Abstract
We determined the prognostic relevance of CD25 (IL-2 receptor-α) expression in 657 patients (≤ 60 years) with de novo acute myeloid leukemia (AML) treated in the Eastern Cooperative Oncology Group trial, E1900. We identified CD25POS myeloblasts in 87 patients (13%), of whom 92% had intermediate-risk cytogenetics. CD25 expression correlated with expression of stem cell antigen CD123. In multivariate analysis, controlled for prognostic baseline characteristics and daunorubicin dose, CD25POS patients had inferior complete remission rates (P = .0005) and overall survival (P < .0001) compared with CD25NEG cases. In a subset of 396 patients, we integrated CD25 expression with somatic mutation status to determine whether CD25 impacted outcome independent of prognostic mutations. CD25 was positively correlated with internal tandem duplications in FLT3 (FLT3-ITD), DNMT3A, and NPM1 mutations. The adverse prognostic impact of FLT3-ITDPOS AML was restricted to CD25POS patients. CD25 expression improved AML prognostication independent of integrated, cytogenetic and mutational data, such that it reallocated 11% of patients with intermediate-risk disease to the unfavorable-risk group. Gene expression analysis revealed that CD25POS status correlated with the expression of previously reported leukemia stem cell signatures. We conclude that CD25POS status provides prognostic relevance in AML independent of known biomarkers and is correlated with stem cell gene-expression signatures associated with adverse outcome in AML.
Introduction
Increasingly, recurrent genetic aberrations govern the prognostication of the acute leukemias, including the large group of patients who present with acute myeloid leukemia (AML) without apparent cytogenetic abnormalities. A steadily growing catalog of molecular lesions has led to the development of outcome-based classification systems that incorporate information on perturbed pathogenetic pathways and inform us about potential therapeutic targets.1–4 In the context of this evolving molecular risk assessment, antigen-expression profiles have been identified as surrogates for certain leukemic genotypes. Furthermore, a few single antigens per se have been found to be predictive of clinical response. In most cases, however, the prognostic power of antigens has not been disassociated from underlying genetic determinants.
The prognostic impact of antigens, such as CD1a and CD13, in T-lineage acute lymphoblastic leukemia (ALL)5 has been linked to molecular subgroups with the activation of specific transcription factors and with unique gene expression signatures, reflecting distinct stages of maturation.6 In acute promyelocytic leukemia (APL), expression of the T cell–affiliated antigen, CD2, in association with S-isoform PML/RARα,7 confers inferior prognosis.8 CD2POSS-isoformPOS APL is distinguishable from other isoforms on the basis of gene-expression profiling (GEP)9 and has been posited to be derived from an immature leukemia stem cell (LSC).10
Surface expression of CD34, the sialomucin expressed universally by hematopoietic stem and progenitor cells,11 has been found to be a negative prognosticator in elderly patients with AML when present in > 10% of blasts, together with an elevated white blood cell (WBC) count, elevated lactate dehydrogenase, and advanced age.12 In an effort to assign patients < 60 years of age with AML to optimum postremission treatment, the same investigators13 found that the percentage of CD34POS blasts was significantly lower in patients with cytogenetically defined favorable-risk than with intermediate- or unfavorable-risk AML. In other studies, CD34 surface-marker expression correlated with the expression of BAALC and ERG, biomarkers with established prognostic relevance. Rockova et al found that high transcript levels of CD34 and ERG predicted independently for inferior overall survival (OS) in patients ages < 60 years with cytogenetically intermediate-risk AML.14
In a similar cohort of patients, Langer et al defined a close association between high BAALC expression and overexpression of genes involved in multidrug resistance or encoding stem cell markers CD34 and CD133.15 In contrast, low BAALC expressors in older patients with AML with a normal karyotype showed a down-regulation of genes expressed in less-differentiated precursor cells (including CD34),16 supporting the concept that BAALC is a marker of early hematopoietic progenitor cells. These data suggest a bystander role of the CD34 antigen, reflecting LSC features characterized by high BAALC and ERG expression, rather than an independent role in defining a biologically and prognostically relevant AML subset. In addition, the observation that NPM1-mutated (NPM1MUT) LSC are found both in CD34POS and CD34NEG fractions17 suggests that reduced CD34 expression does not define the relatively favorable prognosis of NPM1MUT AML.18 These data raise the question whether CD34 per se has relevance to prognosis in AML without linkage to gene expression patterns.
By contrast, we have observed that expression of CD25, the α-chain of the interleukin-2 receptor (IL-2RA), in BCR/ABLPOS B-lineage ALL is associated with adverse outcomes independent of established risk factors.19,20 Notably, rare patients with CD25NEGBCR/ABLPOS disease experienced an OS similar to that of BCR/ABLNEG B-lineage ALL, suggesting that CD25 has prognostic relevance independent of the presence or absence of BCR/ABL.
On the basis of our experience with CD25 in ALL, here we investigated whether CD25 affects outcome in de novo AML. Previously published data from small retrospective studies have suggested an unfavorable impact of CD25.21–23 In the largest group studied to date,22 CD25 was an independent adverse factor for the achievement of complete remission (CR), OS, and relapse-free survival in 65 patients with AML who were ≤ 60 years of age treated on protocols from the Hemato-Oncologie voor Volwassenen Nederland (HOVON) or Haemato Oncology Foundation for Adults in The Netherlands. CD25 expression was positively associated with the presence of internal tandem duplications in FLT3 (FLT3-ITD),22,23 a known adverse prognostic factor in AML,24 suggesting that CD25 might function as a surrogate marker for FLT3-ITD. In addition, the increased expression of CD34 in patients with CD25POS AML22 suggested the adverse effect of CD25 on outcome parameters might merely reflect a high degree of primitive differentiation of CD25POS blasts, consistent with biologic studies showing that CD25 is highly expressed by chemotherapy-resistant, cell-cycle quiescent LSC.25
We recently reported the analysis of somatically acquired gene mutations in a large phase 3 AML trial, E1900, which allowed us to develop and validate an integrated risk classification for patients ≤ 60 years of age in which we used cytogenetic and targeted mutational analysis to improve prognostication in AML.26 Here, we investigated the prognostic impact of CD25 in the same E1900 cohort to assess whether this antigen independently contributes to outcome in AML in the context of integrated mutational and cytogenetic analyses. In addition, we asked whether GEP would identify a specific gene cluster for CD25POS AML.
Methods
Patient samples
Diagnostic bone marrow and/or peripheral blood samples from 657 de novo patients with AML who were ≤ 60 years of age enrolled in the phase 3 Eastern Cooperative Oncology Group (ECOG) trial, E1900, were collected by the ECOG Leukemia Translational Studies Laboratory.27 Morphology and cytogenetics were evaluated centrally. “Normal karyotype AML” was defined by ≥ 20 normal bone marrow metaphases; negative interphase FISH for aberrations of chromosomes 5, 7, 8, and split MLL; and negative FISH and PCR for leukemia transcripts AML1/ETO, PML/RARα, BCR/ABL, and CBFβ/MYH11. Intermediate-risk AML included patients with a normal or indeterminate karyotype. Favorable and unfavorable cytogenetic risk groups were defined as reported.28 The integrated risk classification26 was used as the final classifier for outcome analysis. All patients provided written informed consent according to the Declaration of Helsinki for these studies, which were approved by the investigators' institutional review boards.
Flow cytometric determination of CD25 and other stem cell antigen expression
Expression of CD25, CD34, CD133, CD123, CD122, CD132, and P-glycoprotein (MRK-16; Kamiya, Seattle, WA) on gated leukemic myeloblasts was assessed in 4-color flow cytometry with the use of a FACSCalibur flow cytometer with CellQuest software (Becton Dickinson).29 Expression of all but CD133 and CD123 is reported as percentage of antibody-binding leukemic myeloblasts. For CD133 and CD123, mean fluorescence intensity (MFI) of specific antibody staining divided by the MFI of the isotype control was used (MFI ratio). The presence of a distinct CD25POS myeloblast population (range, 19%-99%) defined CD25 positivity (CD25POS). Undifferentiated, CD65(s)LOW,30 and CD11bPOS AML were characterized as previously reported.31
Mutational analyses and GEP
Mutational data for 18 genes have been reported previously and were included in this analysis.32 Gene expression data were obtained with the Roche-NimbleGen 60mer microarrays (Design name: 2006-10-26_Human_60mer_1in2; design ID = 4806).33 Raw data were processed using the RMA algorithm found in the oligo package (version 1.8.3) in BioConductor.34 Data have been deposited to NCBI's Gene Expression Omnibus and are accessible through GEO Series accession no. GSE24505 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24505).
Statistical analysis
Patient baseline characteristics were compared by the use of the Fisher exact test if they were categorical and Wilcoxon rank sum tests if they were continuous. Early death rate was defined as death within 28 days of starting induction therapy. OS was defined as time from randomization to death from any cause. OS probabilities were estimated by use of the Kaplan-Meier method, and statistical significance of associations was assessed with the log-rank test. Univariate models for OS included CD25 status and treatment arms. Multivariate models were further controlled for age, sex, presenting WBC count, cytogenetic risk, ECOG performance status, platelets, percentage of bone marrow and peripheral blood blasts, hemoglobin, and history of secondary AML. A classification tree with 10-fold cross-validation was used to explore the mutation profiles that are likely to have CD25 expression. All P values were based on 2-sided tests. Significance tests evaluating the associations between CD25 status, and mutations were adjusted for multiplicity using Resampling (http://www.resample.com/).
Antigen expression data are described by use of descriptive statistics of observed values, such as medians and 25th and 75th quartiles (interquartile range [IQR]) of the data. The percentages of antigen expressing blast cells or density of expression (for CD133 and CD123) were considered continuous variables and compared with the nonparametric Wilcoxon rank sum test.
Supervised analysis of gene expression microarrays was performed using a moderated t test followed by Benjamini-Hochberg adjustment for multiple testing. Differentially expressed genes were chosen at a fold-change > 2 and adjusted P < .05.
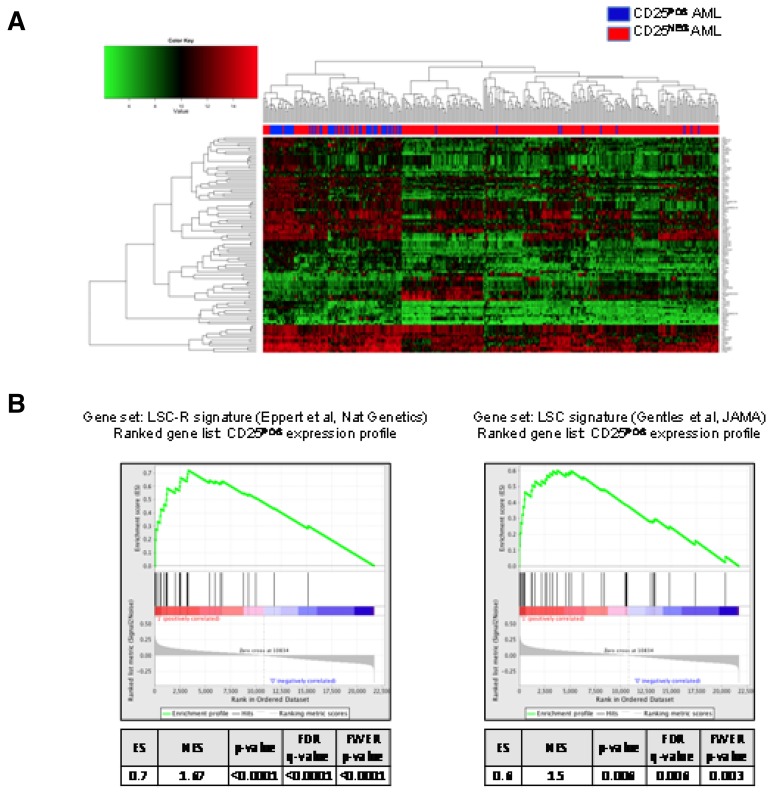
For gene set enrichment analysis, previously reported LSC signatures34,35 were downloaded and used as gene sets to perform gene set enrichment analysis.36 GSEA Version 2.7 (http://www.broadinstitute.org/gsea/index.jsp) was used to examine the association between the CD25 gene expression profiles and the LSC signatures. Gene sets with < 10 or > 500 genes were excluded, and significantly enriched gene sets after 1000 permutations at a FDR of < 0.25 are reported. All statistical analyses were performed using R 2.14 (http://www.r-project.org).
Results
Associations of CD25 expression with baseline characteristics and response in the entire E1900 cohort
Among eligible patients enrolled in E1900, 87 (13%) had CD25POS blasts (median, 59%; IQR, 43%-90%). Regarding myeloid maturation stage, the incidence of undifferentiated (P = .52) and differentiated AML (P = .23) did not differ by CD25 expression. CD11bPOS AML was more common in CD25POS patients (29% vs 16.6% of immunophenotypes, respectively, P = .012). Patients with CD25POS AML presented with variable morphology, including minimally differentiated (10%), without maturation (33%), with maturation (34%), or myelomonocytic (20%) by World Health Organization criteria.37 Expression of CD34 (CD25NEG: median, 95%; IQR, 2.99; CD25POS: median, 49%; IQR, 21.99; P = .89), CD133 (CD25NEG: median MFI ratio, 4.5; IQR 1.4, 10.9; CD25POS: median MFI ratio, 5.2; IQR 1.5, 12.9; P = .53), or P-glycoprotein (median, 26%; IQR 10.3, 58.8; CD25POS: median, 24%; IQR 12, 48; P = .89) was not significantly correlated with CD25. However, the intensity of staining (a reflection of antigen density) for CD123, IL-3Rα, was greater in CD25POS (median MFI ratio, 85; IQR, 50 127) than CD25NEG blasts (median MFI ratio, 27.5; IQR 13, 48.5; P < .0001). CD25POS leukemic myeloblasts lacked expression of the IL-2Rβ (CD122), although they weakly expressed the IL-2Rγ chain (CD132).
CD25POS patients did not differ in age from CD25NEG patients but presented with greater WBC counts (P < .0001) and greater percentages of circulating blasts (P = .001; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The distribution of cytogenetic risk classes was significantly different between the 2 cohorts (P < .0001) in that the majority of CD25POS patients presented with intermediate-risk cytogenetics (92%).
Forty-four percent of CD25POS patients received 45 mg/m2/d standard-dose daunorubicin (SDD), and 51% received 90 mg/m2/d high-dose daunorubicin (HDD) during induction therapy (P = .25). Irrespective of the dose of daunorubicin (P = .27), the CR rate was lower in CD25POS patients (overall: 47.1%; SDD, 36.7%; HDD, 60.5%) than in CD25NEG patients (overall: 67.4%; SDD, 62.5%; HDD, 72.1%) in univariate (P = .0005) and multivariate analyses (P = .0005). The early death rate was greater in CD25POS (6.9%) than CD25NEG patients (2.6%, P = .04). CD25POS patients receiving SDD had a greater early death rate than CD25NEG patients (10.2% vs 1.4%, P = .003) in univariate logistic models, but this was not the case with patients receiving HDD (2.6% vs 3.8%, P = .72).
At 4.5 years' median follow-up, in patients who were still living CD25 positivity was associated with worse OS in univariate (hazard ratio 2.31, 95% confidence interval 1.80-2.96, P < .0001) and multivariate analyses (hazard ratio 2.74, 95% confidence interval 2.06-3.63, P < .0001; supplemental Figure 1) when we controlled for prognostic baseline characteristics and the dose of daunorubicin. There was no correlation between the percentage of CD25POS blasts and OS (P = .24). Eleven CD25POS patients underwent protocol-defined autologous stem cell transplantation (SCT). These patients had a median OS of 0.9 years compared with 142 CD25NEG transplantation cases in whom median survival has not yet been reached (P = .001).
Frequency of mutational alterations in CD25POS AML and risk-altering effect of CD25 expression in previously defined risk groups
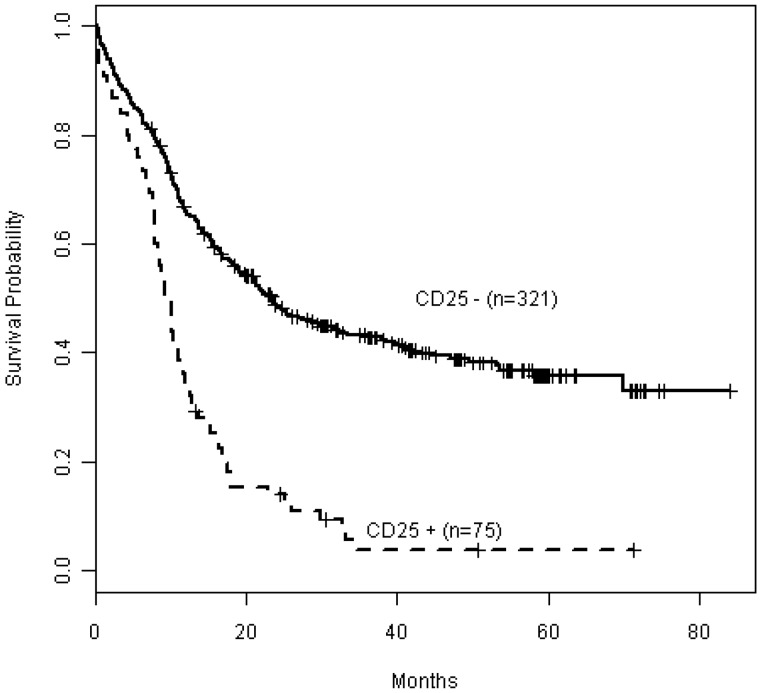
We have previously reported data from detailed mutational profiling in a subset of 396 E1900 patients; this subgroup was comparable with the entire E1900 cohort with respect to baseline characteristics, including cytogenetic risk groups.26 Table 1 summarizes the baseline demographics and clinical characteristics of this cohort with detailed mutational analysis by CD25 status. WBC count (P = .03) and distribution of cytogenetic risk classes (P < .001) differed significantly between CD25NEG and CD25POS patients, as seen in the entire cohort (supplemental Table 1). Among patients with detailed mutational profiling data, 75 (19%) were CD25POS. As seen in the entire E1900 patient population, CD25 expression also affected adversely OS in this subgroup (P < .001; Figure 1). The cohort with mutation data included 1 patient with core-binding factor leukemia (favorable-risk); 4 patients with unfavorable cytogenetics (including deletion 5q, and 7q, in 1 patient each, and 2 with complex karyotypes); and 6 patients with unsuccessful karyotyping. The remaining 64 patients had intermediate-risk cytogenetics (85%), including 54 with normal karyotype (72%). Importantly, CD25 expression was strongly associated with adverse outcome in these cytogenetically intermediate-risk patients (P < .001; supplemental Figure 2).
Table 1.
Baseline demographic and clinical characteristics by CD25 expression in the 396 patients with detailed mutational analysis
| CD25NEG (n = 316) | CD25POS (n = 80) | P | |
|---|---|---|---|
| Age, y | 47 (18-60) | 46 (18-60) | .39 |
| WBC, 1000/mm3 | 18.5 (0.6-212.8) | 24.8 (1.0-181.0) | .03 |
| Hemoglobin, g/dL | 9.2 (5-30) | 9.2 (5-11.7) | .51 |
| Peripheral blasts, % | 45 (0-98) | 52.5 (0-92) | .55 |
| Platelets, 1000/mm3 | 51 (0.7-650) | 48 (1.4-261) | .54 |
| Marrow blast | 68 (3-100) | 65 (8-99) | .61 |
| Female sex, no. patients, % | 146 (46) | 43 (54) | .26 |
| Cytogenetics favorable, no. patients, % | 67 (23) | 4 (5) | < .001 |
| Cytogenetics intermediate, no. patients, % | 171 (58) | 63 (85) | |
| Cytogenetics unfavorable, no. patients, % | 59 (20) | 7 (10) |
All values are median (range) unless otherwise noted.
AML indicates acute myeloid leukemia; and WBC, white blood cell count.
Figure 1.
Kaplan-Meier estimates of OS by CD25 expression in the patient cohort with detailed mutational data (n = 396). CD25NEG (CD25−) patients are depicted in the solid curve (n = 321), whereas CD25POS (CD25+) patients are depicted as a dashed curve (n = 75).
Somatic mutations were present in 94.7% of CD25POS patients (supplemental Figure 3). CD25 expression correlated with the presence of FLT3-ITD (FLT3-ITDPOS; 76%, P < .001), DNMT3A (DNMT3AMUT; 44%, P < .001) and NPM1 (NPM1MUT; 46%, P = .001) mutations, and with partial tandem duplications of the MLL gene (MLL-PTDPOS; 11%, P = .016, with adjustment for multiple comparisons). CD25 expression was negatively associated with core-binding factor leukemias, IDH or CEBPA mutations, and with MLL translocations (defined as split MLL by FISH; Table 2). All FLT3-ITDPOS patients without a FLT3 wild-type (FLT3WT) allele were in the CD25POS group (P < .001). For CD25POS cases, 82% of DNMT3A mutations occurred in codon R882. There was no correlation of CD25 with WT1 (12%), RUNX1 (9.3%), or TET2 (8%) mutations. Mutations in other genes were detected in ≤ 4 patients.
Table 2.
Frequencies and distribution of genetic abnormalities by CD25 expression in the subgroup with E1900 patients with mutational analysis
| Mutations | CD25NEG (n = 321), no. (%) | CD25POS (n = 75), no. (%) | P |
|---|---|---|---|
| DNMT3A | 55 (18) | 33 (44) | < .001 |
| DNMT3A R882 | 36 (12) | 27 (36) | < .001 |
| DNMT3A other | 21 (7) | 6 (8) | .801 |
| IDH1 or IDH2 | 51 (16) | 5 (7) | .043 |
| TET2 | 28 (9) | 5 (7) | .695 |
| FLT3-ITD | 63 (20) | 57 (76) | < .001 |
| FLT3-TKD mutated | 23 (7) | 4 (5) | .79 |
| NPM1 | 83 (26) | 35 (46) | .001 |
| PHF6 | 7 (2) | 2 (3) | .688 |
| KIT | 22 (7) | 1 (1) | .096 |
| CEBPa | 32 (10) | 2 (3) | .040 |
| WT1 | 21 (7) | 9 (12) | .149 |
| KRAS | 6 (2) | 2 (3) | .654 |
| NRAS | 37 (12) | 3 (4) | .055 |
| TP53 | 6 (2) | 2 (3) | .654 |
| PTEN | 4 (2) | 2 (3) | .326 |
| RUNX1 | 13 (4) | 7 (9) | .090 |
| CBF | 42 (13) | 1 (1) | .002 |
| del(5q) | 11 (3) | 1 (1) | .476 |
| EVI1pos* | 7 (2) | 1 (1) | 1.000 |
| MLL-PTD | 11 (3) | 8 (11) | .016 |
| Split MLL* | 25 (8) | 0 (0) | .007 |
| del(7q) of monosomy 7† | 8 (3) | 3 (1) | 1.000 |
| Trisomy 8† | 13 (4) | 6 (8) | .227 |
| Complex karyotype‡ | 29 (9) | 2 (3) | .091 |
CBF indicates core-binding-factor leukemias (carrying transcripts AML1/ETO or CBFβ/MYH11) that were detected by OCR and FISH; ITD, internal tandem duplication; PTD, partial tandem duplication; TKD, tyrosine kinase domain.
EVI1 and chromosome translocation resulting into a split of the MLL gene (split MLL) were assessed by FISH.
del(7q)/monosomy 7 and trisomy 8 were assessed by standard cytogenetics or FISH.
Complex karyotypes were defined as ≥ 3 clonal cytogenetic aberrations.
After classification via the use of integrated genetic profiling,26 we found that all but 1 CD25POS patient with invalid cytogenetics could be risk classified because they presented with unfavorable-risk mutational genotypes, is, FLT3-ITDPOSDNMT3AMUT in 4 and FLT3-ITDPOSMLL-PTDPOS in 1. Of the resulting 74 CD25POS patients with detailed mutational data and assigned risk (combining cytogenetics and mutational profiling), only 1 patient had favorable-risk AML (1%), whereas 39% of CD25POS patients had intermediate-risk disease and 60% had unfavorable-risk disease. This risk distribution differed significantly (P < .001) from that found in CD25NEG patients, which could be classified relatively equally into favorable-risk (33%), intermediate-risk (37%), and unfavorable-risk (31%) subgroups.
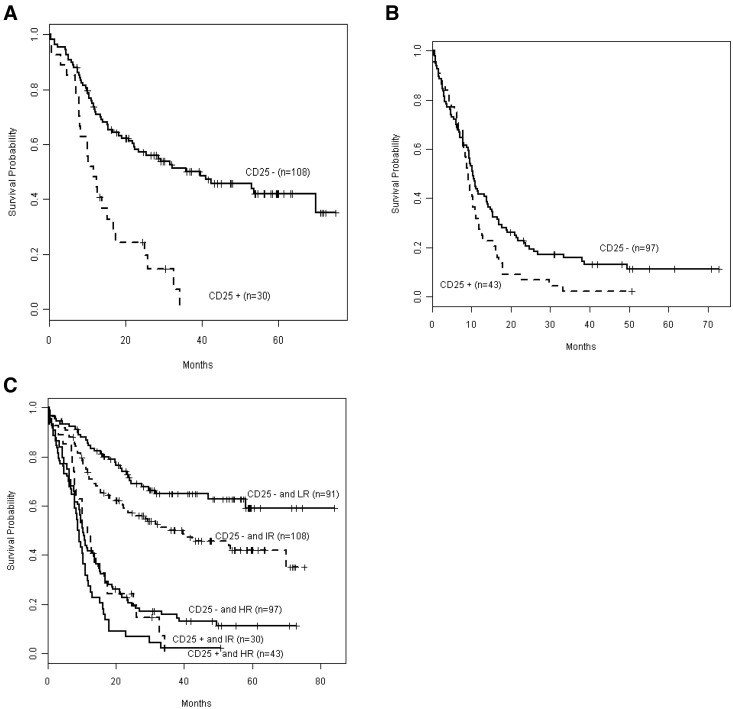
Next, we integrated CD25 status with outcome in genetically defined intermediate- and unfavorable-risk subgroups (Figure 2). CD25 expression was associated with adverse outcome both in patients with integrated intermediate risk (P < .001; Figure 2A) and unfavorable risk (P = .038; Figure 2B). A comparison of OS by CD25 in favorable (low), intermediate, and unfavorable (high) risk (Figure 2C) showed that OS in CD25NEG unfavorable-risk, CD25POS intermediate-risk, and CD25POS unfavorable-risk patients was not significantly different (P = .091). These 3 groups mark a patient cohort with particularly poor prognosis, and their OS is significantly worse than that of CD25NEG favorable-risk patients (P < .001) as well as CD25NEG intermediate-risk patients (P < .001).
Figure 2.
Kaplan-Meier estimates of OS according to integrated-risk status, based on cytogenetic and mutational classification, in CD25POS versus CD25NEG patients. Data are shown for the OS of patients with intermediate risk (A; CD25NEG n = 108, CD25POS n = 30) or (B) with unfavorable (high) risk (CD25NEG n = 97, CD25POS n = 43). (C) The composite shows that OS in CD25POS intermediate risk (IR) patients was not significantly different from that in CD25POS or CD25NEG high-risk (HR) patients (P = .091). Solid lines depict CD25NEG patients, and dashed lines depict CD25POS patients. Only 1 CD25POS patient was low risk (LR).
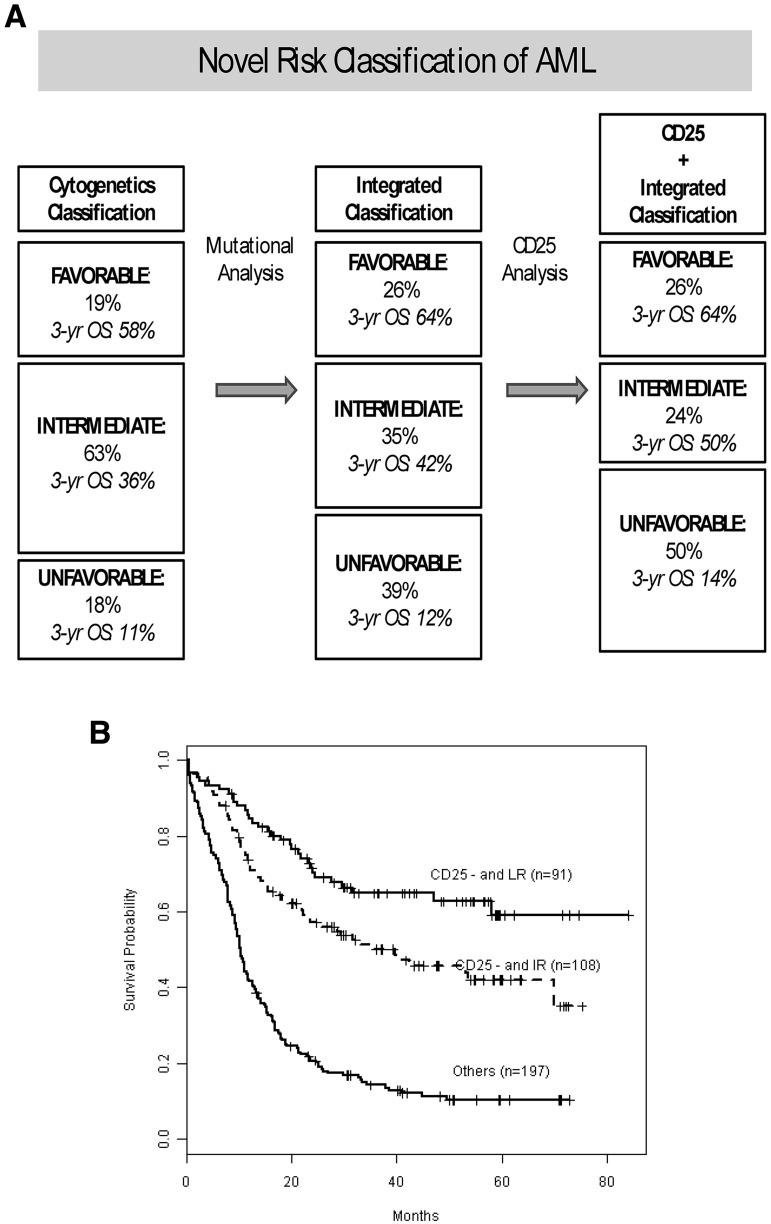
Cytogenetic and mutational risk classification remains important prognostically because CD25NEG favorable-risk patients did significantly better than CD25NEG intermediate-risk patients (P = .013). We then investigated how CD25 expression further improves the integrated prognostic risk classification that we have previously published26 for this cohort of E1900 patients with detailed mutational data. Figure 3A shows our previously published risk-stratification algorithm,26 which we have now updated for CD25 expression. CD25 positivity had no effect on the stratification of favorable-risk patients because of the rare incidence of CD25 expression in favorable-risk patients. By contrast, 11% of patients with intermediate risk, by combined cytogenetic and mutational risk classification, were reclassified as unfavorable risk on the basis of their CD25 expression. The corresponding survival curves are presented in Figure 3B. Patients with integrated favorable risk who were CD25NEG (n = 91) had a 3-year survival of 65%; patients with integrated intermediate risk and who were CD25NEG (n = 108) had a 3-year survival of 50% (increased from 42%); unfavorable-risk patients (with integrated cytogenetic and/or mutational unfavorable-risk or CD25POS; n = 197) had a 3-year survival of 14%.
Figure 3.
Effect of CD25 cell-surface marker expression on the integrated (combined cytogenetic and mutational) risk classification of AML. (A) Effect of CD25 on our previously published risk allocation that was determined by mutational profiling and redefined a substantial proportion of patients who by cytogenetic analysis (left) carried intermediate risk to favorable risk or unfavorable risk categories (middle).26 When we incorporated CD25 expression into the integrated (combined cytogenetic and mutational status) risk stratification algorithm, an additional 11% of intermediate-risk patients were reallocated to the unfavorable-risk class. The favorable-risk group was unaffected by CD25 because of the rare occurrence of CD25 in favorable-risk patients. In each risk category, the percentage of patients in that cohort and their 3 years' OS are given. (B) Kaplan-Meier estimates of OS for the final risk groups, stratified by integrated risk (cytogenetic and mutational) and CD25 expression status.
Effect of CD25 expression in FLT3-ITDPOS AML
Given the high concordance between CD25 expression and the presence of the FLT3-ITD allele, we investigated the prognostic relevance of CD25 in the presence and absence of FLT3-ITD mutations. Using a cross-validated classification-tree, we estimated that the likelihood of an AML case expressing CD25 was extremely low (5.6% of 248 patients) for FLT3WT patients, moderate (32% of 96 patients) for FLT3-ITDPOSDNMT3AWT patients, and high (58% of 52 patients) for FLT3-ITDPOSDNMT3AMUT patients (P < .001). Regardless, however, we found that CD25 was prognostically relevant both in FLT3-ITDPOSDNMT3AWT and FLT3-ITDPOSDNMT3AMUT patients (P < .001 in both groups).
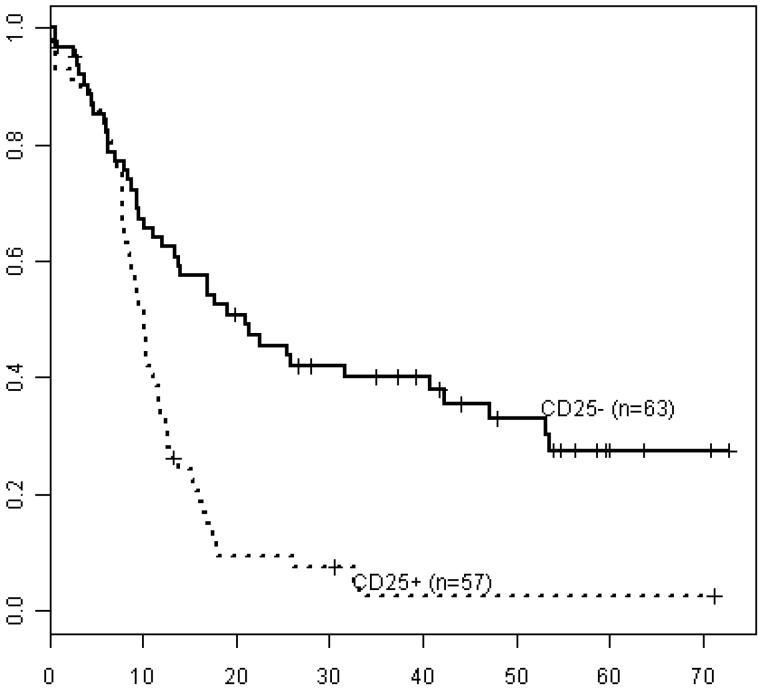
CD25POSFLT3-ITDPOS patients had shorter OS (median OS 10 months, 3-year survival 4%) than CD25NEGFLT3-ITDPOS patients (median OS 25 months, 3-year survival 42%; Figure 4, P < .001), irrespective of cytogenetic risk. Notably, OS in CD25NEGFLT3-ITDPOS patients did not significantly differ from OS in CD25NEGFLT3WT patients (median OS 23 months, 3-year survival 43%; P = .25; Figure 5). In addition, within the subset of patients with cytogenetically defined intermediate-risk disease, CD25POSFLT3-ITDPOS patients had significantly worse OS than CD25NEGFLT3-ITDPOS patients (P < .001), whereas OS in CD25NEGFLT3-ITDPOS and CD25NEGFLT3WT patients was not significantly different (P = .58).
Figure 4.
Kaplan-Meier estimates of OS for FLT3-ITD−positive patients in all integrated risk groups by CD25 status. Data are shown for FLT3-ITD–positive/CD25NEG (CD25−; solid line, n = 63) vs FLT3-ITD−positive/CD25POS patients (CD25+; dashed line, n = 57).
Figure 5.
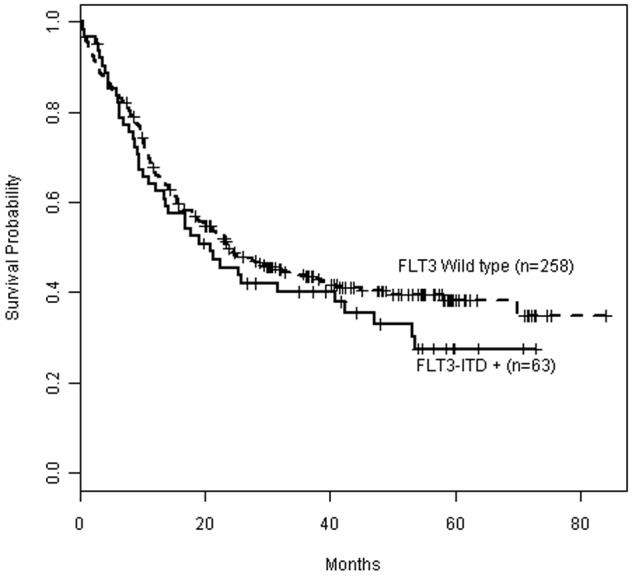
Kaplan-Meier estimates of OS by FLT3 mutation status in CD25NEG patients. Data are shown for overall survival in CD25NEG patients with FLT3 wild type (dashed line, n = 258) compared with CD25NEG patients positive for FLT3-ITD (solid line, n = 63).
We noted that 20 of 74 CD25POS patients (27%) were FLT3-ITDPOS and concurrently carried mutations both in NPM1 and DNMT3A. In comparison, only 12 of 321 (4%) CD25NEG patients (P < .001) had this complex genotype. CD25POS patients with mutations in these 3 genes had poorer OS than CD25POS patients not carrying these mutations (P < .001). In contrast, OS in CD25NEG FLT3-ITDPOSDNMT3AMUTNPM1MUT patients did not differ from that in other CD25NEG patients (P = .623).
Prognostic effect of CD25 in relation to daunorubicin dose intensification
Our previous analysis had demonstrated that dose-intensified daunorubicin induction selectively improved outcome in E1900 patients with DNMT3A mutation, NPM1 mutation, or split MLL abnormalities (by FISH).26 Among the 396 patients with mutational analysis, 195 were treated with SDD and 201 with HDD. CD25NEG patients treated with SDD (n = 157) had a 3-year survival of 41% and median OS of 19 months; when treated with HDD (n = 164), their 3-year survival was 45% with a median OS of 26 months (P = .077). CD25POS patients treated with SDD (n = 38) had a 3-year survival of 0% and OS of 9 months, compared with 8% and 12 months when treated with HDD (n = 37; P = .002).
When we considered the effects of daunorubicin dose with respect to CD25 expression on outcome in the context of somatic mutations, we found that outcome in CD25NEG patients without DNMT3A mutation, NPM1 mutation, or split MLL abnormality by FISH was comparable with HDD (median OS 24 months, 3-year survival 40%) and SSD (median OS 25 months, 3-year survival 45%; P = .931; supplemental Figure 4A). However, HDD significantly improved OS in CD25NEG patients with at least 1 mutation in DNMT3A or NPM1 or with split MLL abnormality (median OS not yet reached, 3-year survival 54%) compared with SDD (median OS 17 months, 3-year survival 36%; P = .006; supplemental Figure 4B). These observations confirmed those seen in the entire E1900 cohort.26 Within the subset of CD25POS patients, HDD treatment resulted in improved survival (median OS 17 months, 3-year survival 0%) over SDD (median OS 8 months, 3-year survival 0%), particularly in cases without DNMT3A or NPM1 mutation or split MLL abnormality (P = .002; supplemental Figure 4C). There was a trend toward improved outcome with HDD in CD25POS patients with DNMT3A or NPM1 mutations (none of the split MLL cases was CD25POS; median OS 10 months, 3-year survival 13%) compared with SDD (median OS 9 months, 3-year survival 0%; P = .067, supplemental Figure 4D).
Gene expression signature in CD25POS AML
GEP analysis was available for 323 patients, of which 61 were CD25POS and 262 were CD25NEG. These 61 CD25POS patients with GEP data did not differ significantly in median age (41 vs 48 years); sex (60% female vs 59%); the incidence of favorable (2% in both groups), intermediate (79% vs 69%) or unfavorable cytogenetics (6% in both groups); or incidence of FLT3-ITDPOS cases (69% vs 64%) from all CD25POS E1900 cases, respectively. Direct comparison of the gene expression profiles of these 2 groups revealed a 77-gene signature that distinguished between the subsets (Figure 6A; supplemental Table 2). Notably, this signature did not include genes in the IL-2 signaling pathway other than CD25 (IL-2RA) itself. By contrast, gene set enrichment analysis revealed that the CD25POS gene expression signature was significantly enriched in genes for leukemia stem cell profiles reported to be associated with increased engraftment potential in mice and with unfavorable clinical outcome (false discovery rate < 0.0001 and = 0.006, respectively; Figure 6B).34,35 We next defined a 71-gene expression signature in the 51 CD25POS patients with intermediate-risk cytogenetics. Although there was significant overlap between the 2 signatures (42 genes, P < .0001), including the up-regulation of HOX genes, there were also a set of unique genes that were differentially expressed in CD25POS patients with intermediate-risk cytogenetics. These unique genes included IL-3R (CD123) and ERG, which were up-regulated in intermediate-risk CD25POS patients.
Figure 6.
GEP of CD25POS AML in all integrated risk groups. (A) 2-dimensional hierarchical clustering of differentially expressed genes between CD25POS (blue) and CD25NEG (red) AML cases. Each column represents a patient, and each row represents a gene. (B) Gene set enrichment analysis using previously reported leukemia stem cell signatures as gene sets showing significant enrichment of the 2 reported signatures in CD25 expression profiles.
Discussion
Using a large cohort of AML patients ≤ 60 years of age who were treated on a single ECOG trial, we confirm data from smaller, retrospective studies21–23 and demonstrate that CD25 antigen expression is associated with adverse outcome in AML. In addition, we reveal that CD25 expression adds independent adverse prognostic relevance to the integrated cytogenetic-mutational risk classification26 in patients with genetically defined intermediate-risk or unfavorable-risk disease.
CD25POS myeloblasts were found predominantly in cytogenetically intermediate-risk AML (92%), mostly with normal karyotype. In accordance with previous observations,22,23 we found a high frequency of FLT3-ITDPOS cases in CD25POS AML (76% of cases), but we failed to confirm an association of CD25 expression with the putative stem cell markers CD34, CD133, or P-glycoprotein. In addition, CD25POS cases failed to demonstrate immunophenotypic features of undifferentiated (CD65NEG) AML more frequently than CD25NEG cases. Interestingly, CD11bPOS AML, a phenotype associated with shortened survival, at least in elderly patients,31 was more common in CD25POS disease. Unexpectedly, we found a significant association of CD25 with expression of CD123, the IL-3Rα, a marker shown to be expressed in LSC.11
We observed that CD25 expression was associated with a reduced response to induction chemotherapy, irrespective of the dose of daunorubicin. The adverse effect on CR rate of CD25 expression is in contrast to other poor risk markers, including FLT3-ITDPOS 24 or DNMT3AMUT 38,39 cases, which are associated with an increased risk of relapse but not with a reduced CR rate in patients with AML.
CD25 expression was uncommon in patients with cytogenetically or mutationally defined favorable-risk disease (1% incidence). Although only 5% of CD25POS patients presented with unfavorable cytogenetics, 54.7% of CD25POS cases had intermediate-risk cytogenetics and mutationally defined unfavorable-risk disease. These data demonstrate CD25POS patients with intermediate-risk cytogenetics have a greater likelihood of harboring unfavorable-risk mutations compared with CD25NEG cytogenetically intermediate-risk patients. It is important to emphasize, however, that CD25 expression was associated with adverse outcome in both genetically defined intermediate- and unfavorable-risk disease, providing clear evidence that CD25 independently impacts outcome in the majority of patients with AML.
We previously demonstrated with integrated (combined cytogenetic and mutational) profiling that 35% of E1900 patients with detailed mutational analysis have an intermediate risk of relapse compared with 63% on the basis of cytogenetic risk alone.26 The incorporation of CD25 status into this risk classification reduced the proportion of intermediate-risk patients further to 24% by reallocating 11% of intermediate-risk patients into the unfavorable-risk category, whereas the fraction of favorable patients remained the same. The remaining CD25NEG intermediate-risk patients had a 3-year OS of 50% compared with 42% in the integrated intermediate-risk group (Figure 3A). Taken together, these data show that CD25 identifies a subgroup of AML patients with cytogenetically and mutationally intermediate risk who have unfavorable outcomes and should be considered for stem cell transplantation and for novel investigative therapies. Subsequent studies are needed to validate this observation and to determine whether CD25 expression has additive prognostic value in other AML subsets, including older patients.
The most significant effect of CD25 on outcome was seen in the FLT3-ITDPOS category. FLT3-ITDPOS E1900 patients lacking a FLT3 wild-type allele, a genotype conferring particularly poor prognosis,40 were exclusively CD25POS. Importantly, CD25NEGFLT3-ITDPOS patients fared equally well as CD25NEGFLT3WT patients (Figure 5), suggesting that lack of CD25 expression outweighs the well-known adverse prognostic effect of the FLT3-ITD mutation. This finding suggests that CD25 status should be considered an important covariate in the selection of patients for therapeutic trials with FLT3-kinase inhibitors.
Previous studies in the E1900 cohort demonstrated an overall benefit for dose-intensified daunorubicin induction in the entire E1900 cohort and particularly in patients with DNMT3A mutation, NPM1 mutation, or MLL involving translocations.26,27 However, our current data suggest that HDD at induction may provide incremental benefit to all CD25POS patients, irrespective of underlying mutations, whereas benefit from HDD in CD25NEG patients may be limited to patients with DNMT3A mutation, NPM1 mutation, or MLL involving translocations.
Although HDD followed by autologous SCT conferred excellent prognosis in intermediate-risk E1900 patients,41 SCT did not benefit the subset of CD25POS patients who were treated with transplantation compared with CD25NEG patients. Additional studies are needed to validate this observation and to determine the role of SCT in CD25POS patients.
Although we demonstrate that CD25 surface expression has independent prognostic relevance in AML, the biologic basis for this observation has not been elucidated. We did not detect expression of the high-affinity IL-2R (IL-2Rα + β + γ chains) on the surface of CD25POS blasts. The lack of a functional IL2R and the absence of an IL-2 signaling signature in CD25POS AML argue against a role for the IL-2 signaling pathway in the pathogenesis of CD25POS AML or in impacting outcome in CD25POS AML. Terwijn et al reported that minimal residual disease levels after induction chemotherapy were significantly greater in CD25POS than CD25NEG AML.22 Similarly, the presence of a high frequency of LSCs at diagnosis has been correlated with high levels of minimal residual disease after chemotherapy.42
We hypothesize that CD25 expression in AML serves as a surrogate for the presence of chemoresistant LSCs. Consistent with this notion, the molecular signature of CD25POS myeloblasts included the expression of genes previously described in LSC signatures.34,35 Human LSCs have been shown to migrate to an endosteal region rich in osteoblasts after xenotransplantation into immunocompromised mice.43 Localization to this endosteal niche was associated with resistance to chemotherapeutic agents.43 Interestingly, CD25POS human LSCs were highly enriched in the endosteal niche, were predominantly in the G0 phase of the cell cycle, and displayed low Ki67 staining consistent with increased quiescence.25 Hence, CD25POS AML cells appear to represent a quiescent, chemotherapy-resistant population of LSCs residing preferentially in a privileged compartment within the bone marrow.
It is possible that signals from this microenvironment might be involved in up-regulation of IL-2RA in these cells. When GEP analysis was performed in CD25POS intermediate-risk patients, we observed increased expression of IL-3R (CD123) and of ERG in this subgroup. High-density expression of CD123 on leukemic CD25POS myeloblasts is of biologic and therapeutic interest, particularly in view of previous biologic studies showing that CD123 expression is a typical feature of LSCs in AML but not of normal stem cells.11 The finding of ERG up-regulation raises the intriguing question as to whether CD25 positivity in normal karyotype/intermediate-risk AML may serve as a surrogate marker for ERG gene expression; the relative impact of CD25 and ERG on outcome in AML will require studies in large, intermediate-risk AML cohorts.
Taken together, our data demonstrate an independent, clinically relevant role for CD25 expression in AML and provide an algorithm to use CD25 surface expression as an additional biomarker to improve prognostication in AML. Although the role of CD25 expression in AML pathogenesis, chemoresistance, and self-renewal requires further investigation, we believe our data provide a rationale for incorporating CD25 assessment into current prognostic schema in AML.
Supplementary Material
Acknowledgments
The authors thank Kerrie O'Shea and Xerxes Vevai for their excellent technical assistance.
This work was supported by grants from the National Cancer Institute CA143798 and CA138834 awarded to M.G.; CA021115 and CA114737 awarded to E.P.; and the Leukemia and Lymphoma Society Special Fellow Award and Doris Duke Charitable Foundation Clinical Scientist Development Award to M.E.F. J.P.P. is supported by an ASH Trainee Research Award; O.A.-W. is an ASH Basic Research Fellow; A.M. is funded by a Leukemia and Lymphoma Society SCOR and TRP award, is a Burroughs Wellcome Clinical Translational Scholar, and Scholar of the Leukemia Lymphoma Society; A.M. and R.L.L. are supported by a grant from the Starr Cancer Consortium; and R.L.L. is an Early Career Award Recipient of the Howard Hughes Medical Institute and is a Scholar of the Leukemia Lymphoma Society and is supported by a grant of the Gabrielle's Angel Fund.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.E.F., J.P.P., O.A.-W., J.R., A.M., R.L.L., and E.P. designed the experiments, conducted experimental procedures, and analyzed the data; M.G. and Z.S. performed statistical analyses; R.P.K. reviewed cytogenetics; H.F., J.M.R., and M.S.T. reviewed patient data; and M.G., Z.S., M.E.F., A.M., R.L.L., and E.P. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elisabeth Paietta, PhD, Montefiore Medical Center-North Division, Albert Einstein College of Medicine, Bronx, NY 10466; e-mail: epaietta@earthlink.net.
References
- 1.Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350(16):1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 2.Mrózek K, Marcucci G, Paschka P, et al. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007;109(2):431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 4.Santamaría CM, Chillón MC, García-Sanz R, et al. Molecular stratification model for prognosis in cytogenetically normal acute myeloid leukemia. Blood. 2009;114(1):148–152. doi: 10.1182/blood-2008-11-187724. [DOI] [PubMed] [Google Scholar]
- 5.Marks DI, Paietta E, Moorman AV, et al. T-cell acute lymphoblastic leukemia in adults: clinical features, immunophenotype, cytogenetics and outcome from the large randomized prospective trial (UKALL XII/ECOG 2993). Blood. 2009;114(25):5136–5145. doi: 10.1182/blood-2009-08-231217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Vlierberghe P, Ambesi A, Perez-Garcia A, et al. ETV6 mutations in early immature T-cell leukemias. J Exp Med. 2011;208(13):2571–2579. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paietta E, Goloubeva O, Neuberg D, et al. A surrogate marker profile for PML/RARα expressing acute promyelocytic leukemia and the association of immunophenotypic markers with morphologic and molecular subtypes. Cytometry B Clin Cytom. 2004;59B(1):1–9. doi: 10.1002/cyto.b.20001. [DOI] [PubMed] [Google Scholar]
- 8.Lin P, Hao S, Medeiros LJ, et al. Expression of CD2 in acute promyelocytic leukemia correlates with short form of PML/RARα transcripts and poorer prognosis. Am J Clin Pathol. 2004;121(3):402–407. doi: 10.1309/XC8P-9M8N-KQDT-38LB. [DOI] [PubMed] [Google Scholar]
- 9.Schoch C, Kohlmann A, Schnittger S, et al. Acute myeloid leukemias with reciprocal rearrangements can be distinguished by specific gene expression profiles. Proc Natl Acad Sci U S A. 2002;99(15):10008–10013. doi: 10.1073/pnas.142103599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimwade D, Outram SV, Flora A, et al. The T-cell affiliated CD2 gene lies within the open chromatin environment in acute promyelocytic leukemia cells. Cancer Res. 2002;62(16):4730–4735. [PubMed] [Google Scholar]
- 11.Will B, Steidl U. Multi-parameter fluorescence-activated cell sorting and analysis of stem and progenitor cells in myeloid malignancies. In: Paietta E, editor. Bailliere's Best Practice & Research: Clinical Haematology, Immunophenotyping in Haematologic Malignancies: State of the Art. Amsterdam, The Netherlands: Elsevier; 2010. pp. 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Röllig C, Thiede C, Gramatzki M, et al. A novel prognostic model in elderly patients with acute myeloid leukemia: results of 909 patients entered into the prospective AML96 trial. Blood. 2010;116(6):971–978. doi: 10.1182/blood-2010-01-267302. [DOI] [PubMed] [Google Scholar]
- 13.Pfirrmann M, Ehninger G, Thiede C, et al. Prediction of post-remission survival in acute myeloid leukaemia: a post-hoc analysis of the AML96 trial. Lancet. 2012;13(2):207–214. doi: 10.1016/S1470-2045(11)70326-6. [DOI] [PubMed] [Google Scholar]
- 14.Rockova V, Abbas S, Wouters BJ, et al. Risk stratification of intermediate-risk acute myeloid leukemia: integrative analysis of a multitude of gene mutation and gene expression markers. Blood. 2011;118(4):1069–1076. doi: 10.1182/blood-2011-02-334748. [DOI] [PubMed] [Google Scholar]
- 15.Langer C, Radmacher MD, Ruppert AS, et al. High BAALC expression associated with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: a Cancer and Leukemia Group B (CALGB) study. Blood. 2008;111(11):5371–5379. doi: 10.1182/blood-2007-11-124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwind S, Marcucci G, Maharry K, et al. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116(25):5660–5669. doi: 10.1182/blood-2010-06-290536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martelli MP, Pettirossi V, Thiede C, et al. CD34+ cells from AML with mutated NPM1 harbor cytoplasmic mutated nucleophosmin and generate leukemia in immunocompromised mice. Blood. 2010;116(19):3907–3922. doi: 10.1182/blood-2009-08-238899. [DOI] [PubMed] [Google Scholar]
- 18.Falini B, Nicoletti I, Martelli MF, Mecucci C. Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): biologic and clinical features. Blood. 2007;109(3):874–885. doi: 10.1182/blood-2006-07-012252. [DOI] [PubMed] [Google Scholar]
- 19.Paietta E, Racevskis J, Neuberg D, et al. Expression of CD25 (interleukin-2 receptor alpha chain) in adult acute lymphoblastic leukemia predicts for the presence of BCR/ABL fusion transcripts: results of a preliminary laboratory analysis of ECOG/MRC intergroup study E2993. Leukemia. 1997;11(11):1887–1890. doi: 10.1038/sj.leu.2400836. [DOI] [PubMed] [Google Scholar]
- 20.Geng H, Brennan S, Milne TA, et al. Integrative epigenomic analysis of adult B-acute lymphoblastic leukemia identifies biomarkers and therapeutic targets. Cancer Discov. doi: 10.1158/2159-8290.CD-12-0208. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakase K, Kita K, Otsuji A, et al. Diagnostic and clinical importance of interleukin-2 receptor alpha expression on non–T-cell acute leukemia cells. Br J Haematol. 1992;80(3):317–326. doi: 10.1111/j.1365-2141.1992.tb08139.x. [DOI] [PubMed] [Google Scholar]
- 22.Terwijn M, Feller N, van Rhenen A, et al. Interleukin-2 receptor alpha-chain (CD25) expression on leukemic blasts is predictive for outcome and level of residual disease in AML. Eur J Cancer. 2009;45(9):1692–1699. doi: 10.1016/j.ejca.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 23.Cerny J, Woods L, Yu H, et al. Expression of CD25 on acute myeloid leukemia (AML) blasts is an independent risk factor associated with refractory disease, which may be overcome by stem cell transplantation [abstract]. Blood (ASH Annual Meetings Abstracts) 2011;118(21) Abstract 3560. [Google Scholar]
- 24.Kindler T, Lipka DB, Fischer T. FLT3 as a therapeutic target in AML: still challenging after all these years. Blood. 2010;116(24):5089–5102. doi: 10.1182/blood-2010-04-261867. [DOI] [PubMed] [Google Scholar]
- 25.Saito Y, Kitamura H, Hijikata A, et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci Transl Med. 2010;2(17):17ra9. doi: 10.1126/scitranslmed.3000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel JP, Gonen M, Figueroa ME, et al. Prognostic and therapeutic relevance of integrated genetic profiling in AML. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez HF, Sun Z, Yao X, et al. Anthracycline dose intensification in acute myeloid leukemia: results of ECOG Study E1900. N Engl J Med. 2009;361(13):1249–1259. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slovak ML, Kopeckcy KJ, Cassileth PA, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96(13):4075–4083. [PubMed] [Google Scholar]
- 29.Cripe LD, Uno H, Paietta E, et al. Zosuquidar, a novel modulator of P-glycoprotein, does not improve the outcome of older patients with newly diagnosed acute myeloid leukemia: a randomized, placebo-controlled, trial of the Eastern Cooperative Oncology Group (ECOG 3999). Blood. 2010;116(20):4077–4085. doi: 10.1182/blood-2010-04-277269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paietta E, Neuberg D, Bennett JM, et al. Low expression of the myeloid differentiation antigen CD65s, a feature of poorly differentiated AML in older adults: study of 711 patients enrolled in ECOG trials. Leukemia. 2003;17(8):1544–1550. doi: 10.1038/sj.leu.2402999. [DOI] [PubMed] [Google Scholar]
- 31.Paietta E, Andersen J, Yunis J, et al. Acute myeloid leukaemia expressing the leucocyte integrin CD11b: a new leukemic syndrome with poor prognosis. Result of an ECOG database analysis. Br J Haematol. 1998;100(2):265–272. doi: 10.1046/j.1365-2141.1998.00561.x. [DOI] [PubMed] [Google Scholar]
- 32.Figueroa ME, Reimers M, Thompson RF, et al. An integrative genomic and epigenomic approach for the study of transcriptional regulation. PloS One. 2008;3(3):e1882. doi: 10.1371/journal.pone.0001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carvalho B, Irizarry RA. oligo: Tools for low-level analysis of oligonucleotide arrays. R package version 183. [Accessed August 7, 2012]. http://bioconductor.org/packages/devel/bioc/manuals/oligo/man/oligo.pdf.
- 34.Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010;304(24):2706–2715. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eppert K, Takenaka K, Lechman ER, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17(9):1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 38.Thol F, Damm F, Ludeking A, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29(21):2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 39.Hou H-A, Kuo Y-Y, Chou W-C, et al. DNMT3A mutations in acute myeloid leukemia: stability during disease evolution and clinical implications. Blood. 2012;119(2):559–568. doi: 10.1182/blood-2011-07-369934. [DOI] [PubMed] [Google Scholar]
- 40.Whitman SP, Archer KJ, Feng L, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: A Cancer and Leukemia Group Study. Cancer Res. 2001;61(19):7233–7239. [PubMed] [Google Scholar]
- 41.Fernandez HF, Sun Z, Litzow MR, et al. Autologous transplantation gives encouraging results for young adults with favorable-risk acute myeloid leukemia, but is not improved with gemtuzumab ozogamicin. Blood. 2011;117(20):5306–5313. doi: 10.1182/blood-2010-09-309229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Rhenen A, Feller N, Kelder A, et al. High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin Cancer Res. 2005;11(18):6520–6527. doi: 10.1158/1078-0432.CCR-05-0468. [DOI] [PubMed] [Google Scholar]
- 43.Ishikawa F, Yoshida S, Saito Y, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25(11):1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.