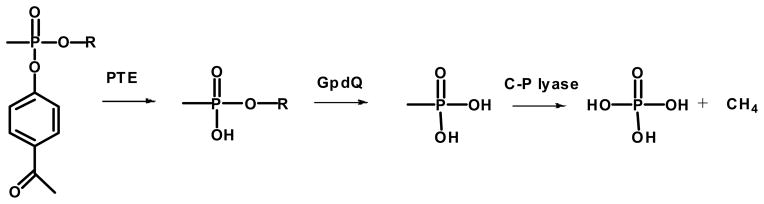
Abstract
Phosphotriesterase (PTE) from soil bacteria is known for its ability to catalyze the detoxification of organophosphate pesticides and chemical warfare agents. Most of the organophosphate chemical warfare agents are a mixture of two stereoisomers at the phosphorus center and the SP-enantiomers are significantly more toxic than the RP-enantiomers. In previous investigations PTE variants were created through the manipulation of the substrate binding pockets and these mutants were shown to have greater catalytic activities for the detoxification of the more toxic SP-enantiomers of nerve agent analogs for GB, GD, GF, VX, and VR than the less toxic RP-enantiomers. In this investigation alternate strategies were employed to discover additional PTE variants with significant improvements in catalytic activities relative to the wild type enzyme. Screening and selection techniques were utilized to isolate PTE variants from randomized libraries and site specific modifications. The catalytic activities of these newly identified PTE variants towards the SP-enantiomers of chromophoric analogs of GB, GD, GF, VX, and VR have been improved up to 15,000 fold relative to the wild-type enzyme. The X-ray crystal structures of the best PTE variants were determined. Characterization of these mutants with the authentic G-type nerve agents has confirmed the expected improvements in catalytic activity against the most toxic enantiomers of GB, GD, and GF. The values of kcat/Km for the H257Y/L303T (YT) mutant for the hydrolysis of GB, GD, and GF were determined to be 2 × 106 M−1 s−1, 5 × 105 M−1 s−1, and 8 × 105 M−1 s−1, respectively. The YT mutant is the most proficient enzyme reported thus far for the detoxification of G-type nerve agents. These results support a combinatorial strategy of rational design and directed evolution as a powerful tool to discover more efficient enzymes for the detoxification of organophosphate nerve agents.
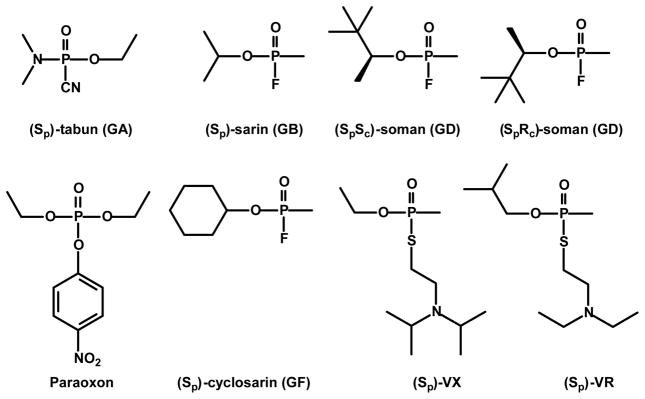
Phosphotriesterase (PTE) from Psuedomonas diminuta catalyzes the detoxification of a wide range of organophosphate insecticides and chemical warfare agents, including tabun (GA), sarin (GB), soman (GD), cyclosarin (GF) and VX.1–3 The structures of these compounds are illustrated in Scheme 1. The catalytic proficiency of PTE for the hydrolysis of the insecticide paraoxon is close to the diffusion-controlled limit (kcat/Km ~ 108 M−1 s−1),4 and the stereoselectivity of the wild-type enzyme for chiral organophosphate substrates has been determined.5–8 Wild-type PTE prefers to hydrolyze the RP-enantiomers of GB, GD, GF and their chromophoric analogues.5,9 However, the SP-enantiomers of these nerve agents are substantially more toxic than the RP-enantiomers.10 PTE mutants with an inverted stereoselectivity have been constructed by alteration of the active site residues, and these modified enzymes catalyze the hydrolysis of the toxic SP-enantiomers faster than the wild-type enzyme.5,11,12
Scheme 1.
In addition to the rational redesign of the PTE active site, directed evolution of catalytic activity towards toxic organophosphates has been undertaken by several laboratories. Chen and coworkers created a randomized PTE library by DNA shuffling and saturation mutagenesis. They identified two distal mutations, K185R and I274N, which are apparently important for improvement in the catalytic activity towards the insecticides paraoxon, parathion, chlorpyrifos and coumaphos.13,14 Tawfik and colleagues utilized DNA shuffling to construct randomized PTE libraries that contained variants with enhanced protein stability and increased enzymatic activity. One of these variants had three mutations located outside of the substrate binding pocket (K185R, D208G and R319S) that increased net protein expression by ~20-fold.15 The organophosphate degrading enzyme OpdA from Agrobacterium radiobacter P230 is ~90% identical to PTE. The Ollis group mutated OpdA and libraries of these variants were created by error-prone PCR and DNA shuffling. Some of these mutants were shown to have improved protein expression and better activity towards methyl paraoxon, methyl parathion and demeton-S.16 Many of the amino acid changes that originated from randomized mutations were localized to the surface of the protein and these alterations may have also contributed to an increase in protein stability.17,18
In this paper, multiple directed evolution techniques were utilized to obtain mutant forms of PTE that are significantly enhanced in their ability to catalyze the hydrolysis of the most toxic forms of selected organophosphate nerve agents. We initiated this investigation using highly sensitive chromophoric assays to identify alterations in PTE that enable the most toxic stereoisomers of the G-agent analogues to be hydrolyzed more efficiently. An in vivo selection technique was subsequently exploited that incorporates the phosphonate assimilation pathway in E. coli to identify beneficial PTE variants in larger, more randomized libraries. These in vivo assays utilized the co-expression of glycerophosphodiesterase (GpdQ) from Enterobacter aerogenes and PTE in E. coli. This selection method requires the enzymatic hydrolysis of phosphonate esters by PTE when these cells are grown in the absence of phosphate.19,20 We have identified mutants of PTE with rate enhancements of more than four-orders of magnitude relative to the wild-type enzyme for specific enantiomers of organophosphate nerve agent analogs. The analog results have been validated by tests using the authentic nerve agents.
Materials and Methods
Materials
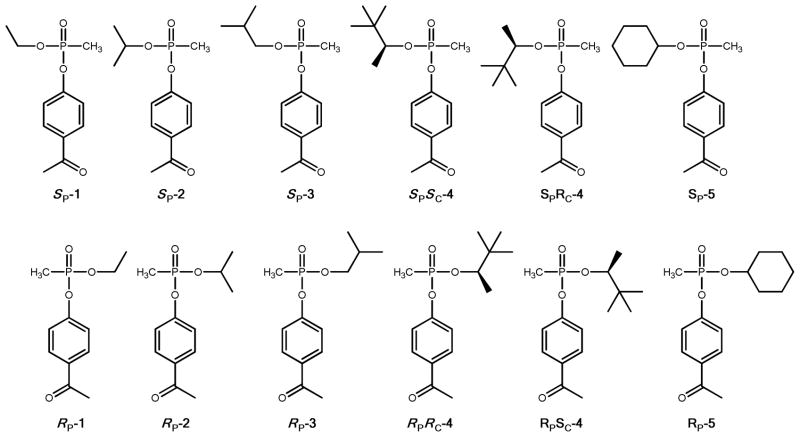
The preparation and isolation of the chiral organophosphonates used in this investigation have been previously described.5 The structures of these compounds are illustrated in Scheme 2. These compounds are toxic and should be used with the appropriate safeguards.
Scheme 2.
Plasmid Construction
For single protein expression experiments the gene for PTE was inserted between the NdeI and EcoRI restriction sites of pET20b (EMD Millipore). The plasmid GpdQ-pETDuet was constructed by inserting the gene for glycerophosphodiesterase (GpdQ) from Enterobacter aerogenes into the first T7 promoter site on the pETDuet (EMD Millipore) vector.19 The PTE/GpdQ-pETDuet plasmid was created by inserting the PTE gene into the second T7 promoter site on the GpdQ-pETDuet plasmid using the NdeI and AvrII restriction sites.
Site-Directed Mutagenesis
The A80V, K185R and I274N mutations in PTE were constructed using the QuikChange site directed mutagenesis method from Agilent. Oligonucleotide pairs that contained the mutated codons at the specified sites were used as primers to amplify the genes for the wild-type enzyme and the following mutant enzymes: QF, YT and GWT. The complete identity of mutants is provided in Table 1. The mutations were added to each template sequentially to make the following mutant proteins: RN, QFRN, YTRN, GWT-d1 and GWT-d2.
Table 1.
Identification of mutants created for this investigation.
| Abbreviation | Mutational sites of mutants |
|---|---|
| RN | K185R/I274N |
| QF | H254Q/H257F |
| GWT | H254G/H257W/L303T |
| QF-RN | H254Q/H257F/K185R/I274N |
| YT-RN | H257Y/L303T/K185R/I274N |
| GWT-d1 | H254G/H257W/L303T/K185R/I274N |
| GWT-d2 | H254G/H257W/L303T/K185R/I274N/A80V |
| GWT-d3 | H254G/H257W/L303T/K185R/I274N/A80V/S61T |
| GWT-f1 | H254G/H257W/L303T/M317L/K185R/I274N |
| GWT-f2 | H254G/H257W/L303T/M317L |
| GWT-f3 | H254G/H257W/L303T/M317L/I106C/F132I/L271I/K185R/I274N |
| GWT-f4 | H254G/H257W/L303T/M317L/I106C/F132I/L271I/K185R/I274N/A80V |
| GWT-f5 | H254G/H257W/L303T/M317L/I106C/F132I/L271I/K185R/I274N/A80V/R67H |
Construction of Single and Double Substitution Libraries
Single (M317X) and double substitution libraries (W131X/F132X, S308X/Y309X, and L271X/S308X) were constructed from the gene for GWT-d1 using the QuikChange (Agilent) protocol. The primers contained the degenerate codon NNS at the targeted position where N = A, T, G and C, and S = C and G). The M317X, W131X/F132X, F306X/Y309X and S308X/Y309X libraries were created with a single set of primers. The I106X/S308X library was made using two sets of degenerate primers for the randomization of Ile-106 and Ser-308. Bacterial colonies from the unfractionated I106X library were collected and plasmids from this library were isolated for a second round of mutagenesis using primers for the randomization of Ser-308. The double substitution libraries, C59X/S61X and I255X/W302X, were constructed using the GWT-d2 mutant as the starting template. A single set of degenerate primers randomized the Cys-59 and Ser-61sites. Two pairs of primers with degenerate codons for the Ile-255 and Trp-302 sites were utilized sequentially.
Construction of a Multi-Site Randomized Library
A multi-site PTE library was constructed using the gene for GWT-f1 as the initial template. Six residues in the substrate binding pocket were partially randomized, including Ile-106, Phe-132, Leu-271, Phe-306, Ser-308 and Tyr-309. The substitutions for the Ile-106 site were alanine, cysteine, glycine, isoleucine, and valine. The variations for Phe-132 were alanine, cysteine, phenylalanine, glycine, histidine, isoleucine, leucine, proline, arginine, serine, threonine, and tyrosine. The randomized residues for the Leu-271 site consisted of phenylalanine, isoleucine, leucine, tyrosine and glutamine. The substitution of residues for Phe-306 were phenylalanine, isoleucine, and methionine. For Ser-308, the substitutions were alanine, cysteine, glycine, serine, threonine, glutamate, leucine, glutamine, valine and asparagine. The variants for Tyr-309 consited of alanine, cysteine, aspartate, phenylalanine, glycine, leucine, isoleucine, asparagine, serine, threonine, valine, tyrosine, and tryptophan.
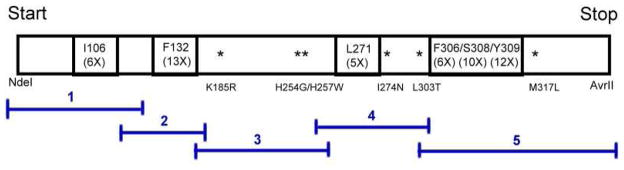
The multi-site partially randomized PTE library was constructed by combining five separate segments of the gene for PTE as illustrated in Scheme 3 using primerless PCR for 15 cycles and then amplified by PCR for 55 cycles using primers specific for the 5′- and 3′-termini. The potential size of this multi-site library is 1.9 × 105 variants. In this scheme the numbers below the residue identifier indicates the number of amino acids that were allowed during the library construction. The amplified PTE library was digested with NdeI and AvrII restriction enzymes and ligated into the GpdQ-pETDuet plasmid using T4 DNA ligase. The ligation mixture was purified using the QIAquick Kit (Qiagen) and then transformed into freshly made E. coli Top10 competent cells (Life Technologies). The transformants were incubated at 37 °C for 1 hour and then plated on LB-ampicillin agarose. Approximately 5.7 × 105 colony forming units (CFU) were collected and grown in LB medium for 6 hours at 37 °C. The plasmids from the PTE library were extracted using the Promega Wizard Plus Miniprep kit.
Scheme 3.
Construction of PTE Library by Error-Prone PCR
Random error-prone PCR was performed using the Gene-Morph II system (Agilent). The gene for GWT-f4 served as the template and approximately ~1.5 mutations per 1,000 bp were introduced. The amplified PTE library and the GpdQ-pETDuet plasmid were digested with NdeI and AvrII and then ligated using T4 DNA ligase. The library was transformed into fresh E. coli Top10 competent cells (Life Technologies). The transformants were incubated at 37 °C for 1 hour and then plated on LB-ampicillin agarose. Approximately 6 × 105 CFU were collected and then grown in LB medium for 6 hours at 37 °C.
Growth Assays
The PTE/GpdQ-pETDuet plasmids were transformed into E. coli BL21 (DE3) competent cells and grown on LB plates containing 0.1 mg/mL ampicillin. The colonies from fresh transformations were inoculated and grown overnight in Super Broth (32 g/L tryptone, 20 g/L yeast extract, 5 g/L NaCl, and 5 mM NaOH) at 30 °C. Overnight cultures were washed four times with sterile water to remove all traces of the rich medium. A 20-mL phosphate-free minimal medium (MOPS minimal medium) culture was inoculated with 2% fresh and previously washed overnight culture as previously described.19 The minimal medium was supplemented with 0.1% glucose as the carbon source, and 0.1 mg/mL thiamine.19 Various organophosphonate compounds were added to the cultures to a final concentration of 1.0 mM. For control experiments, BL21(DE3) cells with the empty vector (PTE-/GpdQ-) were grown in the presence and absence of inorganic phosphate. BL21(DE3) cells containing the PTE/GpdQ-pETDuet vector were induced with 0.5 mM IPTG for protein expression. Cell growth was monitored by measuring the OD600 for 5–10 days at 30 °C.
Screening Single and Double Substitution Libraries
The products of the QuikChange mutagenesis experiments were transformed into E. coli BL-21(DE3) competent cells and inoculated in 96-deep well culture blocks containing 1.0 mL Super Broth supplemented with 0.5 mM CoCl2 and 0.1 mg/mL ampicillin. Variants of the single and double substitution libraries were screened with paraoxon and p-nitrophenol containing versions of SP-4, and SP-5 in the presence of 2% Bug Buster. The hydrolysis of paraoxon, SP-4 and Sp-5 were measured by monitoring the formation of p-nitrophenol at 400 nm (ε400= 17,000 M−1 cm−1) in 50 mM CHES, pH 9.0, buffer using a plate reader at 30 °C.
In vivo Selection on Phosphate Free Minimal Medium Plates
The PTE/GpdQ-pETDuet library was transformed into E. coli BL21(DE3) cells and plated on LB-ampicillin agarose. The cells were collected and grown in Super Broth at 30 °C for 12 hours. The cells were washed for four times with sterile water to remove any traces of the rich medium. The cells were plated on MOPS phosphate-free minimal medium agarose with 0.4 mg/mL ampicillin and 0.5 mM IPTG and grown for 7–10 days at 30 °C in the presence of 1.0 mM SP-519 The colonies with sizes larger than background were selected. The first round of selection used the GWT-f2 mutant as the background and the second round used the GWT-f4 mutant as the background. Approximately 6 × 105 colonies from the multi-site-randomized PTE and the error prone PTE libraries were plated for selection.
Expression, Growth and Purification of Phosphotriesterase Mutants
The mutant phosphotriesterase plasmids were transformed into E. coli BL-21(DE3) cells. The transformed cells were grown in Luria-Bertani (LB) broth overnight at 37 °C. The overnight cultures were used to inoculate 1 L Terrific Broth (TB) containing 25 Qg/mL ampicillin and 1.0 mM CoCl2 at 30 °C. The expression of PTE was induced by the addition of 1.0 mM IPTG when the OD600 reached 0.4. The cells were harvested at 4 °C by centrifugation at 6,000 rpm for 10 min after allowing the culture to reach stationary phase after 36–42 hours at 30 °C. The mutant proteins were purified according to previously reported protocols.18
Protein Crystallization, Data Collection, and Structure Determination
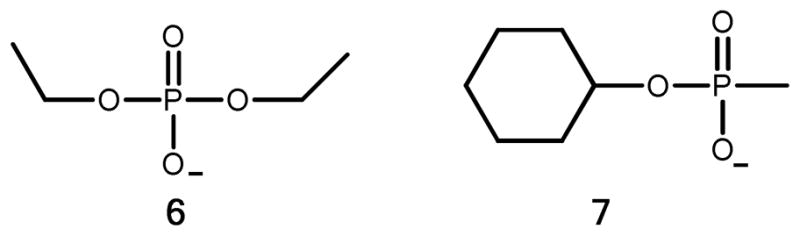
Vapor diffusion techniques were used to grow single crystals of five PTE mutants. The mutants (RN, QFRN, GWT-d2, GWT-f5, and GWT-d3) crystallized after one day incubation at 22 °C. In a typical hanging drop experiment, 2 μL of protein and 2 μL of precipitating agent were mixed and placed on a siliconized coverslip over 500 μL of the precipitating agent. Showers of small crystals of the GWT-f3 (12 mg/mL) variant were obtained using PEG MME 5000 as a precipitant. The small crystals were crushed, serially diluted in a stabilizing solution (24% PEG MME 5000, 4% dioxane, 1.0 mM CoCl2, and either 0.1 M imidazole pH 7.0 or 0.1 M HEPES pH 7.5) and used as microseeds to grow crystals of the other PTE variants. Crystals of GWT-d2 (12 mg/mL), GWT-f5 (8 mg/mL), and GWT-d3 (10 mg/mL) were grown using the imidazole seeding solution. GWT-d2 crystallized in 26% PEG MME 5000, while GWT-f5 crystalized in 22% PEG MME 5000 with 100 mM diethylphosphate (6), and GWT-d3 crystallized in 18% PEG MME 5000. Crystals of RN (11.5 mg/mL) were generated with the HEPES seeding solution with 100 mM diethylphosphate and 25% PEG MME 5000 as the precipitant. One large crystal of QFRN (11 mg/mL) was produced in 0.1 M MES pH 6.0 with 18% PEG MME 5000. GWT-f5 and GWT-d3 were crystallized from the seeding solutions in the presence of 100 mM cyclohexyl methylphosphonate (7) using similar conditions (26% PEG MME 5000, 0.1 M imidazole pH 7.0, 1 mM CoCl2, and 24% PEG MME 5000, 0.1 M imidazole pH 7.0, 1 mM CoCl2, respectively). The crystals were immersed in the well solution containing 13% ethylene glycol for 30 seconds and then immediately frozen in liquid nitrogen. The frozen crystals were transferred to a cassette and shipped to the Stanford Synchrotron Radiation Lightsource (SSRL) for remote data collection. Mutants RN, GWT-f5, and GWT-d3 were collected on SSRL Beamline 11-1; the rest were collected on SSRL Beamline 7-1. The structures of compounds 6 and 7 are presented in Scheme 4.
Scheme 4.
The data sets were indexed and scaled with the HKL2000 package,22 and phases were determined by molecular replacement with Phaser version 2.1.23 The search model was a previously refined PTE structure (PDB ID: 1HZY). Initial refinement of the structures was performed using Refmac5 from the CCP4 suite.24.25 Difference electron density and omit maps were manually fit with the XtalView package.26 After the cofactors (cobalt and inhibitor) were added, further refinement was done with CNS.27 All structures were superimposed with Sequoia.28 Data collection and refinement statistics are summarized in Table 2.
Table 2.
Diffraction data collection and refinement statistics
| GWT-d2 | QF-RN | GWT-d3 | GWT-d3-(7) | GWT-f5-(6) | GWT-f5-(7) | RN-(6) | |
|---|---|---|---|---|---|---|---|
| Data Collection | BL 7-1 | BL 7-1 | BL 11-1 | BL 7-1 | BL 11-1 | BL 7-1 | BL 11-1 |
| Wavelength (Å) | 0.9794 | 0.9794 | 0.97945 | 0.9794 | 0.97945 | 0.9794 | 0.97945 |
| Space Group | P2(1)2(1)2 | P2(1)2(1)2 | P2(1)2(1)2 | P2(1)2(1)2 | P2(1)2(1)2 | P2(1)2(1)2 | P2(1)2(1)2 |
| Unit cell (Å and °) | 85.1, 85.2, 87.7 | 85.6, 85.4, 88.3 | 85.2 85.5, 88.1 | 85.5 86.1, 88.7 | 85.5 86.4, 88.1 | 85.5 85.3, 87.9 | 85.5, 86.0, 88.0 |
| Resolution (Å) | 50.0 – 2.00 | 50.0 - 1.95 | 50.0 - 1.89 | 50.0 - 1.95 | 50.0 - 1.77 | 50.0 – 2.1 | 50.0 - 1.60 |
| last shell (Å)* | 2.07-2.00 | 2.02-1.95 | 1.95-1.89 | 2.02-1.95 | 1.83-1.77 | 2.18-2.10 | 1.66-1.60 |
| Observations | 418098 | 467416 | 768808 | 515722 | 1250181 | 303570 | 1586065 |
| Unique Observations | 43780 | 47614 | 52186 | 48549 | 64191 | 72161 | 86462 |
| Redundancy | 9.5 (9.2) | 9.8 (8.8) | 14.7 (14.5) | 10.6 (10.4) | 19.5 (14.1) | 4.2 (4.2) | 18.3 (13) |
| Completeness (%) | 99.9 (100) | 99.9 (99.7) | 99.9 (100) | 99.7 (99.9) | 99.6 (95.8) | 100 (100) | 100 (99.7) |
| Mean I/σI | 23.0 (6.7) | 22.8 (6.8) | 23.4 (10.5) | 19.1 (6.1) | 44.6 (17.3) | 23.3 (6.0) | 38.2 (8.8) |
| Rsym (%)† | 10.8 (38.2) | 11.6 (35.5) | 10.8 (32.3) | 11.8 (46.1) | 6.5 (19.5) | 6.3 (39.5) | 7.5 (31.4) |
| Refinement | |||||||
| Residues in molecule A | D35-T361 | D35-T361 | D35-T361 | D35-T361 | R36-A259, N265-T361 | D35-T361 | D35-T361 |
| Residues in molecule B | D35-I260, N265-R363 | R36-I260, S267-T361 | R36-I260, L262-T361 | D35-T361 | D35-I260, N265-T361 | D35-I260 A266-T361 |
D35-G261 N265-T361 |
| Solvent atoms | 378 | 439 | 372 | 368 | 559 | 316 | 608 |
| Rwork/Rfree (%)‡ | 21.6/24.6 | 20.1/23.2 | 23.3/25.3 | 22.0/24.7 | 21.6/23.3 | 20.3/23.5 | 20.9/22.8 |
| r.m.s.d. bond lengths (Å) | 0.014 | 0.015 | 0.015 | 0.013 | 0.013 | 0.016 | 0.012 |
| r.m.s.d. bond angles (°) | 1.67 | 1.61 | 1.66 | 1.63 | 1.63 | 1.63 | 1.59 |
| Ramachandran plot (%) | |||||||
| Most favored regions | 88.5 | 89.8 | 89.3 | 89.9 | 89 | 89.4 | 89.3 |
| Additional allowed regions | 10.6 | 9.3 | 10 | 9.4 | 10.3 | 9.9 | 10 |
| Generously allowed regions | 0.9 | 0.9 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 |
| Disallowed regions | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PDB ID | 3UR2 | 3UPM | 3URA | 3URN | 3URB | 3URQ | 3UR5 |
Values in parentheses are the statistics for the highest resolution shell of data.
Rsym = Σ | Ihkl − <I> |/S<I>, where <I> is the average individual measurement of Ihkl.
Rwork = (S |Fobs−Fcalc|)/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is calculated the same as Rwork, but from the data (5%) that were excluded from the refinement.
Kinetic Measurements and Data Analysis
The kinetic constants for each chromogenic substrate were determined by monitoring the production of p-acetylphenol at 294 nm (ε294= 7,710 M−1 cm−1) in 50 mM CHES, pH 9.0, buffer using a plate reader at 30 °C. Protein concentration in crude samples was determined by measuring the absorbance at 280 nm. The kinetic constants (kcat and kcat/Km) were obtained by fitting the data to equation 1, where v is the initial velocity, kcat is the turnover number, [A] is the substrate concentration, Et is the enzyme concentration, and Km is the Michaelis constant.
| (1) |
Kinetic constants for sarin (GB), soman (GD), and cyclosarin (GF) were determined by monitoring the release of free fluoride at 25 °C in 50 mM bis-tris-propane buffer, pH 7.2, using a fluoride electrode. Initial screenings were conducted using a single fixed 3.0 mM substrate concentration. Kinetic constants were determined for those mutants showing the highest specific activity in the initial screenings.
Stereospecificity for hydrolysis of nerve agents was obtained by following the complete hydrolysis of 0.5 mM racemic mixtures of GB or GF. Reactions were conducted in 50 mM bis-tris-propane buffer, pH 7.2, and the reaction followed by the release of fluoride. Substantial stereospecificity was observed as bi-phasic curves. Complementation assays were used to determine the stereopreference for variants by mixing equal units of the variant being tested with a variant with a known stereopreference. Variants with the same preference display biphasic curves when mixed while variants of opposite preference complement each others slow phase resulting in a single hydrolytic phase being observed. The enantiomeric preference for GB was determined by the ability of each variant to complement the hydrolysis by the YT variant. The enantiomeric preference for variants with GF was determined by the ability to complement the hydrolysis catalyzed by GWT. The enantiomeric preference of the GWT variant for GF was previously confirmed by polarimetry.9
Results
Expression of PTE with K185R and I274N Mutations
The K185R and I274N mutations were added to the parental GWT mutant of PTE (GWT-d1). The net expression of soluble protein for the GWT-d1 mutant was greater than that for GWT and approximately 10-fold more protein could be obtained following purification (data not shown). The addition of the K185R and I274N mutations to PTE variants helped to increase the expression and protein yields by two- to ten-fold in each case.
Screening Single and Double Substitution Libraries with Sp-5
The GWT mutant was previously shown to have the highest activity for the hydrolysis of the toxic SP-enatiomers of 4 and 5.5 Mutant libraries were constructed using GWT-d1 as the starting template to identify more active PTE variants for the hydrolysis of SP-5. Nine amino acid residues in the substrate binding pocket were considered as potential “hot spots” for the construction of these PTE libraries. The single-substitution library M317X was constructed first, followed by four double-substitution libraries (W131X/F132X, F306X/Y309X, S308X/Y309X, and I106X/Y308X). Approximately 60 colonies from the M317X library and ~550 colonies from each of the double substitution libraries were picked and subsequently screened with SP-5.
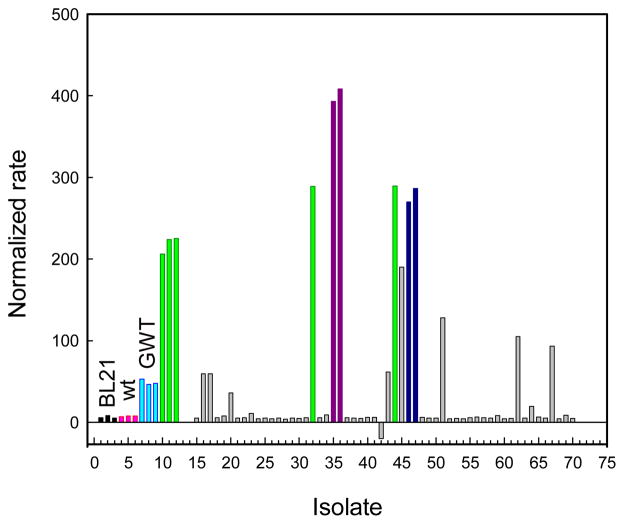
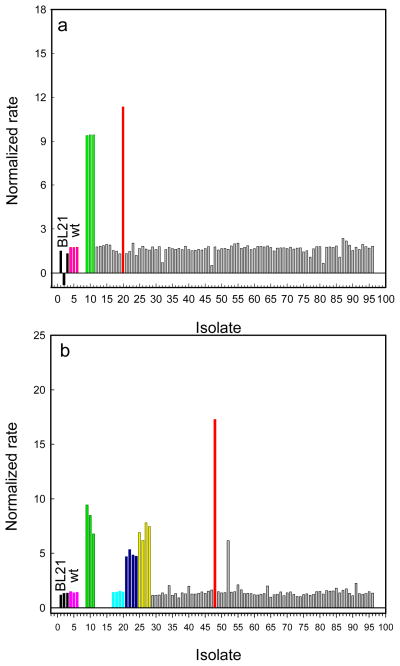
The variants with catalytic activities higher than background from the first round of screening were isolated and then re-screened with the same substrate. No improvement for the hydrolysis of SP-5 was found in the double-substitution libraries, W131X/F132X, F306X/Y309X, S308X/Y309X and I106X/Y308X. Figure 1 illustrates the screening of the M317X single substitution library. The best mutant identified in this screen contained a leucine substitution for Met-317 and is denoted as GWT-f1.
Figure 1.
Screening of the M317X mutant library against Sp-5 using GWT-d1 as the parental template. The bars in light green, purple and dark blue represent the relative catalytic activity of the GWT-d1, GWT-f1 and GWT-d1-M317F mutants, respectively. Those mutants represented by the gray bars were not characterized or sequenced.
Screening Double Substitution Libraries with Sp-enantiomers of 4
Twelve amino acid residues in the substrate binding pocket were chosen for double substitution libraries using the GWT-d1 mutant as the starting template. The W131X/F132X, F306X/Y309X, S308X/Y309X and I106X/Y308X libraries were screened with SPSC-4 and SPRC-4, but enhanced variants were not identified. The A80V mutation was added to GWT-d1 to improve the net protein expression of PTE.13 This mutant was denoted as GWT-d2 and shown to have better protein expression levels and a higher specific activity towards the hydrolysis of SPSC-4 and SPRC-4 than the GWT-d1 parent.
Double substitution libraries that randomize the residues close to the substrate binding pocket were constructed. The I255X/W302X and C59X/S61X libraries were constructed using the GWT-d2 mutant as the template. For each double substitution library, approximately 600 colonies were selected and screened with the two SP-enantiomers of 4. From this screen a mutant was identified that had more activity for the hydrolysis of SPSC-4 and SPRC-4. This mutant, designated as GWT-d3, contained a threonine substitution for Ser-61.
Co-Expression of Phosphotriesterase and Phosphodiesterase
Mutant PTE libraries were co-expressed with a phosphodiesterase capable of hydrolyzing the reaction product to methylphosphonate,19 and this compound is capable of supporting bacterial growth in a phosphate-free medium (Scheme 5).29,30 The GWT-f1 mutant was partially randomized at six sites simultaneously. The total library contained 1.9 × 105 potential variants. Eight colonies from this library were selected to verify that the targeted sites were randomized (data not shown). The PTE/GpdQ-pETDuet plasmid library was transformed into E. coli BL21(DE3) cells. Approximately 5.8 × 105 CFU were plated on phosphate-free minimal medium with 1 mM SP-5 as the sole phosphorus source. The colonies that contained beneficial mutations for the hydrolysis of SP-5 were identified as being larger in size than a background colony of the parent GWT-f1 mutant. Approximately 30 of these colonies were selected for growth in 96-well blocks and subsequently assayed for catalytic activity with SP-5. The screening of the partially randomized multi-site library with SP-5 is shown in Figure 2a. The first nine samples include the empty vector control, wild-type PTE and the GWT-f1 parent. A single variant was found to have more activity than the GWT-f1 parent. This mutant (GWT-f3) contained three additional changes in amino acid sequence: I106C/F132I/L271I. The A80V mutation was added to the GWT-f3 mutant to create the GWT-f4 variant.
Scheme 5.
Figure 2.
(A) Screening of the 6-site randomized library using GWT-f1 as the parental template with Sp-5. The bars in light green and red represent the relative catalytic activity of the GWT-f1 and GWT-f3 mutants, respectively. (B) Screening of the error-prone PCR library using GWT-f4 as the parental template with Sp-5. The bars in light green, cyan, dark blue, yellow and red represent the GWT-f4, GWT-f4-G129D, GWT-f4-I228F, GWT-f4-G254W and GWT-f5 mutants, respectively.
The GWT-f4 mutant served as the template for error-prone PCR (epPCR). Random mutagenesis of the GWT-f4 gene was conducted using the Mutazyme II DNA polymerase. Ten colonies from this library were selected to establish an average mutation rate of ~1.5 mutations per 1000 bp. The epPCR generated PTE/GpdQ-pETDuet library was transformed into E. coli BL21(DE3). Approximately 6 × 105 CFU were plated on phosphate-free minimal medium plates with 1 mM of SP-5 as the sole phosphorus source. Colonies larger in size than the parental strain (GWT-f4) were assayed with SP-5 and the results are shown in Figure 2b. The first 20 samples include the empty vector, wild-type PTE, GWT-f3, GWT-f3-G129D, GWT-f3-I288F, and GWT-f3-H254W. The new variant, GWT-f5, contained a single mutation, relative to GWT-f4, at Arg-67 with a change to histidine.
Kinetic Properties of the PTE Mutants
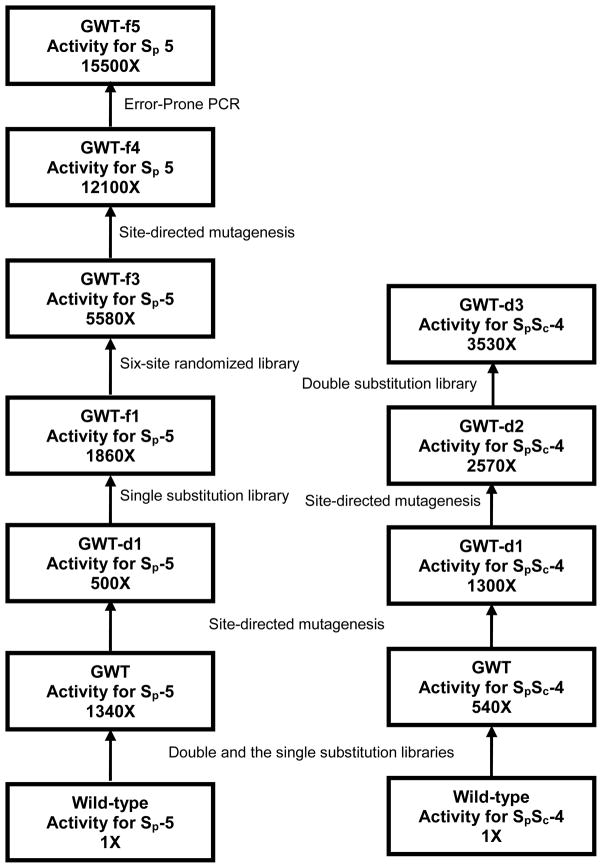
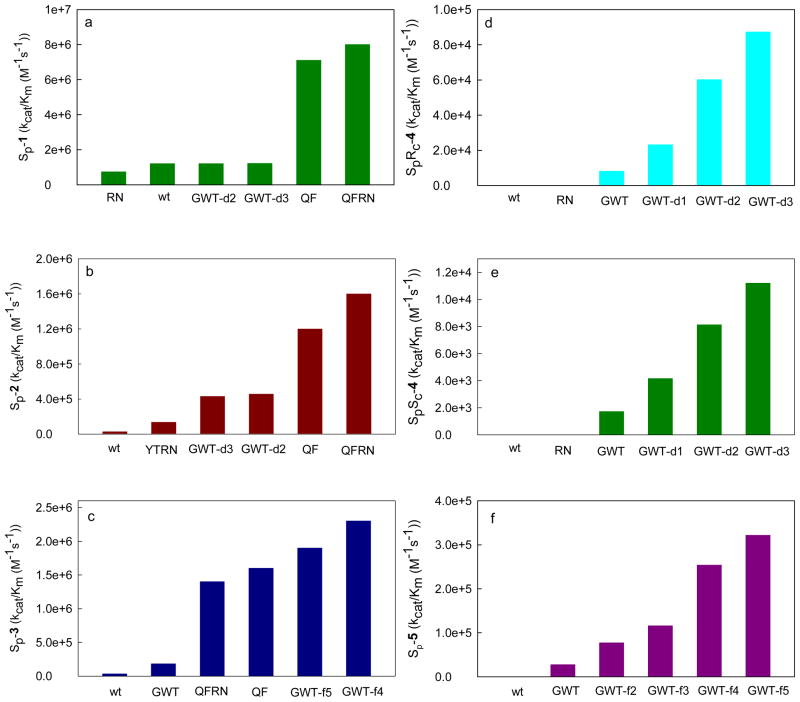
The kinetic parameters of the purified wild-type and mutants of PTE with the entire set of chiral organophosphonate compounds shown in Scheme 2 are provided in Tables 3 and 4. The best mutant (based on the value of kcat/Km) for the hydrolysis of SP-1 is QFRN (kcat/Km = 8.0 × 106 M−1 s−1). This is also the best mutant for the hydrolysis of SP-2 (kcat/Km = 1.6 × 106 M−1 s−1). For the hydrolysis of SP-3 the best mutant identified to date is GWT-f4 (kcat/Km of 2.3 × 106 M−1 s−1). For the two soman analogues (SPSC-4 and SPRC-4) the best mutant is GWT-d3 (kcat/Km = 8.7 × 104 M−1 s−1 and 1.1 × 104 M−1 s−1, respectively). GWT-f5 is the best mutant for the hydrolysis of the cyclosarin analogue, SP-5 (kcat/Km of 3.2 × 105 M−1 s−1). The outline for the discovery of PTE variants with enhanced activity for the hydrolysis of the Sp-enantiomers of compound 4 and 5 by directed evolution is shown in Figure 3.
Table 3.
The values of kcat (s−1) for the wild-type and mutants of PTEa.
| WT | QF | YT | RN | QFRN | YTRN | GWT | GWT-d1 | GWT-d2 | GWT-d3 | GWT-f1 | GWT-f2 | GWT-f3 | GWT-f4 | GWT-f5 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RP-1 | 1.5e2 | 1.7e2 | 7.3e0 | 9.0e1 | 2.0e2 | 1.2e1 | 1.4e1 | 1.4e1 | 1.3e1 | 9.6e0 | 1.3e1 | 2.2e2 | 7.9e1 | ND | ND |
| SP-1 | 6.7e2 | 3.2e1 | 4.1e2 | 8.2e2 | 4.5e1 | 1.0e3 | 1.9e2 | 2.9e2 | 5.3e2 | 4.0e2 | 3.6e2 | 4.4e2 | 6.6e2 | 1.1e3 | 7.2e2 |
| Rp-2 | 1.0e2 | 4.8e1 | 1.8e1 | 6.6e1 | 6.6e1 | 2.0e1 | ND | 2.1e1 | 2.2e1 | 5.9e1 | 9.8e0 | 3.3e1 | ND | ND | ND |
| Sp-2 | 4.0e1 | 7.2e0 | 3.7e2 | 2.0e1 | 1.1e1 | 7.0e2 | 9.2e1 | 8.6e1 | 1.3e2 | 2.0e2 | 3.0e2 | 2.3e2 | 6.2e2 | 1.1e3 | 5.9e2 |
| Rp-3 | 9.3e1 | 7.0e1 | 5.1e1 | 4.8e1 | 1.3e2 | 4.3e1 | 2.0e1 | 8.0e0 | 1.3e1 | 2.2e1 | 2.9e1 | 3.4e1 | 1.3e1 | 1.5e1 | ND |
| Sp-3 | 2.2e1 | 6.3e0 | 1.0e2 | 1.6e1 | 1.3e1 | 7.7e2 | 5.0e1 | 5.5e1 | 5.8e1 | 6.0e1 | 8.0e1 | 8.8e1 | 1.4e2 | 2.5e2 | 1.8e2 |
| RpRc-4 | 3.4e0 | 5.5e-1 | 4.1e-1 | 4.5e0 | 1.1e0 | 5.8e-1 | 2.0e0 | 2.4e0 | 4.3e0 | ND | 4.0e0 | ND | ND | ND | 2.9e0 |
| RpSc-4 | 4.5e-1 | 1.7e-1 | ND | 4.2e-1 | 3.3e-1 | 1.9e0 | 2.1e-1 | 1.4e0 | ND | ND | 8.9e-1 | 1.9e0 | ND | ND | 1.9e0 |
| SpRc-4 | 7.7e-1 | 6.3e-1 | 6.3e0 | 1.3e0 | 1.2e0 | 4.3e0 | 1.2e1 | 1.4e1 | 5.0e1 | 6.4e1 | 3.1e1 | 3.1e1 | 1.6e1 | 4.7e1 | 1.7e1 |
| SpSc-4 | 1.6e-2 | 3.3e-1 | 2.1e0 | ND | 5.e-1 | 3.2e0 | 2.9e0 | 6.5e0 | 1.2e1 | 2.4e1 | 5.7e1 | 8.1e0 | 4.2e0 | 5.6e0 | 6.1e0 |
| Rp-5 | ND | 3.8e1 | 5.9e0 | 2.5e1 | 2.2e1 | 1.7e1 | 8.1e-1 | ND | 9.7e-1 | ND | ND | ND | 2.2e0 | ND | 3.3e0 |
| Sp-5 | ND | ND | 5.1e0 | 1.3e-1 | 4.1e-1 | 7.2e0 | 1.9e1 | 3.1e0 | 4.7e1 | 4.4e1 | 2.6e1 | 3.1e1 | 4.4e1 | 1.2e2 | 1.2e2 |
The standard errors, from fits of the data to equation 1, are less than 20% of the stated values.
Table 4.
The values of kcat/Km (M−1 s−1) for the wild-type and mutant for of PTEa.
| WT | QF | YT | RN | QFRN | YTRN | GWT | GWT-d1 | GWT-d2 | GWT-d3 | GWT-f1 | GWT-f2 | GWT-f3 | GWT-f4 | GWT-f5 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rp-1 | 4.9e5 | 1.5e6 | 2.4e3 | 1.7e5 | 3.0e6 | 4.3e3 | 1.5e3 | 2.5e3 | 6.1e3 | 9.1e3 | 1.0e5 | 5.2e4 | 6.1e3 | 7.2e3 | 8.5e3 |
| Sp-1 | 1.2e6 | 7.1e6 | 1.6e5 | 7.4e5 | 8.0e6 | 1.7e5 | 2.2e5 | 1.9e5 | 1.2e6 | 1.2e6 | 7.2e5 | 6.2e5 | 1.3e5 | 2.3e5 | 2.9e5 |
| Rp-2 | 5.8e5 | 8.2e5 | 2.5e3 | 2.8e5 | 1.6e6 | 3.4e3 | 1.8e3 | 3.5e3 | 6.0e3 | 8.1e3 | 1.4e4 | 4.7e3 | 3.4e3 | 4.2e3 | 6.1e3 |
| Sp-2 | 2.7e4 | 1.2e6 | 1.1e5 | 2.6e4 | 1.6e6 | 1.4e5 | 5.9e4 | 8.6e4 | 4.6e5 | 4.3e5 | 1.4e5 | 1.9e5 | 1.2e5 | 2.4e5 | 4.2e5 |
| Rp-3 | 8.5e5 | 3.1e5 | 1.3e4 | 4.0e5 | 1.3e6 | 1.3e4 | 1.5e3 | 3.0e3 | 5.4e3 | 7.6e3 | 3.3e3 | 3.8e3 | 8.2e2 | 2.6e3 | 2.2e3 |
| Sp-3 | 3.4e4 | 1.6e6 | 7.3e4 | 4.8e4 | 1.4e6 | 3.9e5 | 1.8e5 | 5.0e5 | 1.1e6 | 1.2e6 | 6.7e5 | 1.0e6 | 1.1e6 | 2.3e6 | 1.9e6 |
| RpRc-4 | 1.3e3 | 1.9e2 | 5.8e1 | 3.0e3 | 2.8e2 | 3.0e2 | 2.2e2 | 6.8e2 | 6.4e2 | 5.6e2 | 4.2e2 | 4.1e2 | 6.3e2 | 7.9e2 | 8.5e2 |
| RpSc-4 | 2.0e2 | 5.5e1 | 1.6e1 | 4.6e2 | 7.4e1 | 1.9e2 | 1.3e2 | 1.2e2 | 1.5e2 | 1.4e2 | 2.1e2 | 1.4 e2 | 1.4e2 | 2.0e2 | 1.8e2 |
| SpRc-4 | 1.1e2 | 1.6e3 | 1.8e3 | 2.3e2 | 1.8e3 | 1.2e3 | 8.1e3 | 2.3e4 | 6.0e4 | 8.7e4 | 1.3e4 | 1.4e4 | 3.2e3 | 5.0e3 | 3.8e3 |
| SpSc-4 | 3.2e0 | 6.2e1 | 2.5e2 | 1.5e1 | 9.6e1 | 4.8e2 | 1.7e3 | 4.2e3 | 8.1e3 | 1.1e4 | 2.6e3 | 2.5e3 | 1.5e3 | 1.5e3 | 1.2e3 |
| Rp-5 | 1.6e4 | 5.2e3 | 1.9e3 | 1.7e4 | 9.0e3 | 3.5e3 | 2.5e2 | 3.0e2 | 5.4e2 | 5.5e2 | 6.0e2 | 8.6e2 | 4.5e2 | 5.1e2 | 7.7e2 |
| Sp-5 | 2.1e1 | 3.3e2 | 5.8e3 | 2.8e1 | 3.6e2 | 1.4e4 | 2.8e4 | 1.0e4 | 5.2e4 | 1.5e5 | 3.9e4 | 7.7e4 | 1.2e5 | 2.5e5 | 3.2e5 |
The standard errors, from fits of the data to equation 1, are less than 20% of the stated values.
Figure 3.
Outline of the parental lineage for the construction of mutants of PTE that are enhanced for the hydrolysis of Sp-4 and Sp-5.
Structural Analysis of Enhanced Mutants
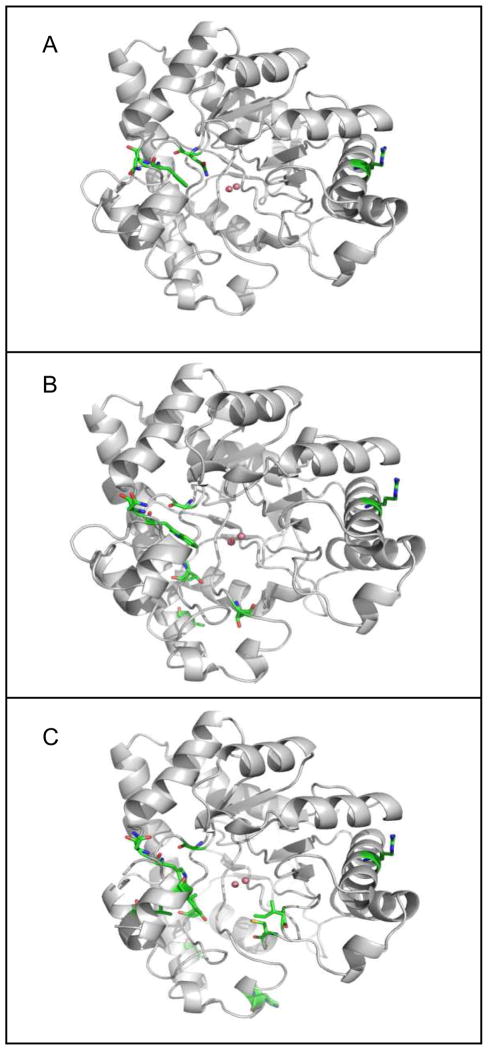
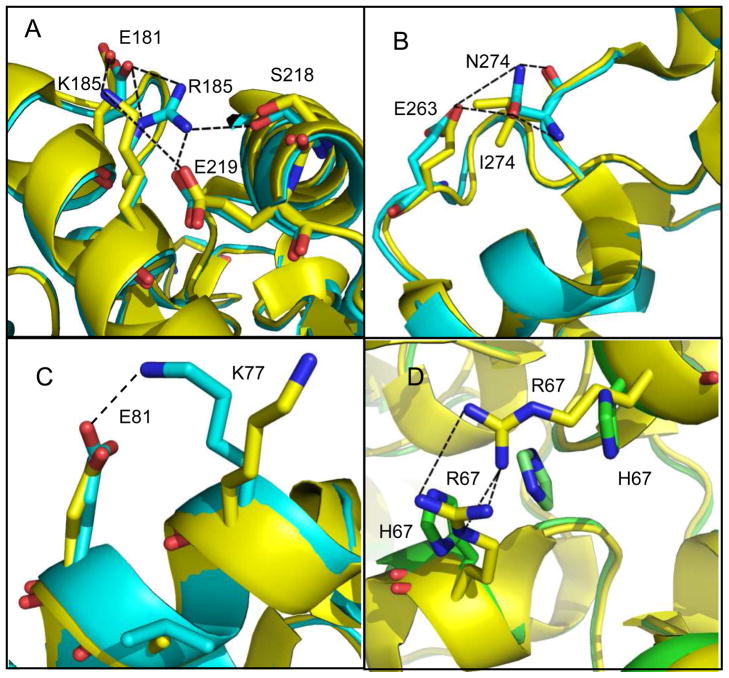
The three-dimensional structures of RN, QF-RN, GWT-d2, GWT-d3, and GWT-f5 were determined. The backbone conformations of the residues in these proteins are quite similar to one another. Superposition of QF-RN, GWT-d3 and GWT-f5 with wild-type PTE (PDB ID: 1HZY) provides a Cα atom rmsd of 0.37, 0.38 and 0.71 Å, respectively. The locations of the mutations within QF-RN, GWT-d3, and GWT-f5 are highlighted in Figure 4. The K185R and I274N mutations are on the surface of the protein. Arg-185 extends outward from a-helix 4 and Asn-274 is positioned in loop 7. The arginine in the K185R mutation forms hydrogen bond interactions with Ser-218 and Glu-219 (Figure 5a). In GWT-d3, the distance between Nω of Arg-185 and the nearest carboxylate oxygen of Glu-219 is 2.8 Å, whereas the distance to the hydroxyl group of Ser-218 is 3.0 Å. The I274N mutation changes a hydrophobic residue for a hydrophilic group. In GWT-d3, the carboxamide group of Asn-274 forms a hydrogen bond interaction with the main chain amide group of itself and an additional hydrogen bond with the side chain carboxylate of Glu-263 (Figure 5b). The mutation of Ala-80 to valine does not alter the orientation of the side chain or backbone. However, the side chain of the neighboring Lys-77 within GWT-f5 and GWT-d3 is displaced up to 5.3 Å relative to the parental GWT structure (PDB: 1P6B and 1P6C). This movement facilitates a hydrogen bond interaction between Lys-77 and Glu-81 (Figure 5c). The R67H mutation in GWT-f5 is positioned at the interface of two subunits. In the parental structure the arginines from different subunits appear to form a hydrogen bond interaction with each other (3.1 Å). In the structure of GWT-f5 there is an imidazole from solvent that is positioned between the side chains of the two histidine residues (Figure 5d).
Figure 4.
The localization of the mutations made in QFRN (A), GWT-d3 (B) and GWT-f5 (C). The mutated residues are highlighted in green.
Figure 5.
Structural perturbations at the mutated sites in GWT-d3 and GWT-f5. (A) The parent enzyme (GWT) in the region of Lys-185 is highlighted in yellow and the mutant, GWT-d3, is highlighted in cyan. (B) The parent enzyme (GWT) in the region of I274 is highlighted in yellow and the mutant, GWT-d3 is highlighted in cyan. (C) The parent enzyme (GWT) in the region of Ala-80 is highlighted in yellow and the mutant, GWT-f5 is highlighted in cyan. (D) The parent enzyme (GWT) in the region of Arg-67 is highlighted in yellow and the mutant, GWT-f5, is highlighted in green. An imidazole is found in a π–stacking arrangement between the two histidine residues at the subunit interface.
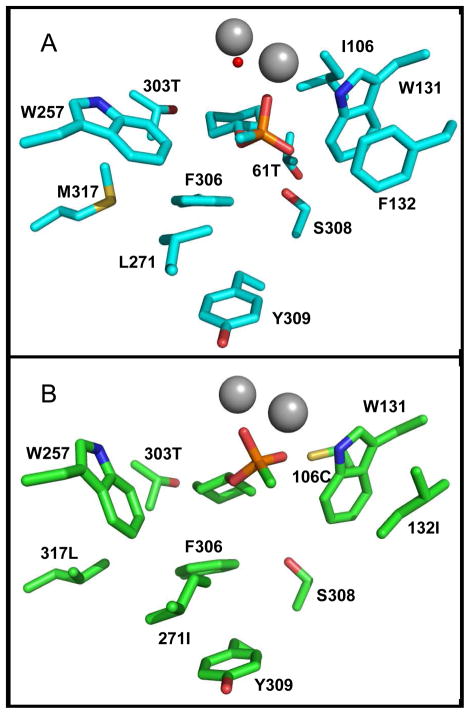
Several changes in structure are observed in GWT-d3 without bound ligands compare to the structures of GWT-d3 complexed with compound 7 and GWT (PDB: 1P6B and 1P6C). The indole ring of Trp-257 moves 1.2 Å closer to the glycine at position 254, the phenyl group of Phe-306 is 0.98 Å closer to the binuclear metal center, and Thr-303 is twisted with the side chain hydroxyl group pointing inward toward the binding pocket surface. However, there are no major differences in the active site residues in the GWT-d3 structure complexed with compound 7 and GWT.
The amino acids residues in the binding pocket of GWT-f5 with compounds 6 and 7 exhibit several alterations compared to the parental GWT structure (PDB: 1P6B and 1P6C). The indole ring of Trp-257 moves 2.0 Å closer to the leucine at residue position 317 because more space is provided by the shorter leucine side chain relative to methionine. The indole ring of Trp-131 is displaced around 1.0 Å, and the mutation at the 132 residue shifts the main chain 1.1 Å (Figure 6a). Comparing the structures of GWT-f5 with compounds 6 and 7, the side chains of the isoleucine at residue position 271 are twisted; Cδ and Cγ2 of Ile-271 are displaced 1.5 Å and 3.0 Å, respectively. The large O-cyclohexyl group of compound 7 is pointed towards the enlarged small pocket, and the small methyl group is between the smaller large pocket and the leaving group pocket.
Figure 6.
The substrate binding pocket of GWT-d3 (A) and GWT-f5 (B) complexed with compound 7.
Screening and Kinetic Analysis of Wild-Type and Mutant Enzymes on GB, GD and GF
For purposes of initial screening, specific activities were determined with 3 mM substrate (Table 5). The YT mutant was very active with all three substrates, and the YT-RN mutant had higher activity than wild-type with GD. Therefore, kinetic constants were determined for these enzyme/substrate combinations (Table 6). The stereospecificity of variants was determined by following the fluoride released during the complete hydrolysis of GB or GF using 0.5 mM of the racemic substrate. Substantial differences in the rates for the individual enantiomers result in biphasic curves (data not shown). The specificity of GWT towards GF is known from polarimetry experiments,9 while the stereopreference of the remaining variants was determined by the ability of the variant to complement the slow phase in the GWT catalyzed reaction (Table 5). YT has the same stereo-preference as GWT for GF and has previously been shown to have the same preference as GWT for GD.5 The stereopreference for the variants toward GB was determined by the ability to complement the slow phase in the YT catalyzed reaction (Table 5).
Table 5.
Activity of wild-type and mutant enzymes with racemic G-agentsa.
| Enzyme | GB | GD | GF | GBb preferred enantiomer | GFb preferred enantiomer |
|---|---|---|---|---|---|
| Wild-type | 303 | 14 | 363 | NAc | RP |
| YT | 843 | 212 | 240 | SP | SP |
| YT-RN | 263 | 115 | 116 | SP | SP |
| QF-RN | 32 | 1.0 | 41 | NA | NA |
| GWT | 20 | 2.0 | 44 | SP | SP |
| GWT-d1 | 57 | 2.0 | 7 | SP | SP |
| GWT-d2 | 52 | 1.0 | 211 | SP | SP |
| GWT-d3 | 48 | 8 | 35 | SP | SP |
| GWT-f3 | 142 | 10 | 94 | SP | SP |
| GWT-f5 | 240 | 19 | 59 | SP | SP |
micromoles/minute/mg protein
Determined with 0.5 mM racemic substrate.
NA; no significant stereopreference under these conditions.
Table 6.
Kinetic constants for hydrolysis of GB, GD and GFa.
| Enzyme | Substrate | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Wild-type | GB | 430 ± 50 | 1800 ± 400 | 2.4 (0.6) × 105 |
| Wild-type | GD | 12 ± 1 | 800 ± 200 | 1.5 (0.4) × 104 |
| Wild-type | GF | 210 ± 30 | 900 ± 300 | 2.3 (0.8) × 105 |
| YT | GB | 520 ± 30 | 260 ± 50 | 2.0 (0.4) × 106 |
| YT | GD | 240 ± 20 | 460 ± 90 | 5 (1) × 105 |
| YT | GF | 130 ± 10 | 170 ± 50 | 8 (2) × 105 |
| YTRN | GD | 100 ± 10 | 300 ± 100 | 4 (2) × 105 |
Racemic mixtures of GB, GD, and GF were used for these measurements.
Discussion
Wild-type PTE catalyzes the hydrolysis of the insecticide paraoxon with kinetic constants that approach ~104 s−1 and ~108 M−1 s−1 for kcat and kcat/Km, respectively.21 However, the catalytic efficiency of the wild-type enzyme for the hydrolysis of organophosphorus nerve agents is significantly smaller, and the enzyme prefers the less toxic RP-enantiomers relative to the more toxic SP-enantiomers.5 The degree of stereoselectivity exhibited by the wild-type enzyme for chiral substrates is dependent to a significant extent on the size of the substituents attached to the central phosphorus core.6 The QF mutant was shown previously to exhibit the highest activity towards the more toxic SP-enantiomers of compounds 1, 2, and 3, which are analogues of VX, GB, and VR, respectively.5 The GWT mutant possessed the highest hydrolytic activity towards the more toxic SP-enantiomers of compounds 4 and 5, which are analogs of soman and cyclosarin, respectively.5 These mutants were selected for further modification in an attempt to increase the catalytic activity of PTE through directed evolution.
The introduction of the K185R and I274N modifications to PTE was initiated in an attempt to increase net protein expression levels and solubility. These two mutations also enhanced the hydrolytic activity for some of the compounds presented in Scheme 2. The QF-RN mutant has higher values of kcat and kcat/Km for most of the compounds tested, relative to the parent QF mutant. QF-RN has the best activity towards the SP-1 and SP-2, analogues of VX and sarin, respectively. Overall, the values of kcat/Km for these two compounds are 7- and 60-fold higher than wild-type PTE and the absolute magnitude of kcat/Km exceeds 106 M−1 s−1 for each compound.
An in vitro screening strategy was utilized to identify mutants that are further optimized starting from the GWT-d1 parent. The GWT-f1 enzyme contains an additional M317L mutation and is ~4-fold better in terms of kcat/Km than the GWT-d1 mutant for the hydrolysis of SP-5 and ~2,000-fold better than the wild-type PTE. The GWT-f1 mutant was subsequently co-expressed with GpdQ in E. coli cells. These cells can grow in a phosphate-free minimal medium with a 165-hour lag phase using SP-5 as the sole phosphorus source. Therefore, the in vivo selection method is practical when SP-5 is used to screen large mutant libraries. GWT-f3 was identified from a mutant library containing six partially-randomized residues within the active site of PTE. The additional changes to the protein sequence included I106C, F132I and L271I. GWT-f3 was ~3-fold better in terms of kcat/Km than the GWT-f1 parent and ~6,000-fold better than the wild-type PTE for the hydrolysis of SP-5.
The A80V mutation was added to GWT-f3 to create the GWT-f4 mutant. This change increased the catalytic efficiency for the hydrolysis of SP-5 about 2-fold. The GWT-f4 mutant was subjected to error-prone PCR mutagenesis and GWT-f5 with a R67H substitution was identified using the selection strategy in phosphate-free medium with SP-5 as the sole phosphorus source. GWT-f5 was ~50% more active for the hydrolysis of SP-5 than the parent enzyme and the value of kcat/Km was ~3 × 105 M−1 s−1. This is the best mutant for the hydrolysis of the GF analogue and it is ~15,000-fold better than the wild-type enzyme. GWT-f5 is also the best mutant for the hydrolysis of SP-3, the analogue of the nerve agent VR. For this compound the value of kcat/Km is ~2 × 106 M−1 s−1 and the rate enhancement, relative to the wild type enzyme, is increased by nearly two orders of magnitude.
We have previously shown that the GpdQ diesterase is unable to hydrolyze the product produced by the action of PTE on SPSC-4 and SPRC-4, analogues of the two most toxic diastereomers of soman.21,31 Therefore, the selection strategy for the identification of beneficial mutations for the hydrolysis of these compounds could not be utilized. Nevertheless, utilization of the in vitro screening approach yielded mutants with substantially enhanced kinetic constants. GWT was previously identified as being a good catalyst for the hydrolysis of the two SP-diastereomers of compound 4. The addition of the RN mutations to this variant gave GWT-d1. These changes enhanced the values of kcat/Km by a factor of ~3 for the hydrolysis of each compound relative to GWT. The addition of the A80V mutation to GWT-d1 further enhanced the hydrolysis rates another 2–3 fold. Finally, the GWT-d2 mutant was modified with two double substitution libraries and the S61T change was identified via screening of individual colonies. GWT-d3 is nominally better than GWT-d2 for the hydrolysis of the SP-diastereomers of compound 4. Overall, GWT-d2 is enhanced ~800-fold, relative to the wild -type enzyme, for the hydrolysis of SPRC-4 and ~3500-fold for the hydrolysis of SPSC-4. The absolute values for kcat/Km for the hydrolysis for SPRC-4 are ~105 M−1 s−1 and 104 M−1 s−1 for the hydrolysis of SPSC-4.
While the analogs 1–5 contain the authentic chiral centers seen in the nerve agents, the leaving groups are very different making verification of the analog data important. The variant GWT-d3 was developed with the GD analog (4) and displays a 4-fold improvement over the GWT parent with authentic GD. This correlates well with the 6-fold improvement predicted by the analog. It is important to note that the wild-type enzyme hydrolyzes the toxic SPSC enantiomer of GD more than 15-fold slower than the other three enantiomers while GWT hydrolyzes this enantiomer with a similar rate to the SPRC enatiomer.5 The measured rate of hydrolysis of racemic GD is relatively low, but the enhancements in the GWT-d3 variant, which has the same stereoselectivity profile as GWT, represents greater than a 60-fold improvement against the toxic enantiomers of GD. Similarly the variants that were developed for the hydrolysis of GF (GWT-f3 and GWT-f5) are specific for the hydrolysis of the SP-enantiomer. Wild-type PTE is known to hydrolyze the SP enantiomer ~20-fold slower than the RP-enantiomer, while the GWT variant prefers the SP-enantiomer by 6-fold.9 The absolute value of the measured activity against the racemic GF is lower for the improved variants, but taking into account the switch in specificity the activity against the SP enantiomer of GF is improved ~3-fold for GWT and GWT-f5 and by ~5-fold for the GWT-f3 variant as predicted from the analog studies.
The data collected with authentic nerve agents did contain some unexpected results. The variants GWT-f3 and GWT-f5 were substantially enhanced over GWT for the hydrolysis of both GB and GD with nearly an order of magnitude improvement seen for both nerve agents. Similarly the GWT-d2 variant is 5-fold better than GWT against GF, and taking into account the different stereopreference, more than an order of magnitude better than wild-type for the hydrolysis of the SP-enantiomer. The best variant identified to date against GB, GD, and GF is the YT variant with kcat/Km values of 2.0 × 106 M−1s−1, 5 × 105 M−1s−1, and 8 × 105 M−1s−1 respectively. Not only is this variant significantly improved over wild-type PTE for all of the nerve agents it also prefers the more toxic SP-enantiomers.
The other enzymes that are known to hydrolyze the G-type nerve agents, organophosphorus acid anhydrolase (OPAA), diisopropylfluorophosphatase (DFPase), and human paraoxonase 1 (hPON1), have catalytic efficiencies that lag behind the YT variant of PTE. The G-agents are hydrolyzed by OPAA with high kcat values ~600 s−1 (GB), 3100 s−1 (GD) and 1700 s−1 (GF), but the Km values for all of the G-agents are estimated to be between 1–10 mM, and it is known that OPAA is selective for the less toxic RP enantiomers of the G-agents.32,33,34 The YT mutant of PTE has approximately an order of magnitude better catalytic efficiency against GB. Similarly, The high Km values of OPAA for GD and GF and the known preference for the hydrolysis of the RP-enantiomers makes the activity of the YT variant active substantially better against the more toxic SP-enantiomers.
Variants optimized for the SP-enantiomers of the G-agents have been reported for both DFPase and hPON1. The improved hPON1 variant shows little stereoselectivity against GB, but the reported kcat/Km value of 5 × 103 M−1 s−1 is significantly below that of the YT variant.35 The best DFPase variant is selective for the SP-enantiomer of GB and has a kcat/Km value of 2.3 × 105 M−1 s−1, but the Km is greater than 10 mM.36 The optimized DPFase variants prefer the SP-enantiomers of GD and GF, but the Km values are in excess of 10 mM. For GF the best variant of DFPase has a kcat/Km value of 2.3 × 105 M−1 s−1 which is somewhat below that measured for the YT variant.36 The optimized hPON1 variant has kcat/Km values for the two SP-enantiomers of GD of 1.2 × 105 M−1 s−1 and 1 × 104 M−1 s−1, both of which are below the values measured for the YT variant of PTE, and the kcat/Km value for SP-GF of 2.9 × 105 M−1 s−1 is ~2.5-fold lower than that of the YT variant.35
The X-ray crystal structures of QF-RN, GWT-d3 and GWT-f5 show that the K185R and I274N mutations provide additional hydrogen bonding interactions with adjacent amino acid resides. Molecular modeling of the RN mutations predicted that Glu-219 and Glu-181 could form hydrogen bonds with the newly introduced Arg-185 and that Glu-263 could potentially form a hydrogen bond with the newly introduced Asn-274 residue (28). The crystal structures of QF-RN, GWT-d3 and GWT-f5 demonstrated that Ser-218 can also form hydrogen bond with Arg-185. These crystal structures demonstrate that the K185R and I274N mutations provide more hydrogen bonding interactions with neighboring amino acid residues which could enhance conformational stability. In the GWT-d3 and GWT-f5 structures with compound 7, the large cyclohexyl group of compound 7 points towards the enlarged small pocket. This suggests that the more toxic of SP-diastereomers of compounds 1–5 may enter active site of PTE with the larger substituents of these compounds pointing towards the enlarged small pocket and preferentially hydrolyzed with higher activities.
Given the dramatic changes in activity, it is somewhat surprising that no major structural changes to the active site are observed in the improved variants. The majority of changes to active site residues only involve the movement of side chains by 1–2 Å. The slight changes to the protein scaffold brought about by the new hydrogen bonding networks and alterations to the active site combine to bring about dramatic changes in the activity and stereoselectivity. The diminutive nature of these changes makes them hard to predict but the strategy outlined in Figure 3 has allowed significantly improved variants to be identified. This combination of site directed mutagenesis, active site library screening, and in vivo selection has allowed the development of variants with altered stereoselectivity and rate enhancements for hydrolysis of compounds 1–5. GWT from the first generation of mutants was demonstrated to have an inverted stereoselectivity and enhanced catalytic activity towards the more toxic Sp-enantiomers of the soman and cyclosarin analogues.5 The structural modifications to GWT within the substrate binding pocket, surface, and dimer interface have substantially enhanced substrate binding and catalytic turnover. The changes in the values of kcat/Km the SP-enantiomers of the organophosphonates are illustrated in Figure 7.
Figure 7.
Bar graphs illustrating enhanced values for kcat/Km (M−1 s−1) for the Sp-enantiomer of compound 1 (A), 2 (B) and 3 (C), SpRc-4 (D), SpSc-4 (E) and 5 (F) as catalyzed by the wild-type and engineered mutants of PTE.
Abbreviations
- PTE
phosphotriesterase
- GpdQ
glycerophosphodiesterase
- OpdA
organophosphate degrading enzyme from Agrobacterium radiobacter
- PEG MME
polyethylene glycol monomethyl ether
Footnotes
This work was supported in part by the NIH (GM 68550). The X-ray coordinates and structure factors for the PTE mutants have been deposited in the Protein Data Bank: 3UR2, 3UPM, 3URA, 3URN, 3URB, 3URQ and 3UR5.
References
- 1.Dumas DP, Durst HD, Landis WG, Raushel FM, Wild JR. Inactivation of organophosphorus nerve agents by the phosphotriesterase from Pseudomonas diminuta. Arch Biochem Biophys. 1990;277:155–159. doi: 10.1016/0003-9861(90)90564-f. [DOI] [PubMed] [Google Scholar]
- 2.Lai K, Grimsley JK, Kuhlmann BD, Scapozza L, Harvey SP, DeFrank JJ, Kolakowski JE, Wild JR. Rational enzyme design: Computer modeling and site-directed mutagenesis for the modification of catalytic specificity in organophosphorus hydrolase. Chimia. 1996;50:430–431. [Google Scholar]
- 3.Raveh L, Segall Y, Leader H, Rothschild N, Levanon D, Henis Y, Ashani Y. Protection against tabun toxicity in mice by prophylaxis with an enzyme hydrolyzing organophosphate esters. Biochem Pharma. 1992;22:397–400. doi: 10.1016/0006-2952(92)90028-h. [DOI] [PubMed] [Google Scholar]
- 4.Caldwell SR, Newcomb JR, Schlecht KA, Raushel FM. Limits of diffusion in the hydrolysis of substrates by the phosphotriesterase from Pseudomonas diminuta. Biochemistry. 1991;30:7438–7444. doi: 10.1021/bi00244a010. [DOI] [PubMed] [Google Scholar]
- 5.Tsai PC, Bigley A, Li Y, Ghanem E, Cadieux CL, Kasten SA, Reeves TE, Cerasoli DM, Raushel FM. Stereoselective hydrolysis of organophosphate nerve agents by the bacterial phosphotriesterase. Biochemistry. 2010;49:7978–7987. doi: 10.1021/bi101056m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen-Goodspeed M, Sogorb MA, Wu F, Hong SB, Raushel FM. Structural determinants of the substrate and stereochemical specificity of phosphotriesterase. Biochemistry. 2001;40:1325–1331. doi: 10.1021/bi001548l. [DOI] [PubMed] [Google Scholar]
- 7.Chen-Goodspeed M, Sogorb MA, Wu F, Raushel FM. Enhancement, relaxation, and reversal of the stereoselectivity for phosphotriesterase by rational evolution of active site residues. Biochemistry. 2001;40:1332–1339. doi: 10.1021/bi001549d. [DOI] [PubMed] [Google Scholar]
- 8.Nowlan C, Li Y, Hermann JC, Evans T, Carpenter J, Ghanem E, Shoichet BK, Raushel FM. Resolution of chiral phosphate, phosphonate, and phosphinate esters by an enantioselective enzyme library. J Am Chem Soc. 2006;128:15892–15902. doi: 10.1021/ja0658618. [DOI] [PubMed] [Google Scholar]
- 9.Harvey SP, Kolakowski JE, Chen TC, Rastogi VK, Reiff LP, DeFrank JJ, Raushel FM, Hill C. Stereospecificity in the enzymatic hydrolysis of cyclosarin (GF) Enzyme Microb Technol. 2005;37:547–555. [Google Scholar]
- 10.Benschop HP, Dejong LPA. Nerve Agent Stereoisomers - Analysis, Isolation, and Toxicology. Acc Chem Res. 1988;21:368–374. [Google Scholar]
- 11.Hill CM, Li WS, Thoden JB, Holden HM, Raushel FM. Enhanced degradation of chemical warfare agents through molecular engineering of the phosphotriesterase active site. J Am Chem Soc. 2003;125:8990–8991. doi: 10.1021/ja0358798. [DOI] [PubMed] [Google Scholar]
- 12.Li WS, Lum KT, Chen-Goodspeed M, Sogorb MA, Raushel FM. Stereoselective detoxification of chiral sarin and soman analogues by phosphotriesterase. Bioorg Med Chem. 2001;9:2083–2091. doi: 10.1016/s0968-0896(01)00113-4. [DOI] [PubMed] [Google Scholar]
- 13.Cho CMH, Mulchandani A, Chen W. Functional analysis of organophosphorus hydrolase variants with high degradation activity towards organophosphate pesticides. Protein Eng Des Sel. 2006;19:99–105. doi: 10.1093/protein/gzj007. [DOI] [PubMed] [Google Scholar]
- 14.Cho CMH, Mulchandani A, Chen W. Altering the substrate specificity of organophosphorus hydrolase for enhanced hydrolysis of chlorpyrifos. Appl Environ Microbiol. 2004;70:4681–4685. doi: 10.1128/AEM.70.8.4681-4685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roodveldt C, Tawfik DS. Directed evolution of phosphotriesterase from Pseudomonas diminuta for heterologous expression in Escherichia coli results in stabilization of the metal-free state. Protein Eng Des Sel. 2005;18:51–58. doi: 10.1093/protein/gzi005. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, Carr PD, McLoughlin SY, Liu JW, Horne I, Qiu X, Jeffries CM, Russell RJ, Oakeshott JG, Ollis DL. Evolution of an organophosphate-degrading enzyme: a comparison of natural and directed evolution. Protein Eng. 2003;16:135–145. doi: 10.1093/proeng/gzg013. [DOI] [PubMed] [Google Scholar]
- 17.Reetz MT, Carballeira JD, Vogel A. Iterative saturation mutagenesis on the basis of B factors as a strategy for increasing protein thermostability. Angew Chem Int Ed Engl. 2006;45:7745–7751. doi: 10.1002/anie.200602795. [DOI] [PubMed] [Google Scholar]
- 18.Machius M, Declerck N, Huber R, Wiegand G. Kinetic stabilization of Bacillus licheniformis alpha-amylase through introduction of hydrophobic residues at the surface. J Biol Chem. 2003;278:11546–11553. doi: 10.1074/jbc.M212618200. [DOI] [PubMed] [Google Scholar]
- 19.Ghanem E, Li Y, Xu C, Raushel FM. Characterization of a phosphodiesterase capable of hydrolyzing EA 2192, the most toxic degradation product of the nerve agent VX. Biochemistry. 2007;46:9032–9040. doi: 10.1021/bi700561k. [DOI] [PubMed] [Google Scholar]
- 20.McLoughlin SY, Jackson C, Liu JW, Ollis DL. Growth of Escherichia coli coexpressing phosphotriesterase and glycerophosphodiester phosphodiesterase, using paraoxon as the sole phosphorus source. Appl Environ Microbiol. 2004;70:404–412. doi: 10.1128/AEM.70.1.404-412.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aubert SD, Li Y, Raushel FM. Mechanism for the hydrolysis of organophosphates by the bacterial phosphotriesterase. Biochemistry. 2004;43:5707–5715. doi: 10.1021/bi0497805. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Ccrystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.McRee DE. XtalView/Xfit A Versatile Program for Manipulating Atomic Coordinates and Electron Density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges N, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallographic and NMR system (CNS): A new software system for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Bruns CM, Hubatsch I, Ridderstrom M, Mannervik B, Tainer JA. Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products. J Mol Biol. 1999;288:427–439. doi: 10.1006/jmbi.1999.2697. [DOI] [PubMed] [Google Scholar]
- 29.Wanner BL. Molecular genetics of carbon-phosphorus bond cleavage in bacteria. Biodegradation. 1994;5:175–184. doi: 10.1007/BF00696458. [DOI] [PubMed] [Google Scholar]
- 30.Kononova SV, Nesmeyanova MA. Phosphonates and their degradation by microorganisms. Biochemistry (Mosc) 2002;67:184–195. doi: 10.1023/a:1014409929875. [DOI] [PubMed] [Google Scholar]
- 31.Benschop HP, Konings CAG, Genderen JV, DeJong LPA. Isolation, anticholinesterase properties, and acute toxicity in mice of the four stereoisomers of the nerve agent soman. Toxicol Appl Pharmacol. 1984;72:61–74. doi: 10.1016/0041-008x(84)90249-7. [DOI] [PubMed] [Google Scholar]
- 32.DeFrank JJ, Beaudry WT, Cheng TC, Harvey SP, Stroup AN, Szafraniec LL. Screening of halophilic bacteria and Alteromonas species for organophosphorus hydrolyzing enzyme activity. Chem Biol Interact. 1993;87:141–148. doi: 10.1016/0009-2797(93)90035-w. [DOI] [PubMed] [Google Scholar]
- 33.Cheng TC, Liu L, Wang B, Wu J, DeFrank JJ, Anderson DM, Rastogi VK, Hamilton AB. Nucleotide sequence of a gene encoding an organophosphorus nerve agent degrading enzyme from Alteromonas haloplanktis. J Ind Microbiol Technol. 1997;18:49–55. doi: 10.1038/sj.jim.2900358. [DOI] [PubMed] [Google Scholar]
- 34.Hill CM, Li WS, Cheng TC, DeFrank JJ, Raushel FM. Stereochemical specificity of organophosphorus acid anhydrolase toward p-nitrophenyl analogs of soman and sarin. Bioorg Chem. 2001;29:27–35. doi: 10.1006/bioo.2000.1189. [DOI] [PubMed] [Google Scholar]
- 35.Gupta RD, Goldsmith M, Ashani Y, Simo Y, Mullokandov G, Bar H, Ben-David M, Leader H, Margalit R, Silman I, Sussman JL, Tawfik DS. Directed evolution of hydrolases for prevention of G-type nerve agent intoxication. Nat Chem Biol. 2011;7:120–125. doi: 10.1038/nchembio.510. [DOI] [PubMed] [Google Scholar]
- 36.Melzer M, Chen JCH, Heidenreich A, Gab J, Koller M, Kehe K, Blum MM. Reversed enantioselectivity of diisopropyl fluorophosphatase against organophosphorus nerve agents by rational design. J Am Chem Soc. 2009;131:17226–17232. doi: 10.1021/ja905444g. [DOI] [PubMed] [Google Scholar]