Abstract
Germinal centers (GCs) are sites of B-cell clonal expansion, hypermutation, and selection. GCs are polarized into dark (DZ) and light zones (LZ), a distinction that is of key importance to GC selection. However, the difference between the B cells in each of these zones in humans remains unclear. We show that, as in mice, CXCR4 and CD83 can be used to distinguish human LZ and DZ cells. Using these markers, we show that LZ and DZ cells in mice and humans differ only in the expression of characteristic “activation” and “proliferation” programs, suggesting that these populations represent alternating states of a single-cell type rather than distinct differentiation stages. In addition, LZ/DZ transcriptional profiling shows that, with the exception of “molecular” Burkitt lymphomas, nearly all human B-cell malignancies closely resemble LZ cells, which has important implications for our understanding of the molecular programs of lymphomagenesis.
Introduction
Germinal centers (GCs) are microanatomic structures that develop in secondary lymphoid organs in response to antigenic stimulation.1–3 GCs are essential to the development and selection of B cells expressing high-affinity immunoglobulins (Ig) because they are the site of somatic hypermutation and class switch recombination. Both of these reactions are initiated by activation-induced cytidine deaminase (AID), an enzyme that deaminates cytidine to uracyl in single-stranded DNA.4 However, AID targeting is not entirely specific to the Ig locus, and can cause mutations in oncogenes5–8 or double-strand DNA breaks that lead to genomic instability and translocation.9–11 These aberrant events are thought to be at the origin of most types of mature B-cell lymphoma.12
Earlier histologic observations, many of which were carried out using human tonsil samples, classified GC B-cells into 2 cell types (centroblasts and centrocytes) based on morphologic criteria, such as size and nuclear contour.13–15 Within a fully developed GC, centroblasts and centrocytes would distribute preferentially to opposite poles of this structure: a centrocyte-rich light zone (LZ), proximal to the lymph node capsule or spleen red pulp; and a centroblast-rich dark zone (DZ), proximal to the T-cell area.2 In contrast, recent studies done in mice using techniques such as 2-photon microscopy and in situ photoactivation have shown that B cells in the LZ and DZ of mouse lymph node GCs are much more similar than traditional models suggest.16–19 These observations raised questions as to the extent to which the polarization observed in human GC B cells was fully recapitulated in the mouse.
We have demonstrated that, in mice, staining for a combination of CD83 or CD86 with CXCR4 defines 2 subsets of GC cells by flow cytometry that corresponded to anatomically defined LZ and DZ populations.17 Here we show that expression of CXCR4 and CD83 also distinguishes LZ and DZ populations in humans. Although the number of genes differentially expressed between LZ and DZ populations is limited, these differences are highly conserved between species, as is the polarization of cell division toward the DZ. Using a common signature of LZ/DZ phenotypes derived from overlaying mouse and human data, we find that, with the exception of a small subset of “molecular” Burkitt lymphomas (mBLs),20 most human mature B-cell lymphomas resemble LZ rather than DZ GC cells.
Methods
Specimens
Tonsils were obtained from routine tonsillectomies performed at the Babies and Children's Hospital of Columbia–Presbyterian Medical Center. Samples were exempt from informed consent for being fully anonymous residual material obtained after diagnosis. All procedures were approved by the institutional ethics committee. Samples were placed on ice immediately after surgical removal. Tonsillar mononuclear cells (MCs) were isolated by mincing of tissues in RPMI medium followed by Ficoll-Isopaque density centrifugation.
Mice and immunizations
C57BL/6 mice were obtained from The Jackson Laboratory. AID−/− mice on a C57BL/6 background were bred and maintained at the Rockefeller University. To generate GCs, mice were immunized subcutaneously with 50 μg 4-hydroxy,3-nitrophenylacetyl conjugated to keyhole limpet hemocyanin (Biosearch) precipitated in one-third volume of alum (Imject Alum; ThermoScientific) and killed on days 10-12 after immunization. Cells were harvested by forcing draining lymph nodes through a 70-μm nylon mesh into RPMI media supplemented with 6% FCS and 1mM EDTA on ice.
Flow cytometry and cell sorting
Cells were resuspended in PBS supplemented with 0.5% BSA and 1mM EDTA (FACS buffer) and stained with the reagents indicated in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) for 30 minutes at 4°C. Mouse cell suspensions were preincubated with anti–mouse CD16/32 (FcBlock, clone 93, eBioscience) for 5 minutes before addition of the primary stain. Stained cell suspensions were analyzed using a BD LSR Fortessa flow cytometer or sorted using a BD FACSAria cell sorter. For DNA content analysis, cells were fixed with Cytofix/Cytoperm (BD Biosciences) for 15 minutes at room temperature and incubated with 200 ng/mL 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) in BD Perm/Wash (BD Biosciences) for 5 minutes at room temperature and washed once with BD Perm/Wash before resuspension in FACS buffer. Samples were read at a maximum of 1500 events per second.
Tissue immunofluorescence
For CXCR4 and CD86 staining, tonsil samples were fixed for 1 hour in PBS/4% paraformaldehyde/10% sucrose followed by overnight incubation in 30% sucrose, then embedded in OCT (TissueTek) and stored at −80°C. Frozen samples were cut into 20-μm sections using a cryostat and stained as described previously.17 For the remaining stains, samples were fixed overnight in 10% buffered formalin and postfixed in 70% ethanol overnight before embedding in paraffin. The 3-μm sections were dewaxed in xylene and rehydrated in decreasing concentrations of ethanol. Antigen was unmasked by heat-induced antigen retrieval in citrate buffer (pH 6.0) in a pressure cooker for 15 minutes. Stainings were performed as described,21 with minor modifications. Endogenous peroxidase and biotin were blocked as described, and samples were further blocked in PBS-0.5%/Tween-3%/BSA-5% goat serum for 1 hour at room temperature. All primary antibody incubations were done overnight at room temperature. For double CD23/CD83 staining, CD23 antibody was directly labeled with Fab-FITC complexes as per the manufacturer's instructions (Zenon labeling kit, Invitrogen) and added to the sample after blocking the first secondary anti–mouse antibody with MOM Ig block (Vector). Antibody details are provided in supplemental Table 2.
Microscopy
Stained tissue sections were imaged either on a Zeiss LSM 510 confocal microscope with 488, 543, and 633 nm excitation lines (Rockefeller University Bio-Imaging Resource Center) using a Plan-Apochromat 20× (NA 0.75) objective or on a Nikon Eclipse E400 fluorescence microscope, equipped with a Cool SNAP EZ Camera (Photometrics) and Nikon NIS-Elements Imaging Software, using a Plan-Apo VC 20× objective (NA 0.75).
RNA extraction
Both mouse and human LZ/DZ cell samples were resuspended in Trizol after FACS sorting, and RNA isolated as per manufacturer's instructions by addition of chloroform. Total RNA was further purified using RNAeasy columns (QIAGEN). RNA integrity was assessed using a BioAnalyzer (Agilent), and samples with RNA integrity number (RIN) > 9 were processed for gene expression profiling.
Microarrays
A total of 50 ng of total RNA was amplified, labeled, and fragmented using the 3′IVT Affymetrix Express Kit (Affymetrix) following the manufacturer's instructions. Labeled and fragmented cRNA was hybridized on Affymetrix U133Plus2 arrays (human samples) or Affymetrix Mouse 430_2 arrays, as per the manufacturer's instructions. Hybridization images were obtained using an Affymetrix GeneChip Scanner 3000 7G, connected to Command Console software. Row analysis was performed using the Affymetrix Expression (Herbert Irving Cancer Research Center Genomics Facility, Columbia University Medical Center). All microarray analyses were performed in quadruplicate (4 different patients or pools of mice).
Microarray analysis
Intensity (CEL) files were extracted using the Affymetrix Console software and normalized using the RMA or MAS Version 5.0 algorithms. Unsupervised clustering analysis of MAS Version 5.0 normalized raw data were performed using a an average linkage method implemented in a clustering algorithm previously described in Klein et al.22 Cluster distances are based on Euclidean metrics, and were calculated by using only genes that were differentially expressed > 1.5-fold among samples. For scatter and volcano plots, RMA-normalized data were processed using Microsoft Excel 2008 to determine mean expression, fold change, and P values (Student t test). Graphs were plotted using Graphpad Prism Version 5.0. Expression heat maps were generated using MultiExperiment Viewer (MeV Version 4.8), part of the TM4 Microarray Software Suite.23 Hierarchical clustering trees in (see Figures 5 and 6; supplemental Figures 6 and 7) were generated using geWorkBench (Version 2.2.2; http://www.geworkbench.org), developed at Columbia University (NIH Roadmap Initiative, 1U54CA121852–01A1) and the National Cancer Institute.24 Data were collapsed to Official Gene Symbol using the maximum of probes, and hierarchical clustering built using Pearson's metrics and total linkage algorithms. Fold changes between zones are calculated for each pair of samples from the same individual (human) or pool (mice; eg, LZ1/DZ1, LZ2/DZ2). Consensus clustering and Class Prediction analyses of tumor samples, as shown in Figures 5 and 6, were performed with tools available at the GenePattern server.25 Consensus clustering was achieved using 1000 iterations and a Total Linkage clustering algorithm.26 The relatedness of primary tumor samples and cell lines to the LZ or DZ phenotypes was estimated using the Weighted Voting class prediction tool27 (classifier based on 30 markers) or the Class Prediction Tool based on the SPLASH algorithm (classifier built the complete set of LZ/DZ differentially expressed genes, 2% delta, full support), part of the BlueGenes and Genes@Work microarray software packages, as described.22,28 The microarray data described in this publication have been deposited in NCBI's Gene Expression Omnibus under accession no. GSE38696 and GSE38697.
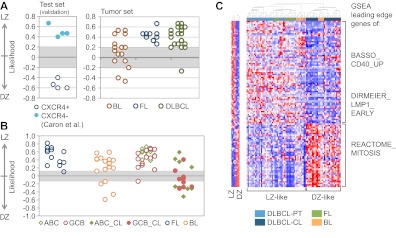
Figure 5.
Most GC-derived B-NHLs share an LZ-related phenotype. (A) Class prediction of GC-derived B-NHLs using the Weighted Voting algorithm.27 The dataset reported by Caron et al,37 where tonsillar GC B-cells were isolated according to their surface expression of CXCR4, was used as a validation set. (B) Class prediction in the same B-NHL samples using the SPLASH algorithm within the Bluegenes tool.22 In this analysis, DLBCL cell lines were included. All DLBCL primary cases and cell lines are coded based on their cell-of-origin classification (ABC/GCB).39 (C) Hierarchical clustering of the same GC-derived B-NHL based on the expression of the “compound pathway signature,” as described in the main text. Note the presence of 2 main clusters (DZ-like, LZ-like). Red represents highly expressed genes; and blue, lower-expressed genes. The behavior of the signature in normal GC LZ/DZ B-cells is plotted in the accompanying heat map (left panel). DLBCL-PT indicates primary DLBCL cases; and DLBCL-CL, DLBCL cell lines.
Figure 6.
LZ/DZ-related pathways underlie the distinction between molecular subgroups in aggressive mature B-NHL. (A) Consensus clustering (1000 bootstraps) of the aggressive mature B-NHL case series described by Hummel et al,20 according to the expression pattern of the common human/mouse signature (1.5-fold cutoff; supplemental Figure 4; supplemental Table 4). Shown is the clustering image with k = 2 (no significant improvement of the CDF was observed with higher k values). (B) Distribution of mBL and non-mBL cases, as defined by Hummel et al,20 among the 2 different subgroups identified by the consensus clustering analysis shown in panel A. The P value shown refers to the significance of the distribution of the 3 subclasses in Hummel et al20 (mBL, intermediate, non-mBL) among each class (DZ-like or LZ-like; χ2 test). (C) GSEA plots illustrating the enrichment for mBL and non-mBL classifier gene signatures (as defined by Hummel et al20) in human LZ and DZ GC B-cell gene expression data. Nominal and adjusted P values and false discovery rate are all below the detection level (< .001) for both comparisons.
Ig gene sequencing
DNA was extracted from sorted tonsil LZ or DZ cells and amplified as previously described.29 Pooled PCR products were cloned using TOPO TA cloning kit (Invitrogen), and bacterial colonies were PCR-amplified and sequenced. Sequences were analyzed using the NCBI IgBLAST tool.
Statistical analysis
The distribution of the number of mutations per V-region and of silent/replacement ratios was compared between LZ and DZ for all sequences pooled or for each individual patient using the Student t test. Silent/replacement ratio was used rather than replacement to silent ratio to avoid division by zero (many individual sequences had zero silent mutations). The significance of the distribution of tumor cases into classes was calculated by χ2 test using GraphPad Prism Version 5.0 software. The statistics provided for gene set enrichment analysis (GSEA; including P value, normalized P value, and false discovery rate) were calculated by the GSEA software.30 Statistical treatment of microarray data are described above.
Results
Expression of CXCR4 and CD83 identifies LZ and DZ B-cell populations in human tonsil GCs
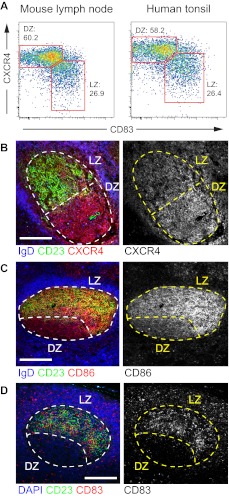
In the mouse, LZ and DZ GC cells can be distinguished by flow cytometry by their expression of activation markers CD83 and CD86 and of chemokine receptor CXCR4. LZ cells are CD83hiCXCR4lo or CD86hiCXCR4lo, and DZ cells are CD83loCXCR4hi or CD86loCXCR4hi.17 To determine whether the same populations could be discerned in human GCs, we stained mononuclear cell suspensions obtained from pediatric human tonsil samples with antibodies to these markers (Figure 1A; supplemental Figure 1A). As in the mouse, staining of human GC B cells with antibodies to CXCR4 and CD83 results in a continuum of expression of the 2 markers with a clear inflection point between CXCR4hiCD83lo (DZ) and CXCR4loCD83hi (LZ) cells that is sufficient to delineate 2 cell populations with an approximately 2:1 ratio (Figure 1A, and compare with Victora et al.17 Separation of these populations using CD86 instead of CD83 was less clear in humans, mostly because human CXCR4hi cells did not down-regulate CD86 to the same extent as their mouse counterparts (supplemental Figure 1B). To confirm that expression of these markers corresponded to anatomic location in humans, we stained tonsil or reactive lymph node sections with antibodies to CXCR4, CD83, and CD86. Histologic examination confirmed higher expression of CXCR4 in the DZ and higher expression of CD83 and CD86 in the LZ (Figure 1B-D).
Figure 1.
CXCR4 and CD83 define human LZ and DZ B-cell populations. (A) Flow cytometric profile of day 10 mouse lymph node GCs and human tonsil GCs stained for markers CXCR4 and CD83. LZ and DZ gates and percentages are shown. Gating as shown in supplemental Figure 1A. (B-D) Each plot is representative of at least 4 independent experiments. Immunofluorescent staining of frozen tonsil (B-C) or paraffin-embedded reactive lymph node (D) samples showing the anatomic distribution of CXCR4 (B), CD86 (C), and CD83 (D) in human GCs. Light zones are defined by CD23 staining (green). Note that CD23 and CD86 staining appears to overlap in the LZ because of insufficient resolution of confocal microscopy to discern molecules expressed on juxtaposed membranes. The GC perimeter is defined in frozen and paraffin-embedded sections by counterstainings with IgD and DAPI, respectively (both in blue). Scale bars represent 100 μm. Histology data are representative of 2 independent experiments. Image acquisition parameters are described in “Microscopy.”
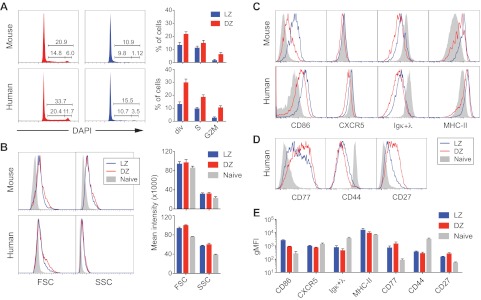
Human LZ and DZ populations defined by expression of CD83 and CXCR4 resembled their mouse counterparts in terms of DNA content. Whereas close to 30% of mouse and human DZ cells are in the S/G2/M phases of the cell cycle, only 10% to 15% of LZ cells were after G1 (Figure 2A). As previously reported in mice,17 the LZ compartment is nearly devoid of cells in G2/M (Figure 2A), suggesting that B cells leave the LZ before completion of the cell cycle. In addition, as in the mouse, LZ and DZ cells were equivalent in terms of size and complexity, as measured by forward and side scatter, respectively (Figure 2B). This finding was unexpected given the traditional view of large centroblasts and small centrocytes.
Figure 2.
Phenotype of LZ and DZ B cells in mice and humans. (A) Cell-cycle (DNA content) profiles of human and mouse LZ and DZ cells (gated shown in supplemental Figure 1A). Multiple experiments (n = 5 for mouse and n = 3 for human) are quantified in the graphs on the right (error bar represents SD). (B) Forward scatter (FSC) and side scatter (SSC) profiles of human and mouse LZ, DZ, and naive B cells. Naive B cells are defined as CD19+IgD+CD83− in human tonsil and as B220+FAS−CD38+ in mouse lymph node. Right panels: quantification of multiple experiments (n = 4 for mouse and n = 3 for human; bar represents SD). (C) Expression of selected markers by human and mouse LZ, DZ, and naive B cells. (D) Expression of selected B-cell markers by human LZ, DZ, and naive B cells. (E) Reproducibility of surface molecule expression between human samples. Graph represents quantification of multiple experiments (n = 2 for CD86, MHC class II; n = 3 for Igκ+λ; and n = 4 for the remaining markers; bar represents SD).
To extend the comparison to other markers, we measured expression of surface molecules known to differ between mouse LZ and DZ B cells.17 Human LZ cells expressed slightly higher levels of surface immunoglobulin and chemokine receptor CXCR5 and substantially higher levels of CD86 than DZ cells (Figure 2C). Also conserved was the presence in the DZ of a subpopulation of cells with slightly lower expression of MHC class II (Figure 2C). Other molecules up-regulated in mouse LZ cells,17 including CD69, CCR6, and CD23, could not be detected in human GCs, regardless of compartment (supplemental Figure 1C). We then analyzed human LZ and DZ populations for their expression of the cell-surface glycolipid CD77 and the adhesion receptor CD44, 2 markers classically thought to distinguish between centrocytes and centroblasts.31–33 Neither marker was capable of distinguishing between the 2 cell populations (Figure 2D). Surprisingly, CD27, a marker traditionally associated with memory B cells,34 was expressed to substantially higher levels in DZ cells (Figure 2D). Expression of all markers analyzed by flow cytometry was highly reproducible between different human samples (Figure 2E).
Using a set of primers that captures all heavy chain V segments,29 we sequenced VDJ rearrangements from clones of B cells sorted from LZ and DZ to determine whether there were differences in the number, position and nature of mutations in the 2 zones. As expected from the rapid exchange of cells between the LZ and DZ compartments observed in vivo in the mouse,17 no substantial differences in terms of mutations were found between human LZ and DZ cells (supplemental Table 3).
Gene expression in human and mouse light and dark zone B cells
To investigate whether the transcriptional programs associated with localization to LZ or DZ are conserved between mouse and human GCs, we performed gene expression profiling by microarray on LZ and DZ cells sorted from human tonsils (supplemental Figure 1A). As a comparison, we also performed microarray analysis on LZ and DZ cells sorted from mouse skin-draining lymph nodes 12 days after subcutaneous immunization with 4-hydroxy,3-nitrophenylacetyl conjugated to keyhole limpet hemocyanin (supplemental Figure 1A). Gene expression profiles and signatures for mouse LZ and DZ obtained using this immunization strategy were essentially identical to those obtained by in situ photoactivation,17 further confirming the validity of CXCR4 and CD83 as markers of LZ/DZ location in mouse (supplemental Figure 2A-B).
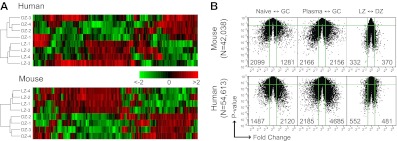
Unsupervised hierarchical clustering of mouse and human samples indicated that differences between the LZ and DZ states were robust enough to be reproducible among different subjects in both species (Figure 3A). However, as reported in mice,17 the number of genes differentially expressed between LZ and DZ B cells was limited, especially compared with the differences in gene expression between naive and GC B cells or between GC B cells and plasma cells (data obtained from Luckey et al35 and Longo et al36; Figure 3B). Thus, although LZ and DZ cells are sufficiently different to be robustly distinguishable by gene expression in both humans and mice, this difference is much smaller in magnitude than differences resulting from true developmental transitions, such as those from naive to GC cell or from GC to plasma cell.
Figure 3.
Gene expression in mouse and human LZ/DZ. (A) Unsupervised clustering of microarray data from LZ and DZ cells sorted from mouse lymph node or human tonsil. (B) Volcano plots showing global differences between naive and GC, plasma cell and GC, and LZ and DZ B cells in human and mouse. Microarray data for naive and total GC B cells and plasma cells data were obtained from Luckey et al35 and Long et al.36 Green lines indicate 2-fold differences (x-axis) and P = .05 (y-axis). Numbers in the plots indicate the number of probes differing by 2-fold or more (P < .05 in each of the comparisons). The total number of probes plotted for each species is indicated on the left.
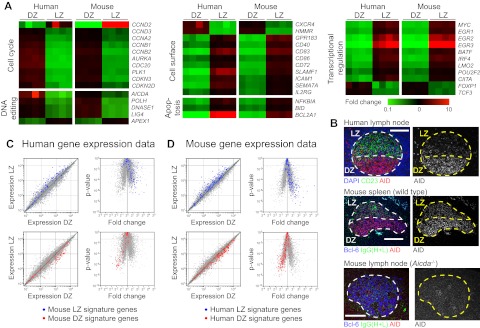
Analysis of individual genes showed that many of the relevant changes in gene expression between mouse LZ and DZ cells17 were also detected in humans (Figure 4A). These included genes involved in cell cycle progression (eg, cyclins D2/D3/A2/B1/B2, and AURKA), cell surface molecules and receptors (eg, CXCR5, GPR183, and SLAMF1), genes involved in DNA editing (eg, AICDA, POLH, and LIG4), regulators of apoptosis and cell death (BCL2LA1, BIM, and NFKBIA), and several transcriptional regulators (eg, MYC, EGR1-3, and BATF). For a small number of genes, changes in expression between LZ and DZ were not conserved between species. These included genes up- or down-regulated in 1 species and that were equally expressed in the other species (eg, CD44, LMO4, and GPR137B), not expressed or not captured by the microarray probes in the other species (eg, CXCR7, LIFR, and S1PR3), or that were regulated in the opposite direction in mice and humans (eg, CD38; supplemental Figure 3A).
Figure 4.
Polarization of gene expression in LZ and DZ B cells. (A) Heat maps showing differential expression in LZ and DZ of selected genes in mice and humans. Colors indicate fold change (Log10 base) between one zone and the opposite zone within the same sample (human) or pool (mouse). (B) Immunofluorescence of human (top) and wild-type mouse (middle) GCs showing the anatomic distribution of AID protein. AID is stained in both species using the same rat monoclonal antibody (mAID-2). Light zones are defined by CD23 staining (green) in humans and Ig (anti-IgG, heavy and light chains, which stains mostly immune complexes deposited on follicular dendritic cells) in mice. The GC perimeter is defined by counterstaining with DAPI in human and Bcl-6 in mouse (both in blue). Immunofluorescent staining of an Aicda−/− lymph node is shown as a control for the specificity of the anti-AID antibody (bottom). Scale bars represent 100 μm. Image acquisition parameters are described in “Microscopy.” (C) Mouse LZ and DZ signatures overlaid on human gene expression data, plotted as expression scatter plots (left) or volcano plots (right). Human gene expression data are shown (gray) with genes contained in mouse LZ or DZ signatures highlighted in blue or red, respectively. Interspecies orthologs defined as genes bearing the same Official Gene Symbol. Volcano plots: dotted line represents 2-fold P < .05. (D) Human LZ and DZ signatures overlaid on mouse gene expression data. Details are as in panel C.
Previous studies have shown that, in humans, AID is expressed at higher levels in the DZ and in CXCR4hi GC cells.21,37 In our gene expression analysis, although AID expression was polarized toward the DZ in both species, the extent of this polarization differed between mice and humans (5.2-fold in humans vs 1.5-fold in mice). Immunohistochemical staining of mouse and human GCs with an anti-AID monoclonal antibody reactive with both species confirmed this trend (Figure 4B).
To compare mouse and human LZ and DZ populations at a global scale, we defined stringent LZ and DZ gene expression signatures (full support, 2% delta, see “Microarray analysis”) for mice and humans (supplemental Table 4). Global comparison of human and mouse signatures (matched by Official Gene Symbol) revealed remarkable interspecies conservation of changes in gene expression between LZ and DZ (Figure 4C-D). Of the 69 genes in the mouse LZ signature that differed reproducibly between human LZ and DZ samples (P < .05 in the human data), 64 (93%) were also up-regulated in human LZ (Figure 4C). Similarly, of the 54 genes in the mouse DZ signature that met this criterion, 50 (93%) were also up-regulated in human DZ (Figure 4C). Conversely, of the 125 genes in the human LZ signature that differed reproducibly between LZ and DZ in mouse (P < .05), 115 (92%) were also up-regulated in mouse LZ (Figure 4D), and of the 152 genes in the human DZ signature that met this criterion, 130 (86%) were also up-regulated in mouse DZ (Figure 4D). To determine the significance of this similarity, we carried out GSEA30 overlaying mouse signatures on human data and vice-versa (supplemental Figure 3B). This analysis confirmed the strong correspondence between mouse and human GC compartments, with enrichment scores ranging from 0.70 to 0.82 and P values and false discovery rates uniformly below the minimum detectable by the algorithm (supplemental Figure 3B). Thus, we conclude that the CXCR4hiCD83lo and CXCR4loCD83hi populations in human and mouse GCs are equivalent and that these correspond to DZ and LZ B-cell populations, respectively.
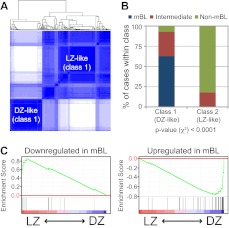
A gene expression signature for LZ/DZ cells common to mice and humans
Based on the assumption that genes whose changes in expression between LZ and DZ B cells are conserved between species are also probably relevant to GC compartmentalization, we constructed a common LZ/DZ signature (supplemental Figure 4). We generated low-stringency signatures for each species (cut-off of 1.33-fold and P < .05) and used the intersection of the mouse and human signatures (matched by Official Gene Symbol) as a common signature. This signature was composed of 473 genes, of which 290 were up-regulated in the LZ and 183 in the DZ. To investigate which functional pathways were overrepresented in this common signature, we used the GSEA Gene Set Overlaps tool to find significant overlaps between this signature and gene sets present in the GSEA database. This analysis showed evidence of up-regulation in LZ of signatures related to CD40/LMP1 activation, NF-κB and c-Myc engagement, and negative regulation of apoptosis (supplemental Table 5). In contrast, genes involved in cell cycle regulation, especially mitosis, dominated the DZ signature (supplemental Table 5). Up-regulation of these signatures was confirmed on the microarray data as a whole by performing GSEA analysis separately on the human and the mouse data (supplemental Table 5). We conclude that the major distinction in gene expression between LZ and DZ B cells in both mice and humans is the up-regulation of activation-related signatures in the LZ and of the mitotic cell cycle program in the DZ.
Gene expression profiles of GC B-cell malignancies resemble those of LZ B cells
The majority of mature B-cell non-Hodgkin lymphomas (B-NHL) are thought to be of GC origin based on phenotypic and genetic evidence.3,38 We therefore sought to assess the relationship of the different types of GC-derived B-NHL to the LZ or DZ GC subpopulations based on the similarity of their gene expression programs. To this end, we used 2 different algorithmic approaches to build gene classifiers capable of assigning samples with high confidence to either a LZ or a DZ phenotype. For both class-prediction methods (Bluegenes/SPLASH algorithm22 and Weighted Voting), we used the human LZ/DZ samples (4 + 4) as a training set. We then applied these classifiers to gene expression profiles of B-NHL obtained from our database of cases of GC-derived B-NHL (including pediatric Burkitt lymphoma [BL], follicular lymphoma [FL], and diffuse large B-cell lymphoma [DLBCL], both primary tumors and derived cell lines). For reference, we also applied these same classifiers to previously published gene expression profiles of normal CXCR4hi and CXCR4lo GC cells from human tonsils.37
Regardless of algorithm, the majority of tumors analyzed were assigned to the LZ class with likelihood similar to that of normal CXCR4− GC cells (Figure 5A-B). All FLs and all but 1 DLBCL were classified as LZ-related, although few cases of DLBCL fell within the low-confidence (< 95%) intervals of each algorithm. Primary cases of BL were less polarized, although approximately 75% of cases were still assigned to an LZ phenotype (Figure 5A-B). This observation is in contrast with a previous study suggesting that all BL cases had a DZ phenotype,37 which is also a common assumption based on the cell morphology and high proliferation rate of these lymphomas.
DLBCL cases were assigned to a LZ phenotype irrespective of their classification as activated B cell–like (ABC) or GC B cell–like (GCB).39 Accordingly, genes in the ABC and GCB signatures39 were randomly distributed across both LZ and DZ phenotypes, suggesting that these signatures measure features substantially different from those involved in LZ/DZ polarization (supplemental Figure 5). The assignment of DLBCL cases to a LZ phenotype was lost when DLBCL-derived cell lines were analyzed. Unlike primary tumors, most cell lines analyzed were assigned to a DZ phenotype (Figure 5B). This assignment was also independent of the tumor's classification as ABC or GCB, and is probably attributable to selection for cells undergoing high rates of division in vitro.
To better understand the basis for the assignment of most GC-derived B-NHL to a LZ phenotype, we explored the behavior of representative LZ- and DZ-related pathways common to mice and humans (ie, the CD40 and LMP1 signaling and cell cycle/mitosis/cytokinesis signatures, see supplemental Table 5) in primary cases of GC-derived B-NHL. For this, we first identified genes within these signatures that were enriched in human LZ or DZ GC B-cells (ie, that constituted the “leading edge” of each of these signatures, as determined by GSEA). We then grouped these genes into a “compound pathway signature” (supplemental Table 4), which we used to perform unsupervised hierarchical clustering. As shown in Figure 5C, clustering of primary cases and cell lines using this approach fully recapitulated the results obtained using the full LZ/DZ signature. This suggests that assignment of most primary tumors to LZ is strongly related to their activation of CD40/LMP1 signatures, whereas the assignment of cell lines and a few cases of BL to DZ is mostly related to their proliferative capacity. Equivalent results were obtained when using gene expression profiles of B cells purified from primary B-NHL tumors, confirming that assignment of primary cases to a LZ phenotype was not because of contaminating cells within the tumor tissue (supplemental Figure 6). We conclude that, with the notable exception of a subgroup of BL, most primary cases of B-NHL (FL, DLBCL) resemble LZ B cells in terms of gene expression program, and this is largely because of their expression of the CD40/LMP1 and related signaling response signatures. Loss of these activation-related programs and increased cell proliferation on in vitro culture is associated with a DZ phenotype.
LZ/DZ signatures underlie the “molecular” definition of Burkitt lymphoma
The distribution of primary BL cases into 2 different phenotypic groups (LZ-and DZ-related; Figure 5A-B) suggests that there is substantial biologic heterogeneity among BL cases. Two previous studies have reported that, within mature aggressive B-cell lymphoma cases with a pathologic diagnosis of BL, global gene expression and genomics analysis could identify 2 different subgroups. At one end of this spectrum were bona-fide mBLs; and at the other end, other aggressive lymphomas also diagnosed as BL based on morphology but with poorer outcomes (non-mBL).20,40 We therefore used our common LZ/DZ signature with a more stringent cut-off (1.5-fold rather than 1.33-fold; supplemental Figure 4, supplemental Table 4) to classify a previously described series of 220 cases of aggressive mature B-NHL.20 For this, we used a “consensus clustering” statistical approach, which allows for the refining of hierarchical clustering analysis by repeated iterations of the clustering algorithm after resampling.26 This approach identified 2 robust groups of samples (best k = 2 as assessed by empirical cumulative distribution; data not shown), which largely corresponded to DZ-like and LZ-like cases based on their expression of genes in the common signature (Figure 6A). DZ-like and LZ-like groups overlapped almost entirely with the mBL and non-mBL categories,20 respectively, whereas “intermediate” cases were distributed roughly equally between LZ-like and DZ like classes (Figure 6B). Accordingly, the previously defined mBL and non-mBL gene signatures20 were strongly enriched in the DZ and LZ gene programs, respectively (Figure 6C). A similar analysis carried out using our compound pathway signature (see “Gene expression profiles of GC B-cell malignancies resemble those of LZ B cells”) showed that this resemblance was largely the result of the down-regulation of CD40/LMP1 signatures and up-regulation of proliferation-related genes in mBLs (supplemental Figure 7). Notably, the c-Myc pathway, which is normally down-regulated in DZ B cells, remained up-regulated in the mBL samples (supplemental Figure 7). We conclude that, of all B-NHLs analyzed, mBLs were the only lymphomas to resemble normal DZ cells, although they differ from the latter in that they express high levels of c-Myc–activated genes.
Discussion
The functional and phenotypic distinction between LZ and DZ B cells has been a central issue in GC biology for several decades. In humans, centrocytes and centroblasts were long thought to be distinguishable by their differential expression of the ganglioside CD77 (centroblasts cells being CD77+ and centrocytes, CD77−).15,31,41,42 However, the gene expression profiles of CD77+ and CD77− GC B cells failed to show relevant differences between these populations, suggesting that CD77 expression is incapable of differentiating between LZ and DZ cells.34,43,44
The identification of CXCR4 as a factor necessary for positioning B cells in the DZ45 led to attempts to use this receptor as a marker of DZ cells in humans.37,46 Notably, Fest et al showed that human tonsil GC B cells separated on the basis of CXCR4 expression have gene expression signatures compatible with the putative functional roles of DZ and LZ GC B cells. However, evidence for the polarization of CXCR4 in human GCs remained correlative, and attempts to use this marker to demonstrate polarization by histology were unsuccessful.37 By combining CXCR4 and CD83 staining in human GCs, we were able to resolve 2 populations of cells within the CXCR4/CD83 continuum, by flow cytometry and histology, which are equivalent to those found in mouse LZ and DZs.
Comparison of the gene expression signatures of LZ and DZ B cells in human and mouse showed remarkable interspecies conservation. For example, cell cycle-associated genes (with the notable exception of cyclin D2) are up-regulated in the DZ, and genes associated with cell activation (eg, EGR1-3, MYC, and NFKBIA) are induced in the LZ. More importantly, global analysis of the changes in gene expression signatures across species showed that genes that changed between the 2 zones in 1 species were likely to be modulated in the same direction in the other species. This not only allowed us to define a conserved set of genes that is likely to be relevant to LZ/DZ segregation but also validates the use of the mouse as a model for the analysis of subtle aspects of gene regulation in human GCs. Moreover, in addition to being conserved across species, these differences are also conserved in different types of GCs. Whereas the mouse GCs we analyzed were synchronized, acute responses to immunization, human tonsil GCs are chronic and probably result from simultaneous or overlapping GC reactions within a GC structure that is recycled47 in response to different pathogens. We conclude that the set of conserved LZ and DZ genes represents the core circuit that defines these regions of the GC, regardless of species, anatomic location, and mode of antigenic stimulation.
Part of the interest in defining LZ and DZ markers for human GCs is to understand the relationship between GC subpopulations and human B-cell malignancies.3,48,49 We find that most GC-derived B-NHL (mainly FL, DLBCL, and a fraction of mostly non-mBL) resemble LZ B cells, largely because of the activity of LZ-associated cellular activation pathways. mBLs are an exception to this rule in that they resemble DZ B cells. However, in contrast to normal DZ B cells, mBLs show “ectopic” activation of gene expression signatures related to c-Myc engagement. The concerted activation of the CD40/LMP1 and c-Myc pathways seen in LZ B cells is therefore uncoupled in mBLs, probably because of the t(8;14) Myc/IgH translocation that is characteristic of this class of lymphoma.
The exquisite polarization of gene programs between LZ and DZ in normal GCs suggests that environmental cues encountered by B cells when moving between GC compartments are determinant for their phenotypic and biologic behavior. For example, signals provided by follicular helper T cells and antigen presented on follicular dendritic cells appear to activate LZ-related pathways.17 These observations also suggest that microanatomic environment is a primary determinant of the LZ/DZ cell program, which lends support to our proposed notion that normal LZ and DZ B cells are alternating states of a same cell population. Accordingly, we find that culturing GC-derived lymphoma cells in vitro leads to loss of LZ-related signatures, suggesting that immune activation signatures require indeed signals from the in vivo tumor environment, and up-regulation of mitosis-related genes, as would be expected from cells selected for continuous growth in culture.
DZ and LZ B cells are usually thought of as 2 distinct populations with a precursor-product relationship.2,3 This view was challenged by the prediction,50 and later demonstration,16–19 that GC B cells are rapidly cycling back and forth between the DZ and LZ. These findings both preclude linear differentiation and suggest that LZ and DZ B cells may simply represent different states of activation of the same cell population. Our data strongly support this single-population model. Human LZ and DZ B cells have similar size and complexity and show similar mutation/selection patterns in VH genes. In both mice and humans, differences in gene expression between LZ and DZ cells are highly restricted, especially compared with differences between GC and naive B cells. Rather than differentiation, the documented differences in gene expression suggest a simple alternation between a proliferative mode, in which DZ B cells also up-regulate genes involved in somatic hypermutation, and a “signal-integration” mode, in which LZ B cells are exposed and respond to the selection signals delivered to them in the LZ by follicular dendritic cells and T cells. We would thus like to propose that LZ and DZ phenotypes should be viewed as distinct biologic states of a same cell, rather than as 2 distinct populations. Exchange between these phenotypes would consequently be better described as an oscillation between 2 states than as true differentiation.
Supplementary Material
Acknowledgments
The authors thank Katia Basso, Mukesh Bansal, and Celine Lefebvre (Columbia University, ICG and C2B2) for conceptual suggestions regarding statistical analysis of gene expression data; Ulf Klein for critical reading of the manuscript; Vladan Miljkovic (Core Genomics Facility, Pathology Department and Irving Cancer Research Center, Columbia University) for technical assistance in microarray sample hybridization; and Klara Velinzon and Yelena Shatalina (Nussenzweig Laboratory) and Kristie Gordon, Chenhong Liu, and Sandra Tetteh (Flow Cytometry Facility, Irving Cancer Research Center, Columbia University) for assistance with cell sorting.
D.D.-S. was supported by the National Institutes of Health (grant K99/R00 CA151827). M.C.N. is an HHMI investigator.
Footnotes
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: G.D.V. and D.D.-S. designed and performed the experiments and wrote the manuscript; A.B.H. assisted with bioinformatic analysis; S.D. assisted with experimental work; and R.D.-F. and M.C.N. supervised the project, designed experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for G.D.V. is Whitehead Institute for Biomedical Research, Cambridge, MA.
Correspondence: Gabriel D. Victora, Whitehead Institute for Biomedical Research, 9 Cambridge Center, Cambridge, MA 02142; e-mail: victora@wi.mit.edu.
References
- 1.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 2.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 3.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8(1):22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 4.Wagner SD, Neuberger MS. Somatic hypermutation of immunoglobulin genes. Annu Rev Immunol. 1996;14:441–457. doi: 10.1146/annurev.immunol.14.1.441. [DOI] [PubMed] [Google Scholar]
- 5.Pasqualucci L, Migliazza A, Fracchiolla N, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A. 1998;95(20):11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280(5370):1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 7.Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412(6844):341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 8.Liu M, Duke JL, Richter DJ, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451(7180):841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 9.Robbiani DF, Bothmer A, Callen E, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135(6):1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein I, Resch W, Jankovic M, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;30(1):95–106. doi: 10.1016/j.cell.2011.07.048. 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakim O, Resch W, Yamane A, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484(7392):69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141(1):27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein H, Bonk A, Tolksdorf G, Lennert K, Rodt H, Gerdes J. Immunohistologic analysis of the organization of normal lymphoid tissue and non-Hodgkin's lymphomas. J Histochem Cytochem. 1980;28(8):746–760. doi: 10.1177/28.8.7003001. [DOI] [PubMed] [Google Scholar]
- 14.Rademakers LH. Dark and light zones of germinal centres of the human tonsil: an ultrastructural study with emphasis on heterogeneity of follicular dendritic cells. Cell Tissue Res. 1992;269(2):359–368. doi: 10.1007/BF00319629. [DOI] [PubMed] [Google Scholar]
- 15.Hardie DL, Johnson GD, Khan M, MacLennan IC. Quantitative analysis of molecules which distinguish functional compartments within germinal centers. Eur J Immunol. 1993;23(5):997–1004. doi: 10.1002/eji.1830230502. [DOI] [PubMed] [Google Scholar]
- 16.Schwickert TA, Lindquist RL, Shakhar G, et al. In vivo imaging of germinal centres reveals a dynamic open structure. Nature. 2007;446(7131):83–87. doi: 10.1038/nature05573. [DOI] [PubMed] [Google Scholar]
- 17.Victora GD, Schwickert TA, Fooksman DR, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143(4):592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315(5811):528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 19.Hauser AE, Junt T, Mempel TR, et al. Definition of germinal-center B cell migration in vivo reveals predominant intrazonal circulation patterns. Immunity. 2007;26(5):655–667. doi: 10.1016/j.immuni.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Hummel M, Bentink S, Berger H, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354(23):2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 21.Cattoretti G, Buttner M, Shaknovich R, Kremmer E, Alobeid B, Niedobitek G. Nuclear and cytoplasmic AID in extrafollicular and germinal center B cells. Blood. 2006;107(10):3967–3975. doi: 10.1182/blood-2005-10-4170. [DOI] [PubMed] [Google Scholar]
- 22.Klein U, Tu Y, Stolovitzky GA, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194(11):1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 24.Floratos A, Smith K, Ji Z, Watkinson J, Califano A. geWorkbench: an open source platform for integrative genomics. Bioinformatics. 2010;26(14):1779–1780. doi: 10.1093/bioinformatics/btq282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 26.Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Machine Learning. 2003;52(1):91–118. [Google Scholar]
- 27.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 28.Califano A, Stolovitzky G, Tu Y. Analysis of gene expression microarrays for phenotype classification. Proc Int Conf Intell Syst Mol Biol. 2000;8:75–85. [PubMed] [Google Scholar]
- 29.Scheid JF, Mouquet H, Ueberheide B, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 31.Fyfe G, Cebra-Thomas JA, Mustain E, Davie JM, Alley CD, Nahm MH. Subpopulations of B lymphocytes in germinal centers. J Immunol. 1987;139(7):2187–2194. [PubMed] [Google Scholar]
- 32.Kremmidiotis G, Zola H. Changes in CD44 expression during B cell differentiation in the human tonsil. Cell Immunol. 1995;161(2):147–157. doi: 10.1006/cimm.1995.1021. [DOI] [PubMed] [Google Scholar]
- 33.Feuillard J, Taylor D, Casamayor-Palleja M, Johnson GD, MacLennan IC. Isolation and characteristics of tonsil centroblasts with reference to Ig class switching. Int Immunol. 1995;7(1):121–130. doi: 10.1093/intimm/7.1.121. [DOI] [PubMed] [Google Scholar]
- 34.Klein U, Tu Y, Stolovitzky GA, et al. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A. 2003;100(5):2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103(9):3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Longo NS, Lugar PL, Yavuz S, et al. Analysis of somatic hypermutation in X-linked hyper-IgM syndrome shows specific deficiencies in mutational targeting. Blood. 2009;113(16):3706–3715. doi: 10.1182/blood-2008-10-183632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caron G, Le Gallou S, Lamy T, Tarte K, Fest T. CXCR4 expression functionally discriminates centroblasts versus centrocytes within human germinal center B cells. J Immunol. 2009;182(12):7595–7602. doi: 10.4049/jimmunol.0804272. [DOI] [PubMed] [Google Scholar]
- 38.Kuppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N Engl J Med. 1999;341(20):1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- 39.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 40.Dave SS, Fu K, Wright GW, et al. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354(23):2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 41.Liu YJ, Zhang J, Lane PJ, Chan EY, MacLennan IC. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol. 1991;21(12):2951–2962. doi: 10.1002/eji.1830211209. [DOI] [PubMed] [Google Scholar]
- 42.Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. Analysis of somatic mutation in five B cell subsets of human tonsil. J Exp Med. 1994;180(1):329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hogerkorp CM, Borrebaeck CA. The human CD77- B-cell population represents a heterogeneous subset of cells comprising centroblasts, centrocytes, and plasmablasts, prompting phenotypical revision. J Immunol. 2006;177(7):4341–4349. doi: 10.4049/jimmunol.177.7.4341. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama Y, Stabach P, Maher SE, et al. A limited number of genes are involved in the differentiation of germinal center B cells. J Cell Biochem. 2006;99(5):1308–1325. doi: 10.1002/jcb.20952. [DOI] [PubMed] [Google Scholar]
- 45.Allen CD, Ansel KM, Low C, et al. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat Immunol. 2004;5(9):943–952. doi: 10.1038/ni1100. [DOI] [PubMed] [Google Scholar]
- 46.Kjeldsen MK, Perez-Andres M, Schmitz A, et al. Multiparametric flow cytometry for identification and fluorescence activated cell sorting of five distinct B-cell subpopulations in normal tonsil tissue. Am J Clin Pathol. 2011;136(6):960–969. doi: 10.1309/AJCPDQNP2U5DZHVV. [DOI] [PubMed] [Google Scholar]
- 47.Schwickert TA, Alabyev B, Manser T, Nussenzweig MC. Germinal center reutilization by newly activated B cells. J Exp Med. 2009;206(13):2907–2914. doi: 10.1084/jem.20091225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lennert K, Morhi N, Stein H, Kaiserling E. The histopathology of malignant lymphoma. Br J Hematol. 1975;31(Suppl):193–203. [Google Scholar]
- 49.Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5(4):251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 50.Kepler TB, Perelson AS. Cyclic re-entry of germinal center B cells and the efficiency of affinity maturation. Immunol Today. 1993;14(8):412–415. doi: 10.1016/0167-5699(93)90145-B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.