyl hydrazide in terms of higher yield and stability. Oxime labeled DNA was found to be stable for at least six months after storage at −80 °C, while dabsyl hydrazone labeled DNA did display a small degree of degradation and might require repurification after prolonged storage. Both hydrazones and oximes were stable at room temperature in biologically relevant pHs utilized for hybridization experiments and at pH 4.5 used for kinetics experiments.
Experiments showed that aldehyde derivatives of the quencher dyes provided much higher yields of the desired labeled DNA than did their methyl ketone counterparts. For example, attempted hydrazone formation with dyes 4 and 5 by reaction with hydrazide modified DNA showed no observable product by HPLC while only a low yield was obtained when forming an oxime with these dyes. Similarly, no bis(oximes) were obtained with the methyl ketone dyes. In contrast, formation of hydrazones or oximes with aldehyde dyes provided the desired DNA adduct as the major product (save for compound 2, which did not form a stable hydrazone) as confirmed by the spectral properties and MALDI-TOF results. This is consistent with literature reports of the equilibrium constant for benzaldehyde-oxime and hydrazone formation being orders of magnitude higher than formation of the corresponding acetophenone oximes or hydrazones.41
Dyes 8 and 9 containing the ethyl ester protecting group provided a complication in labeling DNA, as the deprotection conditions employed (concentrated aqueous ammonia) resulted in two different products, one being the free carboxylic acid (8a, 9a) and the other a primary amide (8b, 9b). It is likely that using different deprotection conditions (e.g. 0.05 M K2CO3 in MeOH) would provide only the carboxylic acid product. However, as discussed below, the absorbance and quenching properties of the dyes are sufficiently similar that the differences in the two products are relatively unimportant.
Absorption properties of labeled DNAs
Hydrazones and oximes of aldehyde quenchers
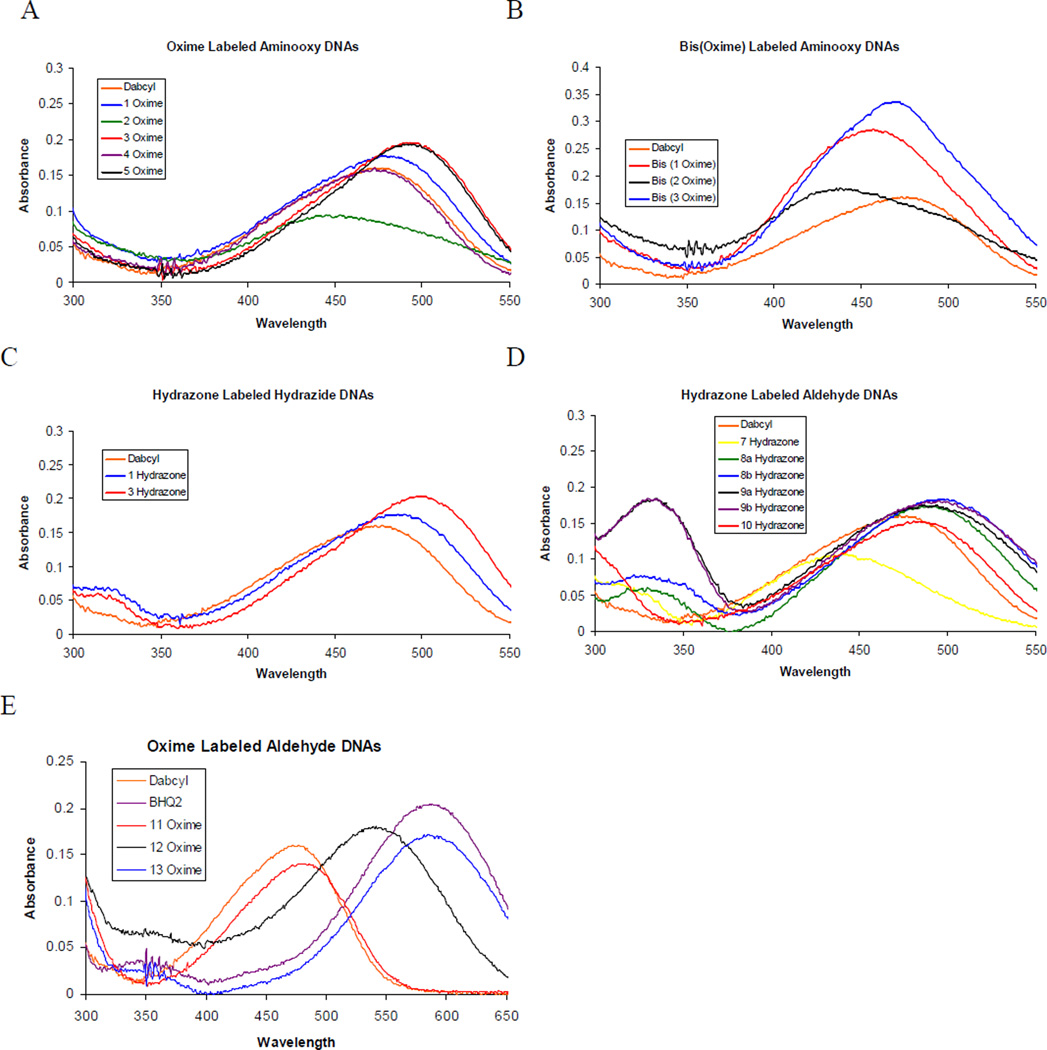
Both hydrazones and oximes of aminooxy or hydrazide modified DNAs with 1 display similar properties to the commercially available amide-linked dabcyl quencher. Absorbance maxima are only slightly red-shifted for the simple dabcyl derivative 1 (less than 10 nm) and display slightly higher extinction coefficients (Figure 3, Table 2). Comparison of the 1-(dimethylamino)-naphthalene moiety as in the oxime of 2 to dabcyl, however, shows a marked blue shift in the absorption maximum of approximately 30 nm with a decrease in the extinction coefficient, spectral properties that are generally not desirable in a fluorescence quencher. Introduction of a methoxy group into the dimethylaniline ring in the form of 3 provides for a red shift of approximately 25 nm and induces a greater increase in the extinction coefficient at the absorbance maximum, both desirable properties. Oximes of acetophenone dyes 4 and 5 provide similar spectral properties to their aldehyde counterparts 1 and 3, with the DNA oxime of 4 possessing very similar properties to the commercially available dabcyl phosphoramidite. Absorbance maxima and other spectral properties for these dye conjugates can be found in Table 2 and the spectra of the quenchers on DNA are provided in Figure 3.
Figure 3.
Absorption spectra of quenchers conjugated to DNA. (a) Oxime-labeled aminooxy DNA (b) Bis(oxime)-labeled aminooxy DNA (c) Hydrazone-labeled hydrazide DNA (d) Hydrazone-labeled aldehyde DNA.
Table 2.
Absorption properties of oxime or hydrazone quenchers on DNA, formed by reaction of carbonyl based dyes with aminooxy or hydrazide modified DNA.
| Dye | Absorption Maxima |
Δwλ ½ max |
|---|---|---|
| Dabcyl | 473 nm | 406 nm – 519 nm (113 nm) |
| 1 DNA Oxime | 477 nm | 408 nm – 526 nm (118 nm) |
| 1 DNA Hydrazone | 486 nm | 417 nm – 532 nm (115 nm) |
| 2 DNA Oxime | 445 nm | 392 nm – 527 nm (135 nm) |
| 3 DNA Oxime | 493 nm | 428 nm – 536 nm (108 nm) |
| 3 DNA Hydrazone | 499 nm | 432 nm – 535 nm (103 nm) |
| 4 DNA Oxime | 473 nm | 405 nm – 516 nm (111 nm) |
| 5 DNA Oxime | 493 nm | 428 nm – 536 nm (108 nm) |
| Bis (1) Oxime | 457 nm | 401 nm – 513 nm (112 nm) |
| Bis (2) Oxime | 438 nm | 384 nm – 520 nm (136 nm) |
| Bis (3) Oxime | 469 nm | 411 nm – 523 nm (112 nm) |
Interesting changes in the absorbance spectrum are observed when two dyes are placed in close proximity via use of the bis(aminooxy) linker on DNA (Figure 3b, Table 2). In the case of two incorporations of 1, the absorbance maximum shifts to 457 nm, an approximately 20 nm blue shift relative to a single 1 DNA oxime. This is likely the result of a dye-dye interaction in the small linker. This shift is accompanied by the expected twofold increase in the molar extinction coefficient. Similar results are seen with the bis(3) oxime DNA, with a similar blue shift to 469 nm (versus 493 nm for the mono 3 oxime DNA) and increase in the absorbance. Interestingly, the bis(2) oxime displayed only a small blue shift in absorbance maximum (from 445 nm for the mono-oxime to 438 nm for the bis-oxime).
Hydrazones of hydrazine-based quencher dyes
The hydrazine dyes reacting with aldehyde-modified DNA provided interesting changes spectral properties (Figure 3d, Table 3). Most valuably, we find that the formation of a hydrazone from the hydrazine dye results in increased conjugation and a general red shift in the optical absorption properties. Comparison of dye 10 with both products of dyes 8 and 9 shows that the latter dyes are red-shifted by approximately 15 nm, with absorption maxima of approximately 495 nm versus 481 nm for 10. Moreover, comparison of the absorption of the Boc-protected dyes 14 – 16 in methanol to the labeled DNA also exhibits the red-shift upon conjugation (although some solvatochromicity likely explains part of the red-shift). While dye 14, for example, absorbs maximally at 412 nm, attachment to DNA in the form of quencher conjugates 8a and 8b yield an absorption maximum of approximately 495 nm, an 80 nm red-shift. The color changes can be readily detected by the naked eye: while dye 15 is bright yellow as a free molecule in solution (absorbing maximally at 383 nm in MeOH), upon hydrazone formation with DNA an orange color is obtained, with an absorption maximum of 441 nm. Even simple 4-nitrophenylhydrazine reacts with aldehyde-modified DNA to shift the absorption maximum from below 400 nm to 461 nm upon hydrazone formation.
Table 3.
Absorption properties of hydrazone and oxime quenchers on DNA, formed by reaction of hydrazine or aminooxy based dyes with aldehyde-modified DNA.
| Dye | Absorption Maxima |
Δwλ ½ max |
|---|---|---|
| Dabcyl | 473 nm | 406 nm – 519 nm (113 nm) |
| BHQ2 | 589 nm | 514 nm – 651 nm (137 nm) |
| 6 DNA Hydrazone | 464 nm | 404 nm – 515 nm (111 nm) |
| 7 DNA Hydrazone | 441 nm | 391 nm – 495 nm (104 nm) |
| 8a DNA Hydrazone | 493 nm | 431 nm – 538 nm (107 nm) |
| 8b DNA Hydrazone | 495 nm | 431 nm – 551 nm (120 nm) |
| 9a DNA Hydrazone | 484 nm | 421 nm – 549 nm (128 nm) |
| 9b DNA Hydrazone | 495 nm | 426 nm – 553 nm (127 nm) |
| 10 DNA Hydrazone | 481 nm | 419 nm – 529 nm (110 nm) |
| 11 DNA Oxime | 482 nm | 417 nm – 527 nm (110 nm) |
| 12 DNA Oxime | 542 nm | 461 nm – 605 nm (144 nm) |
| 13 DNA Oxime | 586 nm | 520 nm – 650 nm (130 nm) |
Surprising similarities are noted between the hydrazones of dyes 8 and 9 in labeled DNA, despite the fact that 9 is a dimeric chromophore while 8 is a monomer. In methanol, protected derivatives 14 and 16 display distinctly different absorption properties, with 14 having an absorption maximum of 412 nm and an extinction coefficient of 35,900, while 16 absorbs maximally at 437 nm with ε = 55,600. When attached to DNA as a hydrazone, however, all derivatives of 8 and 9 display approximately the same absorption spectra in the visible region (Figure 3d). This is likely due to steric crowding that shifts one of the aromatic rings in 9 out of conjugation.
The oxime-labeled DNA derivatives of aminooxy dabsyl (11), aminooxy BHQ1 (12) and aminooxy BHQ2 (13) provided, as expected, similar properties to their parent dyes, as linkers were used for attachment to DNA without any change in the dye scaffold proper (Figure 3e).
Quenching by oxime and hydrazone dye conjugates
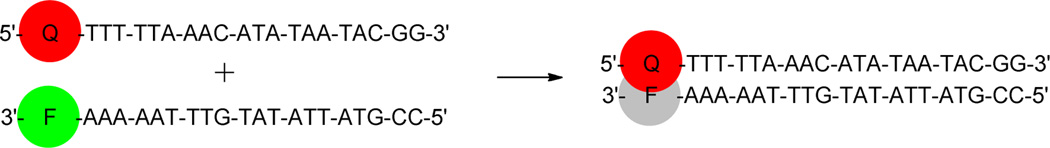
Quenching experiments with the DNA-dye conjugates were performed using fluorescein-labeled DNA complements, monitoring the decrease in emission at 517 nm upon hybridization of the two DNA strands. The experimental duplexes were designed to yield contact quenching (by placing dye quencher and fluorophore together) as opposed to resonance energy transfer (RET), as the former is the method of quenching most often employed in analytical experiments and provides greater quenching efficiency.1–11,15 DNA sequences previously used in other quencher studies were employed here to allow for best comparison with published results and to provide for the simplest control of dye location via attachment to the 5’ end of one strand quenching a fluorophore on the 3’ end of the opposite strand (Figure 4).15,23
Figure 4.
DNA based assay used to assess fluorescence quenching.
Quenching experiments confirmed that the new dyes are as efficient at quenching fluorescein as the commercially available dabcyl and BHQ 2 phosphoramidites (Table 4), confirming that attachment via this new approach retains the optical benefits of these quenchers. Hydrazone and oxime derivatives of 1 are, statistically speaking, as efficient at quenching fluorescein as the parent dabcyl with a general value of nearly 97%. Oximes or hydrazones of 3 provide a slight but not statistically significant increase in quenching efficiency. Two of the dyes do perform slightly worse than dabcyl (see data for 2 and 5), indicating that these are not preferable for use as quenchers in future applications. Ground-state complexes of all dyes with fluorescein were observed and can be found in the Supporting Information (SI) file. As expected, the ground-state complexes between the hydrazone- and oxime-labeled quenchers and fluorescein are similar to the ground-state complex formed between dabcyl and fluorescein, indicative of the same contact quenching mechanism.
Table 4.
Quenching efficiency (Fluorescein, λem = 517 nm) of oxime or hydrazone labeled DNA formed by reaction of carbonyl based quencher with aminooxy or hydrazide modified DNA.
| Dye | Quenching Efficiency |
|---|---|
| Dabcyl | 96.7 ± 0.8% |
| 1 DNA Oxime | 97.0 ± 0.5% |
| 1 DNA Hydrazone | 96.3 ± 0.3% |
| 2 DNA Oxime | 95.0 ± 0.2% |
| 3 DNA Oxime | 96.8 ± 0.2% |
| 3 DNA Hydrazone | 97.1 ± 0.3% |
| 4 DNA Oxime | 96.3 ± 0.2% |
| 5 DNA Oxime | 94.9 ± 0.2% |
| Bis (1) Oxime | 97.3 ± 0.2% |
| Bis (2) Oxime | 96.1 ± 0.4% |
| Bis (3) Oxime | 97.2 ± 0.2% |
Consistent with their lack of spectral advantage (above), the bis(dye) oximes of 1 – 3 provided a negligible increase in quenching efficiency compared to the mono-dye labeled DNA derivatives despite having two chromophores. These results show that a second quencher moiety attached in this way does not provide a benefit in quenching efficiency versus a monomeric dye.
The hydrazones formed by reaction of a hydrazine dye with aldehyde-modified DNA again give similar quenching results as the commercially available dabcyl phosphoramidite (Table 5). Except for 9a as a DNA hydrazone, all derivatives display quenching efficiencies of greater than 96%, statistically similar to the value for dabcyl. The nitroaryl quenchers as (6 and 7) also provide excellent quenching efficiencies of greater than 96% in each case.
Table 5.
Quenching efficiency (Fluorescein, λem = 517 nm) of hydrazone and oxime labeled DNA formed by reaction of hydrazine or aminooxy quencher with aldehyde modified DNA.
| Dye | Quenching Efficiency |
|---|---|
| Dabcyl | 96.7 ± 0.8% |
| BHQ 2 | 96.6 ± 0.3% |
| 6 DNA Hydrazone | 95.6 ± 0.5% |
| 7 DNA Hydrazone | 96.6 ± 0.1% |
| 8a DNA Hydrazone | 96.5 ± 0.2% |
| 8b DNA Hydrazone | 96.7 ± 0.1% |
| 9a DNA Hydrazone | 93.5 ± 0.2% |
| 9b DNA Hydrazone | 96.3 ± 0.3% |
| 10 DNA Hydrazone | 96.7 ± 0.6% |
| 11 DNA Oxime | 96.1 ± 0.1% |
| 12 DNA Oxime | 96.4 ± 0.1% |
| 13 DNA Oxime | 96.5 ± 0.2% |
Like the hydrazone conjugates, the oxime linkages (11 – 13) also showed similar quenching properties as the parent dyes. Statistically speaking, oximes 11 and 13 exhibited the same quenching efficiency as the parent dabcyl or BHQ2 quenchers (Table 5).
Testing hydrazone-linked quenchers in common molecular probes
The ability to easily synthesize aldehyde modified DNA suggested the use of hydrazine quenchers in common fluorometric assays. As a proof of principle, a molecular beacon was prepared using commercial 3’-fluorescein and 5’-formylindole phosphoramidites, placing them in close proximity in the stem region of the beacon.4 Hydrazone formation was easily achieved and two different molecular beacons created using dabsyl hydrazide 10 and the newly created dinitrodiphenylhydrazine 7, which served as a surprisingly effective quencher of fluorescein in the above DNA hybridization experiments.
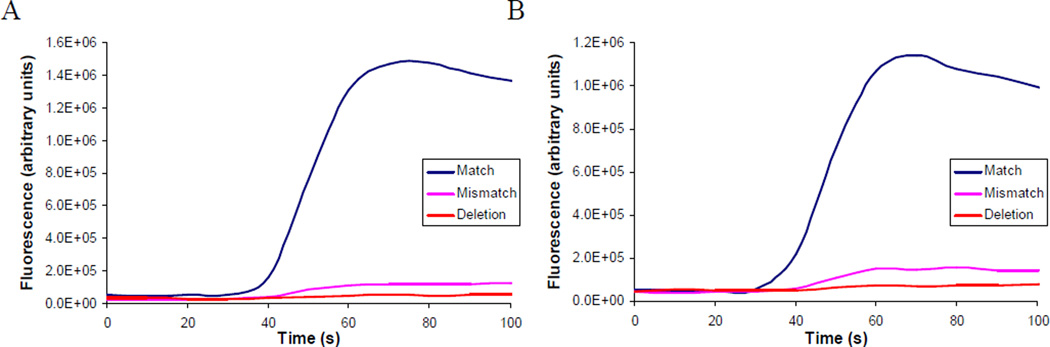
Results showed that the molecular beacon containing quencher 10 increased fluorescence intensity by ca. 27-fold in the presence of the fully matched sequence, corresponding to a quenching efficiency of 96.2%, consistent with the DNA hybridization experiments (Figure 5). Little fluorescence activation was seen in the presence of a singly mismatched DNA or a DNA sequence containing a deletion. The hydrazone conjugate with dye 7 provided yet better results, displaying a >33-fold increase in fluorescence intensity upon hybridization with the fully matched sequence, indicative of a quenching efficiency of approximately 97%, slightly better than found in the DNA hybridization experiments.
Figure 5.
Performance of molecular beacons containing quenchers attached via hydrazone linkages upon exposure to matched, mismatched and deletion target DNA sequences. (A) Beacon containing compound 7 as the quencher and (B) Beacon containing compound 10 as quencher.
Reaction kinetics
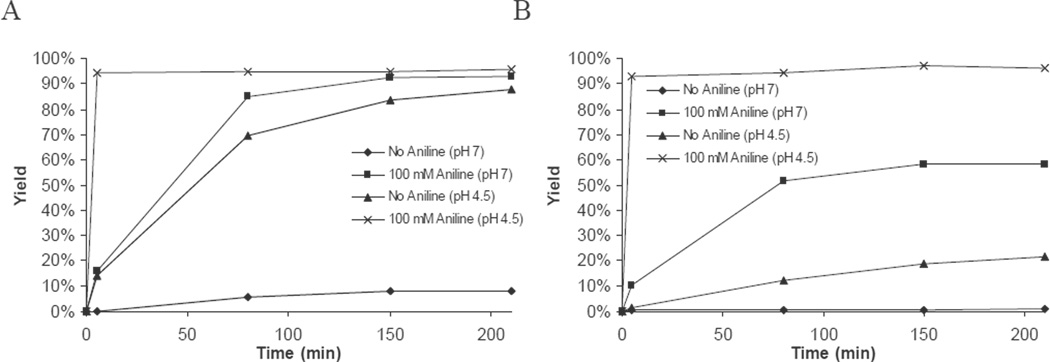
While all hydrazones and oximes above were formed via attachment of the dye to DNA on solid support in a DMF:water mixture, application of quencher attachment directly in solution is often desirable. In this light, experiments were performed to assess oxime and hydrazone formation with dissolved DNAs at pH 4.5 or pH 7 in the presence or absence of aniline (Figure 6). Consistent with previous studies, pH 4.5 (100 mM NH4OAc buffer) proved to be optimal for both hydrazone and oxime formation, while pH 7 (100 mM NaH2PO4) yielded a slower reaction.24–26 Even in the absence of aniline as a catalyst, HPLC analysis indicated 88% conversion to hydrazone labeled DNA after 3.5 hours of reaction at pH 4.5. In the presence of 100 mM aniline at pH 4.5 both the oxime and hydrazone products could be obtained in greater than 90% yield after only five minutes of reaction. As expected, both hydrazone and oxime formation at pH 7 showed considerably slower kinetics, with the uncatalyzed reaction showing less than 8% yield for hydrazone formation after 3.5 hours and oxime formation being less than 1% after this time. Addition of aniline to the pH 7 buffer provided a strong increase in reaction rate, providing the hydrazone in over 90% yield after 3.5 hours while the oxime formation attained approximately 60% yield after this time.
Figure 6.
Kinetics of (a) hydrazone formation or (b) oxime formation between aldehyde modified DNA and 6 (hydrazone) or 11 (oxime) in the presence or absence of aniline at varying pH.
Tests of intracellular quenching
To further test the efficiency and bioorthogonality of oxime and hydrazone formation in quenching of a fluorophore, HeLa cells were labeled by incubation with the aldehyde-containing fluorophore 7-diethylamino-3-formylcoumarin.42 Hydrazone derivatives of this fluorophore have been used previously for intracellular detection of Cu(II)43,44 and cysteine.45 Without any quencher treatment, the bright fluorescence of the aldehyde fluorophore could easily be observed by epifluorescence microscopy (Figure 7a). However, upon treating the cells with aminooxy quencher 11 for three hours in the presence of 10 mM aniline, considerable fluorescence quenching was observed (Figure 7b). Treatment of the cells with the parent non-aminooxy containing quencher dabcyl and 10 mM aniline showed only a small degree of quenching (Figure 7c), confirming the necessity of the aminooxy group in 11 for oxime formation and fluorescence quenching. A similar experiment using a non-aldehydecontaining control fluorophore did not show strong fluorescence quenching by treatment of cells with 11, again indicating the importance of oxime formation (Supporting Information).
Figure 7.
Treatment of HeLa cells with (a) 7-diethylamino-3-formylcoumarin alone, (b) aldehyde fluorophore then three hour incubation with quencher 11 and 10 mM aniline, (c) aldehyde fluorophore then three hour incubation with control dabcyl and 10 mM aniline.
Cellular quenching via the hydrazine quencher 8 with the same aldehyde fluorophore showed even stronger fluorescence quenching, as shown in Figure 8. After three hours’ incubation, very little fluorescence was seen in the treated cells as compared to untreated cells or those exposed to aniline alone, while the presence of the cells was easily observed by brightfield microscopy (Supporting Information).
Figure 8.
Treatment of HeLa cells with (a) 7-diethylamino-3-formylcoumarinalone, (b) aldehyde fluorophore then three hour incubation with quencher 8 and 10 mM aniline, (c) aldehyde fluorophore then three hour incubation with control 10 mM aniline.
Discussion
Our experiments establish compounds 1 – 9 as useful new scaffolds for reactive fluorescence quenchers, offering the convenience of conjugation via oxime or hydrazone formation. We find that dyes 2, 4 and 5 are less effective because of reduced quenching efficiency or poor oxime or hydrazone formation with DNA. However, dyes 1, 3, and 7 – 9 offer excellent properties as quenchers of fluorescein. Dye 3 also provides a new chromophore based on the dabcyl scaffold that has both a red-shift in the absorption maximum and an increase in extinction coefficient relative to the parent dye. Surprisingly, even the carboxylic acid derivative of this dye was apparently unknown in the literature. Incorporation of the methoxy group in the dimethylaniline ring of 3 provides for a moderate red-shift while also increasing the extinction coefficient for the dye in both methanol and on the DNA adducts. As quenching efficiency is slightly increased as well, this may be a desirable dabcyl derivative with improved spectral properties for quenching fluorescein and other dyes emitting in the same region.
To our knowledge, dyes 1 through 10 represent the first examples of aldehyde, ketone or hydrazine based fluorescence quenchers that can be utilized for hydrazone formation with a molecule of interest. Significantly, the formation of the hydrazone results in a red-shift in the absorption of the dyes, and oxime or hydrazone formation also increases the extinction coefficients of the dye when conjugated to DNA. This is also the case for dyes 1 – 5; for example, dye 1 shows a lower extinction coefficient than 4 in methanol, but clearly has a higher extinction coefficient when attached to DNA as a hydrazone or oxime. These results show the beneficial direct electronic interactions that occur upon formation of the dye oxime or hydrazone adducts.
This quencher conjugation approach offers clear advantages of mild reaction conditions, rapid reaction and bioorthogonality of hydrazone formation. Standard post-synthetic labeling of amines with quenchers or other chromophores typically requires many hours to days of reaction, and use of organic co-solvents to dissolve activated chromophores (typically NHS esters). In contrast, formation of the DNA hydrazone of 4-nitrophenylhydrazine (for example) proceeded rapidly in purely aqueous buffer without even the need for catalysis at pH 4.5, reaching 87% yield in less than four hours. Addition of aniline as catalyst provided formation of either oximes or hydrazones in as little as five minutes.
The mild, rapid and selective reaction may have utility beyond simple formation of biomolecular conjugates. For example, experiments in which a fluorescence signal needs to be removed prior to measuring a new signal has previously required photobleaching techniques which may damage cells or other analytes present.46–49 The present experiments suggest a milder approach; rapid attachment of a quencher via bioorthogonal hydrazone or oxime formation might simplify such experiments and avoid high-intensity light exposure. In our preliminary experiments, aminooxy and hydrazine quenchers were shown to efficiently quench an aldehyde fluorophore in a cellular environment. Interestingly, this cellular quenching did not show the need for high amounts of aniline catalysis to quickly attain significant fluorescence quenching at approximately neutral pH inside the cells. Future experiments will explore the generality and applications of this approach.
Supplementary Material
Acknowledgement
This work was supported by the National Institutes of Health (GM068122 and GM067201).
Footnotes
Supporting Information Available. NMR spectra of quencher dyes and intermediates, HPLC purity and MALDI-TOF of DNA strands, and additional spectral data. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Kolpashchikov DM. Binary probes for nucleic acid analysis. Chem. Rev. 2010;110(8):4709–4723. doi: 10.1021/cr900323b. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Cao Z, Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109(5):1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi H, Ogawa K, Alford R, Choyke PL, Urano Y. New strategies for fluorescent probe design in medical diagnostic imaging. Chem. Rev. 2010;110(5):2620–2640. doi: 10.1021/cr900263j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14(3):303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 5.Franzini RM, Kool ET. Efficient nucleic acid detection by templated reductive quencher release. J. Am. Chem. Soc. 2009;131(44):16021–16023. doi: 10.1021/ja904138v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakayama S, Yan L, Sintim HO. Junction probes – sequence specific detection of nucleic acids via template enhanced hybridization processes. J. Am. Chem. Soc. 2008;130(38):12560–12561. doi: 10.1021/ja803146f. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Wang F, Mamon H, Kulke MH, Harris L, Maher E, Wang L, Makrigiorgos GM. Antiprimer quenching-based real-time PCR and its application to the analysis of clinical cancer samples. Clinical Chem. 2006;52(4):624–633. doi: 10.1373/clinchem.2005.063321. [DOI] [PubMed] [Google Scholar]
- 8.Peng X, Chen H, Draney DR, Volcheck W, Schutz-Geschwender A, Olive DM. A nonfluorescent, broad-range quencher dye for Förster resonance energy transfer assays. Anal. Biochemistry. 2009;388:220–228. doi: 10.1016/j.ab.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 9.Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nat. Chem. Biol. 2005;1(4):203–209. doi: 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- 10.Tung C. Fluorescent peptide probes for in vivo diagnostic imaging. Peptide Science. 2004;76(5):391–403. doi: 10.1002/bip.20139. [DOI] [PubMed] [Google Scholar]
- 11.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 12.Johansson MK, Fidder H, Dick D, Cook RM. Intramolecular dimers: a new strategy to fluorescence quenching in dual-labeled oligonucleotide probes. J. Am. Chem. Soc. 2002;124(24):6950–6956. doi: 10.1021/ja025678o. [DOI] [PubMed] [Google Scholar]
- 13.Lakowicz JR. Principles of Fluorescence Spectroscopy. New York, NY: Kluwer Academic/Plenum Publishers; 1999. [Google Scholar]
- 14.Bernacchi S, Mely Y. Exciton interaction in molecular beacons: a sensitive sensor for short range modifications of the nucleic acid structure. Nucleic Acids Res. 2001;29:e62. doi: 10.1093/nar/29.13.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marras SAE, Kramer FR, Tyagi S. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson MK. Choosing reporter-quencher pairs for efficient quenching through formation of intramolecular dimers. In: Didenko VV, editor. Methods of Molecular Biology, Vol 335, Fluorescent Energy Transfer Nucleic Acid Probes: Designs and Protocols. Totowa, NJ: Humana Press Inc.; 2006. [DOI] [PubMed] [Google Scholar]
- 17.Cook RM, Lyttle M, Dick D. Dark quenchers for donor-acceptor energy transfer. 7,019,129. U.S. Patent. 2006 Mar 28;
- 18.Haugland RP, Singer VL, Yue SL. Xanthene dyes and their application as luminescence quenching compounds. 6,399,392. U.S. Patent. 2002 Mar 4;
- 19.Berry DA, Pearson WH. Dark quenchers, probes and other conjugates incorporating the same, and their use. 7,879,986. U.S. Patent. 2006 Feb 3;
- 20.Kopecky K, Novakova V, Miletin M, Kučera R, Zimcik P. Solid-phase synthesis of azaphthalocyanine-oligonucleotide conjugates and their evaluation as new dark quenchers of fluorescence. Bioconj. Chem. 2010;21:1872–1879. doi: 10.1021/bc100226x. [DOI] [PubMed] [Google Scholar]
- 21.Kopecky K, Novakova V, Miletin M, Kučera R, Zimcik P. Synthesis of new azaphthalocyanine dark quencher and evaluation of its quenching efficiency with different fluorophores. Tetrahedron. 2011;67:5956–5963. [Google Scholar]
- 22.Pham W, Weissleder R, Tung CH. An azulene dimer as a near-infrared quencher. Angew. Chem. Int. Ed. Engl. 2002;41(19):3659–3662. doi: 10.1002/1521-3773(20021004)41:19<3659::AID-ANIE3659>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 23.Crisalli P, Kool ET. Multi-path quenchers: efficient quenching of common fluorophores. Bioconj. Chem. 2011;22(11):2345–2354. doi: 10.1021/bc200424r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dirksen A, Hackeng TM, Dawson PE. Nucleophilic catalysis of oxime ligation. Angew Chem. Int. Ed. 2006;45(45):7581–7584. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 25.Dirksen A, Dawson PE. Rapid oxime and hydrazone ligations with aromatic aldehydes for biomolecular labeling. Bioconj. Chem. 2008;19(12):2543–2548. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dirksen A, Yegneswaran S, Dawson PE. Bisaryl hydrazones as exchangeable biocompatible linkers. Angew. Chem. Int. Ed. 2010;49(11):2023–2027. doi: 10.1002/anie.200906756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Key JA, Li C, Cairo CW. Detection of cellular sialic acid content using nitrobenzoxadiazole carbonyl-reactive chromophores. Bioconj. Chem. 2012;23(3):363–371. doi: 10.1021/bc200276k. [DOI] [PubMed] [Google Scholar]
- 28.Dilek O, Bane SL. Synthesis and spectroscopic characterization of fluorescent boron dipyrromethane-derived hydrazones. J. Fluoresc. 2011;21:347–354. doi: 10.1007/s10895-010-0723-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dilek O, Bane SL. Synthesis of boron dipyrromethane fluorescent probes for bioorthogonal labeling. Tetrahedron Lett. 2008;49(8):1413–1416. doi: 10.1016/j.tetlet.2007.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori Y, Niwa T, Toyoshi K. Carcinogenic azo dyes XVIII: Synthesis of azo dyes related to 3’-hydroxymethyl-4-(dimethylamino)azobenzene, a new potent hepatocarcinogen. Chem. Pharm. Bull. 1981;29(5):1439–1442. [Google Scholar]
- 31.Muramoto K, Gotto R, Kamiya H. Analysis of reducing sugars as their chromophoric hydrazones by high-performance liquid chromatography. Anal. Biochem. 1987;162(2):435–442. doi: 10.1016/0003-2697(87)90416-7. [DOI] [PubMed] [Google Scholar]
- 32.Raddatz S, Mueller-Ibeler J, Kluge J, Wäβ L, Burdinski G, Havens JR, Onofrey TJ, Wang D, Schweitzer M. Hydrazide oligonucleotides: new chemical modification for chip array attachment and conjugation. Nucleic Acids Res. 2002;30(21):4793–4802. doi: 10.1093/nar/gkf594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Y, Suzuki Y, Lönnberg T, Komiyama M. Human telomeric DNA sequence-specific cleaving by G-quadruplex formation. J. Am. Chem. Soc. 2009;131(8):2871–2874. doi: 10.1021/ja807313x. [DOI] [PubMed] [Google Scholar]
- 34.Lönnberg T, Suzuki Y, Komiyama M. Prompt site-selective DNA hydrolysis by Ce(IV)-EDTA using oligonucleotide multiphosphonate conjugates. Org. Biomol. Chem. 2008;6(19):3580–3587. doi: 10.1039/b807789d. [DOI] [PubMed] [Google Scholar]
- 35.Kisseljova K, Tsubrik O, Sillard R, Mäeorg S, Mäeorg U. Addition of arylboronic acids to symmetrical and unsymmetrical azo compounds. Org. Lett. 2006;8(1):43–45. doi: 10.1021/ol052403f. [DOI] [PubMed] [Google Scholar]
- 36.Blackburn OA, Coe BJ, Helliwell M. Tetrapalladium(II) bisazobenzene and azoazoxybenzene complexes: synthesis, electronic structures, and optical properties. Organometallics. 2011;30(18):4910–4923. [Google Scholar]
- 37.Li S, Bowerman D, Marthandan N, Klyza S, Luebke KJ, Garner HR, Kodadek T. Photolithiographic synthesis of peptoides. J. Am. Chem. Soc. 2004;126(13):4088–4089. doi: 10.1021/ja039565w. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz DA, Abrams MJ, Hauser MM, Gaul FE, Larsen SK, Rauh D, Zubieta JA. Preparation of hydrazino-modified proteins and their use for the synthesis of 99mTc-protein conjugates. Bioconj. Chem. 1991;2:333–336. doi: 10.1021/bc00011a007. [DOI] [PubMed] [Google Scholar]
- 39.Mezo G, Szabó I, Kertész I, Hegedüs R, Orbán E, Leurs U, Bösze S, Halmos G, Manea M. Efficient synthesis of an (aminooxy) acetylated-somatostatin derivative using (aminooxy)acetic acid as a ‘carbonyl capture’ reagent. J. Pept. Sci. 2011;17(1):39–46. doi: 10.1002/psc.1294. [DOI] [PubMed] [Google Scholar]
- 40.Jencks WP. Studies on the mechanism of oxime and semicarbazone formation. J. Am. Chem. Soc. 1959;81(2):475–481. [Google Scholar]
- 41.More O’Ferrall RA, O’Brien DM, Murphy DG. Rate and equilibrium constants for formation and hydrolysis of 9-formylfluorene oxime: diffusion-controlled trapping of a protonated aldehyde by hydroxylamine. Can. J. Chem. 2000;78:1594–1612. [Google Scholar]
- 42.Ray D, Bharadwaj PK. A coumarin-derived fluorescence probe selective for magnesium. Inorg. Chem. 2008;47:2252–2254. doi: 10.1021/ic702388z. [DOI] [PubMed] [Google Scholar]
- 43.Huang L, Cheng J, Xie K, Xi P, Hou F, Li Z, Xie G, Shi Y, Liu H, Bai D, Zeng Z. Cu2+-selective fluorescent chemosensor based on coumarin and its application in bioimaging. Dalton Trans. 2011;40:10815–10817. doi: 10.1039/c1dt11123j. [DOI] [PubMed] [Google Scholar]
- 44.Ko KC, Wu J-S, Kim HJ, Kown PS, Kim JW, Bartsch RA, Lee JY, Kim JS. Rationally designed fluorescence ‘turn-on’ sensor for Cu2+ Chem. Comm. 2011;47:3165–3167. doi: 10.1039/c0cc05421f. [DOI] [PubMed] [Google Scholar]
- 45.Wu J, Sheng R, Liu W, Wang P, Ma J, Zhong H, Zhuang J. Reversible fluorescent probe for highly selective and sensitive detection of mercapto biomolecules. Inorg. Chem. 2011;50:6543–6551. doi: 10.1021/ic200181p. [DOI] [PubMed] [Google Scholar]
- 46.Borejdo J, Muthu P, Talent J, Gryczynski Z, Calander N, Akopova I, Shtoyko T, Gryczynski I. Reduction of photobleaching and photodamage in single molecule detection: observing single actin monomer in skeletal myofibrils. J. Biomed. Optics. 2008;13(3):034021. doi: 10.1117/1.2938689. [DOI] [PubMed] [Google Scholar]
- 47.Lepock JR, Thompson JE, Kruuv J, Wallach DFH. Photoinduced crosslinking of membrane proteins by fluorescein isothiocyanate. Biochem. Biophys. Res. Commun. 1978;85:344–350. doi: 10.1016/s0006-291x(78)80048-5. [DOI] [PubMed] [Google Scholar]
- 48.Bloom JA, Webb WW. Photodamage to intact erythrocyte membranes at high laser intensities: methods of assay and suppression. J. Histochem. Cytochem. 1984;32:608–616. doi: 10.1177/32.6.6725935. [DOI] [PubMed] [Google Scholar]
- 49.Wolf DE, Edidin M, Dragsten PR. Effect of bleaching light on measurements of lateral diffusion in cell membranes by the fluorescence photobleaching recovery method. Proc. Natl. Acad. Sci. USA. 1980;77:2043–2045. doi: 10.1073/pnas.77.4.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.