Abstract
Background and Aims
Flow cytometry has been used to measure nuclear DNA content in pollen, mostly to understand pollen development and detect unreduced gametes. Published data have not always met the high-quality standards required for some applications, in part due to difficulties inherent in the extraction of nuclei. Here we describe a simple and relatively novel method for extracting pollen nuclei, involving the bursting of pollen through a nylon mesh, compare it with other methods and demonstrate its broad applicability and utility.
Methods
The method was tested across 80 species, 64 genera and 33 families, and the data were evaluated using established criteria for estimating genome size and analysing cell cycle. Filter bursting was directly compared with chopping in five species, yields were compared with published values for sonicated samples, and the method was applied by comparing genome size estimates for leaf and pollen nuclei in six species.
Key Results
Data quality met generally applied standards for estimating genome size in 81 % of species and the higher best practice standards for cell cycle analysis in 51 %. In 41 % of species we met the most stringent criterion of screening 10 000 pollen grains per sample. In direct comparison with two chopping techniques, our method produced better quality histograms with consistently higher nuclei yields, and yields were higher than previously published results for sonication. In three binucleate and three trinucleate species we found that pollen-based genome size estimates differed from leaf tissue estimates by 1·5 % or less when 1C pollen nuclei were used, while estimates from 2C generative nuclei differed from leaf estimates by up to 2·5 %.
Conclusions
The high success rate, ease of use and wide applicability of the filter bursting method show that this method can facilitate the use of pollen for estimating genome size and dramatically improve unreduced pollen production estimation with flow cytometry.
Keywords: Pollen, flow cytometry, unreduced gametes, genome size, nuclei extraction, 2n pollen, cell cycle
INTRODUCTION
Flow cytometry has become an important tool for measuring nuclear DNA content in plant biology, with applications in crop and horticultural science (Leus et al., 2009), genome size research (Leitch and Bennett, 2007), and other areas of population and evolutionary biology (Kron et al., 2007). Publications featuring the DNA content of pollen nuclei represent a small subset of that work, numbering fewer than one a year since Van Tuyl et al.'s (1989) study of Lilium pollen (Table 1).
Table 1.
Plant genera for which flow cytometry has been used to measure the relative DNA content of extracted pollen nuclei
| Genus | Family | No. of species | Primary methods | Tested methods | Reference |
|---|---|---|---|---|---|
| Alstroemeria | Alstroemeriaceae | 1 | C2, O2 | 6 | |
| Begonia | Begoniaceae | 25* | O3, S | C1, O1, O2 | 4, 5 |
| Brassica | Brassicaceae | 1 | S | C1, O1, O2 | 11 |
| Cupressus | Cupressaceae | 2 | C2 | Q | 12 |
| Dendranthema | Asteraceae | 1 | C1 | 2 | |
| Diospyros | Ebenaceae | 4 | C1 | 15, 16 | |
| Hibiscus | Malvaceae | 2 | C1 | 17 | |
| Lilium | Liliaceae | 1† | C1, F | 1, 2, 9, 18 | |
| Petunia | Solanaceae | 1 | C1 | 8 | |
| Rosa | Rosaceae | 1 or 2‡ | B, Q | 7, 13 | |
| Rumex | Polygonaceae | 2 | C1, C3 | 3, 14 | |
| Triticum | Poaceae | 1 | C1 | 2 | |
| Tulipa | Liliaceae | 2 | F | C1? | 10 |
| Zea | Poaceae | 1 | C1 | 2 |
‘Primary methods’ are nuclei extraction methods used for main results; ‘Tested methods’ are extraction methods that were also tried. Methods: B = bead-beating, C = chopping (C1 = simple chopping, C2 = with germinated pollen, C3 = frozen in buffer), F = filter bursting, O = osmotic bursting (O1 = intact pollen, O2 = with chemical/enzymatic protoplast release, O3 = germinated pollen), S = sonication, Q = squashing. References: 1 = Akutsu et al. (2007), 2 = Bino et al. (1990), 3 = Błocka-Wandas et al. (2007), 4 = Dewitte et al. (2006), 5 = Dewitte et al. (2009), 6 = Hirano and Hoshino (2009), 7 = Jacob et al. (2001), 8 = Mishiba et al. (2000), 9 = Nukui et al. (2011), 10 = Okazaki et al. (2005), 11 = Pan et al. (2004), 12 = Pichot and El Maâtaoui (2000), 13 = Roberts (2007), 14 = Stehlik et al. (2007), 15 = Sugiura et al. (1998), 16 = Sugiura et al. (2000), 17 = Van Laere et al. (2009), 18 = Van Tuyl et al. (1989).
* Plus many hybrid cultivars.
† Plus hybrid cultivars involving at least seven species.
‡ Species is not stated in reference 7.
Despite their relative scarcity, these pollen studies have included several different goals and applications, most of which can be placed into three broad categories. Flow cytometric measurement of DNA content has been used to study the development of pollen, ranging from observations about the nuclear replication stages in mature pollen (Van Tuyl et al., 1989; Bino et al., 1990; Sugiura et al., 1998) to multi-stage developmental studies (Pan et al., 2004) and examinations of nuclei development in pollen tubes (Pichot and El Maâtaoui, 2000; Hirano and Hoshino, 2009). In a second application, measurements of pollen DNA have been used to detect individuals and experimental treatments that produce relatively high numbers of unreduced (2n) pollen grains, typically in a horticultural context (e.g. Okazaki et al., 2005; Akutsu et al., 2007; Van Laere et al., 2009). In a third and arguably distinct application, flow cytometry is used to quantify the proportions of different nuclei types produced by individuals with sufficient rigour to allow for statistical comparisons between treatments or individuals. This includes two studies comparing male- and female-determining pollen numbers in Rumex species (Błocka-Wandas et al., 2007; Stehlik et al., 2007), but more often concerns the production of 2n pollen. These studies have included attempts to compare flow cytometric enumeration of 2n pollen with other ploidy indicators, such as pollen size (e.g. Van Tuyl et al., 1989; Dewitte et al., 2009), and to identify developmental stages when 2n pollen production occurs (Pan et al., 2004). A fourth application, the use of pollen nuclei to determine genome size of plants, remains to be developed.
Along with this array of applications comes a variety of nuclei extraction methods (Table 1; for a broader review including non-flow cytometry papers, see Suda et al., 2007a). The most common is the standard method in plant flow cytometry, chopping with a razor blade, first introduced by Galbraith et al. (1983). Variations on this method include first germinating the pollen (Pichot and El Maâtaoui, 2000; Hirano and Hoshino, 2009) and chopping pollen in a frozen block of buffer (Stehlik et al., 2007). Two-thirds of the papers we have identified that measured the DNA content of pollen nuclei at least tested some form of chopping. When other methods were used, they were typically presented as offering some improvement over chopping such as greater efficiency, which implies higher nuclei numbers without significant increases in effort (sonication, Pan et al., 2004; bead-beating, Roberts, 2007). More often, experiments with other methods are presented as attempts to improve on histogram quality, emphasizing increases in nuclei numbers and decreases in debris. Two such methods, for which improvements are claimed, are sonication (Pan et al., 2004) and osmotic bursting, sometimes in combination with overnight germination or enzymatic treatments (Dewitte et al., 2006, 2009; Hirano and Hoshino, 2009).
These attempts to improve nuclei extraction imply that existing methods may not always produce results of sufficient quality. However, the definition of ‘sufficient quality’ varies by application, and minimum standards are not always well defined. For genome size studies in general, 1300 nuclei in the G0/G1 fluorescence peak has been proposed (Greilhuber et al., 2007) and coefficients of variation (CVs) under 5 % are widely recommended for these peaks (Bennett and Leitch, 1995; Doležel et al., 2007; Greilhuber et al., 2007). In comparison, qualitative screening to identify individuals producing large numbers of 2n gametes may be accomplished with relatively low-quality output (histograms), including high CVs and modest nuclei numbers. Similarly, establishing qualitative associations between large or abnormal pollen and polyploidy status may require little more than the ability to identify the number of peaks present and their approximate fluorescence (e.g. Okazaki et al., 2005; Van Laere et al., 2009).
In contrast, higher standards are needed for precise quantification of proportions of 1C, 2C and 4C nuclei, as in statistically rigorous studies of unreduced gamete frequencies or comparisons of 2C nuclei counts with other ploidy indicators. No such set of standards has been described specifically for pollen nuclei, but the precise measurement of relative proportions of different nuclei types has equivalent statistical demands to cell cycle analysis and, in fact, may be considered as cell cycle analysis in developmental studies (e.g. Pan et al., 2004). Consensus standards for clinical cell cycle analysis do exist, and these include a minimum of 10 000 nuclei in the combined peaks of interest, CV < 8 % and aggregate and debris levels under 20 % in the region of analytical interest (Shankey et al., 1993; Ormerod et al., 1998). While the demands of individual studies may differ, these consensus standards for genome size and cell cycle analyses provide useful benchmarks when evaluating methods in pollen flow cytometry.
We have developed protocols for extracting nuclei by forcing pollen against a nylon or polyester mesh that requires no special equipment, similar time and effort as chopping, and that generates high-quality results across a wide range of species. A similar method for bursting pollen with stainless steel mesh was briefly described by Okazaki et al. (2005) and used by Akutsu et al. (2007) and Nukui et al. (2011). None of these papers provided details concerning the relative quality of samples prepared in this way, and the method was used only for qualitative identification of 2n pollen producers. Our objectives here are (1) to describe this method in detail, (2) to test the effectiveness of the method across an extensive and taxonomically diverse set of species, (3) to compare the quality of output with chopping and the nuclei yield with sonication, and (4) to compare genome size estimates from pollen nuclei with those from leaf tissue and test the hypothesis that the two are not significantly different.
MATERIALS AND METHODS
General filter bursting method
The basic nuclei extraction method involves suspending pollen in a buffer and passing this through two filters: a pre-filter, which removes large contaminants while allowing pollen grains to pass, and a second (‘bursting’) filter that collects the pollen. The pollen grains are then rubbed against the bursting filter with a plastic or glass rod, and the nuclei are rinsed through the filter with a staining buffer.
We placed pollen, anthers or whole flowers in a buffer and vortexed them vigorously for a few seconds. The pollen suspension was then passed through the pre-filter and bursting filter, and the flow-through was discarded. For simplicity, we used the same buffer for this step as for the bursting step, although solutions minimizing osmotic bursting may be used if preferred. The bursting filter, with the collected pollen, was then placed on a clean tube and the pollen grains were gently rubbed against the filter for 10–15 s, using a rod with a rounded end. Nuclei were rinsed through the filter with the staining buffer, the pollen was rubbed against the filter for a few seconds and buffer was added a second time. The sample was then allowed to stain for 20–60 min and run according to standard plant flow cytometry protocols (Supplementary Data Video).
We used Partec Celltrics filters (Partec, Münster, Germany), available in 10-, 20-, 30-, 50-, 100- and 150-μm sizes. Filter sizes can be determined by trial and error, starting with a 100- or 150-μm pre-filter and a 10-μm bursting filter, adjusting both if high-quality histograms are not obtained. Preferably, pollen grains and nuclei can be measured under a fluorescence microscope after staining with propidium iodide (PI) or DAPI (4′,6-diamidino-2-phenylindole), and filter sizes selected accordingly. Filters should be large enough to allow passage of pollen through the pre-filter and nuclei through the bursting filter, but not unnecessarily large, to maximize the removal of debris at both stages. The type and amount of staining buffer depends on the requirements of the study, the instrument and the plant species. Our default buffer was LB01 (Doležel et al., 1989), but any nuclei extraction buffer may be used, and changing buffers may improve results in difficult species. PI or DAPI may be used as a nuclear stain. PI should be used for genome size studies because it is not base-pair-specific, but DAPI may produce higher quality histograms and can be used when the accurate measurement of absolute DNA content is not the objective (Doležel et al., 2007). We added two rounds of 0·25 mL buffer for a final sample of about 0·5 mL.
Survey of species
We tested the filter bursting method on 80 plant species (Supplementary Data Table S1). We selected from plants that were readily available to us and attempted to screen as taxonomically broad a sample as possible. We favoured plants from which pollen could be readily collected but we did not entirely avoid those for which pollen collection was restricted due to flower size, morphology or number (e.g. Arabidopsis lyrata, Galium verum, Verbena officinalis). Pollen collection was improved in some species by growing plants in the greenhouse or by field-collecting inflorescences and keeping them overnight as bouquets. For Rumex nivalis, we used desiccated pollen stored in a refrigerator for >5 years (extra pollen collected for Stehlik et al., 2007). Each sample consisted of a single collection made from a single plant. The number of samples tested per species ranged from one to 22, with most species tested fewer than five times (Supplementary Data Table S1). Multiple attempts to improve protocols focused on ways to improve pollen collection (Elymus species and Verbena hastata) and the testing of alternative buffers (Lilium cultivars, Aeschynanthus lobbianus and Chamerion angustifolium).
The basic filter bursting method was applied as described above, with the following specifics. When pollen was measured, we typically used the smallest Partec filter that exceeded the largest observed pollen grain's long axis, or one size larger. For five species with large proportions of pollen close to or exceeding 150 µm, we did not use a pre-filter, but obtained the cleanest pollen sample possible. We used 10 µm as our default bursting filter, but we also used 20 and 30 µm (rarely 50 µm) when microscopy indicated that nuclei approached or exceeded 10 µm (Supplementary Data Table S1). Pollen and nuclei were measured using a Leica DMR microscope (Leica Microsystems, Wetzlar, Germany) and Open Lab software (Ver. 3.5.0, 2002, Improvision Ltd, Lexington, VA, USA). When measured, nuclei were visualized with PI (100 µg mL−1) or DAPI (1 µg mL−1; Vergne et al., 1987) stains and epifluorescence. Samples were run on a BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA), at low (1–30 events s−1) or medium (75–125 events s−1) speed. Run times varied greatly, from 3 to 54 min (rarely exceeding 20 min). Long running times reflected the goal of attaining high-quality histograms (low CVs, high nuclei numbers), rather than maximizing sample numbers. Data were acquired using CellQuest Pro software (Ver. 4.0.2, 2002, BD Biosciences) and the parameter of interest was fluorescence area measured with the FL2 (585/42 nm) detector.
For all parts of this study we measured ungated histograms using ModFit LT for Mac software (3.3.11, 2011, Verity Software House, Inc., Topsham, ME, USA). We used the autoanalysis function with histograms converted to a 256-channel scale to optimize event number per channel, the autodebris and autoaggregate functions activated, and the assumption that no pollen nuclei were in S-phase. From these analyses we obtained fluorescence means, nuclei numbers and CVs for all major nuclei peaks, as well as the histogram's background aggregate and debris percentage (BAD). BAD is defined as the proportion of events identified as either debris or aggregates between the lowest fluorescence G0/G1 peak and the highest putative G2/doublet/2n peak, using ungated histograms (Shankey et al., 1993). Using these measures, we classified histograms into the following classes. ‘Genome size quality’ required at least one pollen peak with at least 1300 nuclei and CV ≤ 5 % (Greilhuber et al., 2007). ‘Cell Cycle quality’ required a minimum of 10 000 nuclei in a set of peaks of interest and total BAD ≤ 20 % (Shankey et al., 1993; Ormerod et al., 1998). ‘Peaks of interest’ were defined differently depending on the pollen type. In species with trinucleate pollen, in which mature pollen grains contain one vegetative and two sperm nuclei (all with 1n chromosomes), we defined peaks of interest as the major 1C nuclei peak plus any minor 2C peak potentially containing 2n pollen nuclei. In species with binucleate pollen, mature pollen grains contain a vegetative (1n) nucleus and a generative nucleus that divides into two sperm nuclei after germination. Observations from microscopy (Brewbaker, 1967) suggesting that typical mature generative nuclei are mitotically arrested in a 2n state have been supported by flow cytometry, in that binucleate pollen generally show both 1C and 2C fluorescence peaks (Bino et al., 1990; Suda et al., 2007a). In binucleate species, we defined the peaks of interest as the major generative nuclei (2C) peak plus any corresponding 4C peak. All of our species were angiosperms, but note that the nuclei types and peaks of interest will be differently defined in gymnosperms (e.g. Pichot and El Maâtaoui, 2000). Shankey et al. (1993) recommend CVs < 8 % for cell cycle analysis, but we retained the criterion CV < 5 %, so that any sample that was of cell cycle quality also met genome size standards. We further identified samples meeting cell cycle criteria while sampling at least 10 000 pollen grains (30 000 nuclei in tri-nucleate species, 10 000 in the generative peak for bi-nucleate species; ‘cell cycle + ’ quality). When samples did not meet the minimum criteria for genome size estimation or cell cycle analysis, they were classified as ‘failed’, even if measurable peaks were detected.
Each taxonomic level (family, genus and species) was assigned to a class based on the best sample obtained within that taxon. Because it was often possible to improve histograms by adjusting protocols, any species failing to meet the minimum genome size criteria but which was tested only once was considered to be borderline with regard to testing of the method. We therefore calculated overall success rates with and without these species.
Comparisons with other nuclei extraction methods
We directly compared sample quality for three nuclei extraction methods: chopping, freeze-chopping (Stehlik et al., 2007) and the method described here (filter bursting). We collected anthers or whole flowers from three individuals from each of five species (Buddleia davidii, Brassica napus, Eupatorium perfoliatum, Solanum lycopersicum and Vicia faba), using greenhouse-grown plants for all except Bu. davidii. A basic pollen amount was determined for each species, based on requirements of our genome size tests (see below), and we used three times that amount for this test. We added 1·1 mL of LB01 buffer to the collected samples, vortexed them for 15 s on moderate to high speed and filtered with the appropriate pre-filter for a final volume of about 1 mL of filtered buffer with pollen. Three sub-samples of 0·3 mL were drawn from the tube, vortexing 2 s before each sub-sample. One sub-sample was added to 0·9 mL of buffer, passed through a bursting filter, and treated according to the filter bursting method described above. A second 0·3-mL subsample was placed in a Petri dish on a frozen block, and chopped for 2 min (1 min for Bu. davidii). An additional 0·9 mL of buffer was then added to the dish and the sample was pipetted through the same size filter as used for filter bursting. A final sub-sample was added to 0·9 mL of the buffer and frozen at –20 °C for 2–3·25 h. The frozen sample was extracted from the tube, chopped with a razor blade until completely thawed (2–2·5 min) and filtered through the same filter as the other methods. For all three methods, small adjustments were made to bring the final filtered volume to 1 mL by passing drops of buffer through the second filter. PI and RNase were added to the filtered samples to obtain 100 µg mL−1 PI and 50 µg mL−1 RNase. Each treatment was replicated three times within a species, once per individual. The samples were stained for 30–36 min (20–36 min in Bu. davidii) and run on medium speed for 3–10 min, with running times kept constant for all samples and treatments within a species. Details of the filter sizes, tissue amounts, staining and running times for each species are presented in Supplementary Data Table S2.
We compared three measures of sample/histogram quality for the three treatments: CV of the 1C peak, total number of nuclei and BAD. We used a restricted maximum-likelihood (REML) ANOVA for each of these response variables with the following factors: Species (fixed), Plant[nested in species] (random), Extraction Method (fixed) and Species × Method interaction. We ln-transformed nuclei number and CV data to correct non-normality of residuals and heterogeneity of variances in the raw data. For this and subsequent ANOVAs, residuals were tested for normality using the Shapiro–Wilk W test and variances were tested for homogeneity using Bartlett's (when residuals were normal) or Brown–Forsythe tests. All analyses were performed with JMP software (Ver. 8.0, 2009, SAS Institute, Inc., Cary, NC, USA).
We calculated filter bursting nuclei yields (number extracted per pollen grain) in Br. napus ‘Hyola 401′ for comparison with published yields for the same species using sonication (Pan et al., 2004). We collected 48 anthers from each of five greenhouse-grown plants. Anthers were placed in LB01 buffer, vortexed and filtered through a 50-μm filter to produce a final volume of 2 mL. This was vortexed for a few seconds before 1 mL was passed through a 10-μm filter and prepared using the usual filter bursting method. The sample was run on medium on a FACSCalibur flow cytometer until no further nuclei could be acquired because the sample was depleted (11–12 min). A second 1 mL was transferred to 69 mL IsotonII diluent (Beckman Coulter, Inc., Indianapolis, IN, USA) for pollen counting. Pollen grains were counted with a Multisizer 3 particle counter (Beckman Coulter, Inc.) and the number of pollen grains in the original 1 mL was calculated as (average of seven 1-mL subsamples) × 70. These samples were tested within 2 min of dilution and stirred continuously during counting. The yield for each filter burst sample was calculated as nuclei (from the 1mL flow cytometry subsample) per total pollen grains (from the 1 mL particle counter subsample). Because each pollen grain contains three nuclei, yield was also expressed as a percentage of total possible nuclei: [(number of nuclei acquired)/(number of pollen grains × 3)] × 100 %.
Genome size estimation with filter burst pollen
We compared genome size estimates obtained with leaf nuclei with those made with pollen nuclei for six species, including three species with binucleate pollen (Bu. davidii, S. lycopersicum and V. faba) and three with trinucleate pollen (Br. napus, E. perfoliatum and Zea mays). These were selected on the basis of availability, sufficient flowering to enable repeated pollen collections and taxonomic breadth (i.e. six families). We measured between 55 and 67 pollen grains and between nine and 24 nuclei per species under the microscope and selected filters accordingly. We experimentally determined basic tissue amounts for each species such that all relevant nuclei peaks (leaf from standard, leaf from test plant, 1C pollen nuclei and 2C pollen in binucleate species) would have approximately equal numbers when standards and test plants were run together. We used LB01 buffer for all species; filters were selected as described above and DNA content standards were selected from the list given by Doležel et al. (2007). For details of these and the following methods see Supplementary Data Table S2.
To allow for replication, plants had to provide three times the number of flowers as needed for a single test, plus additional flowers for the inhibition tests described below. Flower number was limited on at least one individual in some of the species, so to maintain replication across all individuals and to keep pollen amount constant we used less than the ideal pollen amount in some cases and adopted a goal of 1000 nuclei per peak rather than 1300. To estimate genome size from leaf tissue, a piece of young leaf from the test plant was co-chopped with leaf from the standard species. Leaves were chopped with a razor blade in 0·75 mL ice-cold buffer (with 100 µg mL−1 PI, 50 µg mL−1 RNase), filtered through a filter of the same size as selected for pollen bursting, stained for 30–45 min at room temperature and run at low speed. Pollen samples were prepared using the filter burst method described above, except that leaf tissue from the standard was first chopped in staining buffer as described above, and this buffer was then used to wash the pollen nuclei through the bursting filter (2 × 0·25 mL; Supplementary Data Video). Because the standard was chopped in 0·75 mL buffer but only 0·5 mL was then used, 1·5 times as much of the standard tissue amount was used as in the leaf test, in order to keep the number of standard nuclei approximately equal in the two treatments. For each species, these two treatments were applied on five or six plants on a single day, and this was replicated on 3 d. For Z. mays, plants did not flower synchronously, so the tests were spread out over 5 d.
DNA contents of nuclei were calculated relative to the standards, and expressed as pg/1C. Genome size was calculated using leaf and 1C pollen nuclei for trinucleate species, and using leaf, 1C (vegetative) pollen nuclei and 2C (generative) pollen nuclei in binucleate species. For each species separately, an REML ANOVA was performed using 1C DNA content as the response variable and Tissue Type (fixed), Plant (random), and Tissue Type × Plant interaction (random) as sources of variation. There were two Tissue Type levels for trinucleate species (pollen, leaf) and three for binucleate species (vegetative, generative and leaf).
On one test day for each species, an additional three treatments were applied to each plant: leaf tissue chopped without the standard, standard tissue chopped without the test plant and pollen prepared without the standard. These tests were used to determine whether any tissue type appeared to be affecting the staining of other tissues (i.e. inhibition sensu Price et al., 2000). For these treatments, twice the basic tissue amounts were used, in order to keep the total nuclei amounts relatively constant across treatments. We compared the standard and test plant peaks across treatments for evidence of two patterns that might result in differences between DNA content estimates for leaf and pollen: differential impacts of leaf and pollen on the fluorescence of the standard's nuclei, and differential impacts of the standard on pollen and leaf nuclei fluorescence.
RESULTS
General effectiveness of filter bursting across species
We tested 80 species, distributed across 64 genera and 33 families (Supplementary Data Table S1). Representative histograms are shown in Fig. 1. Eighty-one per cent of all species tested included at least one individual meeting the minimum criteria for genome size testing, and 51 % met the requirements for cell cycle analysis. In addition, 41 % of species met cell cycle criteria with generative nuclei numbers corresponding to at least 10 000 pollen grains. Success rates at the genus level reflect these results closely, and rates at the family level are somewhat higher for cell cycle quality (Table 2). When we excluded eight species that failed but were tested only once, the success rate at the species level increased to 90 % for genome size quality and 57 % for cell cycle quality (Table 2). Two species required buffers other than LB01 to achieve their best results: Lilium Asiatic cultivars needed MgSO4 buffer (Arumuganathan and Earle, 1991) with 2·5 % Triton X-100 to achieve near-genome size quality and Malus coronaria required a variant of the de Laat buffer (de Laat and Blaas, 1984; Bino et al., 1992, with 0·25 mmol L−1 PVP-40) for cell cycle quality. Three trinucleate species (Raphanus sativus, Lychnis noctiflora and Symphyotrichum novae-angliae) had differentially staining vegetative and sperm nuclei that generated a doubled 1C peak with a combined CV > 5 %; those of R. sativus could be well enough distinguished on the basis of fluorescence and forward scatter properties that they could be measured separately using CellQuest Pro software.
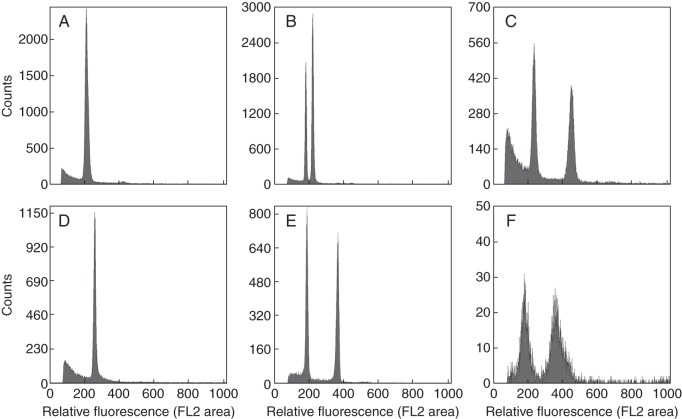
Fig. 1.
Representative ungated histograms showing the range of quality obtained with filter bursting. These six genera were selected because all were tested previously by other authors with other nuclei extraction methods (Table 1). (A) Brassica napus, Cell cycle + quality, 47 306 nuclei (15 769 pollen), CV = 4·5 %, BAD = 11·7 %; (B) Rumex nivalis, Cell cycle + quality, 49 705 nuclei (16 569 pollen), CVs = 2·2 and 2·2 %, BAD =6·1 % – this species has two pollen types (male- and female-determining) that account for the two peaks; (C) Rosa rugosa, Cell cycle + quality, 22 824 nuclei (11 081 pollen), CVs = 4·4 and 3·2 %, BAD = 19·9 %; (D) Zea mays, Cell cycle quality, 16 038 nuclei (5346 pollen), CV = 2·6 %, BAD = 12·2%; (E) Lilium longiforum, Cell cycle + quality, 21 764 nuclei (11 200 pollen), CVs = 3·1 and 2·0 %, BAD =16·6 %; (F) Alstroemeria cultivar, Failed, 1970 nuclei, (849 pollen), CVs = 10·4 and 8·0%, BAD =27·6 %. Pollen numbers in binucleate species are based on the number of generative nuclei.
Table 2.
Summary of quality classifications for species tested using filter bursting
| Number tested |
% at Genome Size quality |
% at Cell Cycle quality |
% at Cell Cycle + quality |
|||||
|---|---|---|---|---|---|---|---|---|
| Taxa | All | Adjusted | All | Adjusted | All | Adjusted | All | Adjusted |
| Species | 80 | 72 | 81 % | 90 % | 51 % | 57 % | 41 % | 46 % |
| Genus | 64 | 59 | 83 % | 90 % | 55 % | 59 % | 44 % | 47 % |
| Family | 33 | 31 | 82 % | 87 % | 67 % | 71 % | 58 % | 61 % |
‘Genome size quality’ = 1300 nuclei in one peak, CV ≤ 5%; ‘Cell cycle quality’ = 10 000 nuclei, BAD ≤ 20 %; ‘Cell Cycle + quality’ = ‘Cell cycle quality’ with nuclei from 10 000 pollen grains; ‘All’ includes all species tested; ‘Adjusted’ excludes species that failed to meet genome size quality but were tested only once.
Comparisons with other extraction methods
Measures of nuclei number, CV and BAD all showed highly significant effects of the extraction method when comparing filter bursting with chopping and freeze-chopping. The effect of method varied among species, however, as we found weak to significant Species × Extraction Method interactions for all measures (Table 3). Nuclei numbers were always highest for filter burst samples, ranging from 5·7 (Br. napus) to 27·6 (Bu. davidii) times higher than chopped samples, and 2·9 (E. perfoliatum) to 12·0 times higher than freeze-chopped (back-transformed least-square means from ANOVA, Table 3, Fig. 2A). Across all species, filter bursting produced 10 times more nuclei than chopping and 7·6 times more than freeze-chopping. Counts from chopped and freeze-chopped samples did not differ significantly overall or for any single species, but filter bursting counts were significantly higher than both of these chopping methods in every species except E. perfoliatum (Tukey HSD test, Fig. 2A).
Table 3.
Fixed effect results from four ANOVAs comparing nuclei number, CV and BAD for three extraction methods (chopping, freeze-chopping and filter bursting)
| Response variable and sources of variation | d.f | d.f. den. | F | P |
|---|---|---|---|---|
| Nuclei number | ||||
| Species | 4 | 10 | 15·132 | 0·0003 |
| Extraction Method | 2 | 20 | 175·627 | < 0·0001 |
| Species × Extraction Method | 8 | 20 | 4·589 | 0·0027 |
| CV (1C peak) | ||||
| Species | 4 | 10 | 3·603 | 0·0456 |
| Extraction Method | 2 | 20 | 89·426 | < 0·0001 |
| Species × Extraction Method | 8 | 20 | 10·716 | < 0·0001 |
| CV (generative peak) | ||||
| Species | 2 | 6 | 3·232 | 0·1116 |
| Extraction Method | 2 | 12 | 28·155 | < 0·0001 |
| Species × Extraction Method | 4 | 12 | 13·570 | 0·0002 |
| BAD | ||||
| Species | 4 | 10 | 9·893 | 0·0017 |
| Extraction Method | 2 | 20 | 26·469 | < 0·0001 |
| Species × Extraction Method | 8 | 20 | 2·398 | 0·0539 |
Numbers in bold are significant values (P < 0·05).
Fig. 2.
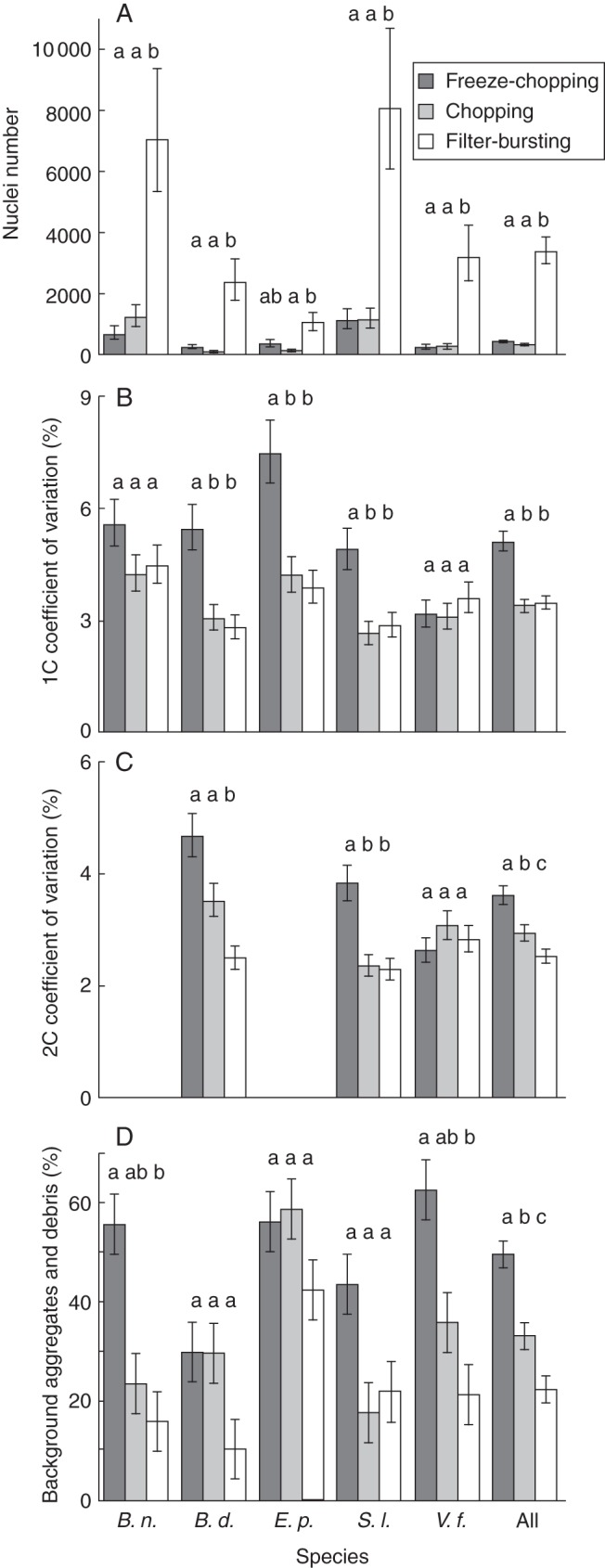
Comparison of sample quality measures for three nuclei extraction treatments: freeze-chopping (dark grey), chopping (light grey) and filter-bursting (white). (A) Total pollen nuclei number across all peaks; (B) CV of 1C pollen peak; (C) CV of 2C (generative) nuclei peak (binucleate species only); (D) BAD. All values shown are least-square means with s.e. from the ANOVAs in Table 3. Bars marked by the same lower-case letter indicate values not significantly different within a species (Tukey's HSD, α = 0·05), but do not have meaning across species. ‘All’ = main treatment effect across all species; B. n. = Brassica napus; Z. m. = Zea mays; E. p = Eupatorium perfoliatum; B. d. = Buddleia davidii; S. l. = Solanum lycopersicum; V. f. = Vicia faba.
For 1C nuclei peaks, CVs for chopping and filter bursting did not differ overall or for any individual species, while freeze-chopping produced significantly higher CVs overall (P < 0·0001; Table 3) and in three of five species (Fig. 2B). For 2C peaks (generative nuclei in binucleate species), all three treatments were significantly different from one another in the overall analysis, with filter bursting producing the lowest and freeze-chopping the highest CVs (P < 0·0001, Table 3, Fig. 2C). The effect differed at the species level, with filter bursting producing significantly lower CVs than freeze-chopping in S. lycopersicum and lower than both chopping and freeze-chopping in Bu. davidii (Fig. 2C).
Across all five species, filter bursting produced significantly lower background aggregates and debris (22 %) than chopping (33 %), which in turn produced significantly less than freeze-chopping (49 %; P < 0·0001, Table 3, Fig. 2D). At the level of individual species, mean values for BAD were lowest for filter bursting in four out of five species, but the differences between chopped and filter burst were never statistically significant, while filter bursting had significantly lower BAD than freeze-chopping in two species (Fig. 2D).
For the five Br. napus plants tested for nuclei yield, the mean extraction rate averaged 1067 (±56) nuclei/1000 pollen grains (±s.e., n = 5, range = 914–1264). This is equivalent to extracting all three nuclei from 35·6 % of pollen grains on average (range 30·5–42·1%). CVs averaged 5·06 (±0·345)%, although only one exceeded 5 %; BAD averaged 13·61 (±1·58)% with a range of 8·91–18·34 %.
Genome size estimates: comparison of pollen and leaf results
Pollen samples were more likely than leaf samples to fall short of our quality control criteria for the genome size tests, even with the relaxed requirement for only 1000 nuclei per peak. For all six species, all leaf tissue histograms met our genome size criteria, while 91 % of pollen samples did. Five pollen samples had peaks with fewer than 1000 nuclei (one each for Bu. davidii and Br. napus, three in V. faba), although all exceeded 860 nuclei. One Br. napus plant had all three pollen replicates with CVs exceeding 5 % (5·05–5·52 %).
Small but statistically significant differences were found between pollen and leaf genome size estimates in most species (Table 4, Fig. 3). Two of three trinucleate species showed significant differences in genome size estimates between pollen and leaf tissue, but all differences were small (≤1·5 %; Fig. 3). In binucleate plants only one species (Bu. davidii) had a significant difference between 1C pollen nuclei (vegetative) and leaf estimates (1·1 %). In comparison, all three binucleate species showed a significant difference between leaf and generative nuclei estimates, although all differences were still modest (between 1 and 2·5 %; Fig. 3).
Table 4.
Genome size estimates for leaf and pollen nuclei and ANOVA results for effect of nucleus type
| Genome size (pg/1C) |
ANOVA |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Pollen type | Standard | Leaf | 1C pollen | 2C pollen | s.e. | d.f. | F | P |
| Brassica napus1 | Trinucleate | Tomato2 | 1·183a | 1·177b | 0·007417 | 1 | 13·470 | 0·0214 | |
| Eupatorium perfoliatum | Trinucleate | Tomato | 2·47a | 2·44b | 0·01125 | 1 | 70·687 | 0·0011 | |
| Zea mays3 | Trinucleate | Tomato | 2·65a | 2·67a | 0·01123 | 1 | 2·596 | 0·1824 | |
| Buddleia davidii | Binucleate | Soybean4 | 1·55a | 1·54b | 1·52c | 0·002464 | 2 | 48·482 | < 0·0001 |
| Solanum lycopersicum2 | Binucleate | Soybean | 1·06a | 1·05a | 1·03b | 0·007100 | 2 | 97·766 | < 0·0001 |
| Vicia faba5 | Binucleate | Rye6 | 13·62a | 13·60a | 13·48b | 0·05665 | 2 | 34·034 | < 0·0001 |
Genome size estimates are least-square means from the ANOVA for that species. Standard error (s.e.) is the same for all genome size estimates within a species. Estimates with the same letter within a species are not significantly different, Tukey's HSD, α = 0·05. Numbers in bold are significant values (P < 0·05).
1B. napus ‘Hyola 401’; 2S. lycopersicum ‘Stupické polní rané’, 1·96 pg/2C, Doležel et al. (1992); 3Z. mays ‘CE-777’; 4Glycine max ‘Polanka’, 2·50 pg/2C, Doležel et al. (1994); 5V. faba ‘Inovec’; 6Secale cereale ‘Dankovské’, 16·19 pg/2C, Doležel et al. (1998).
Fig. 3.
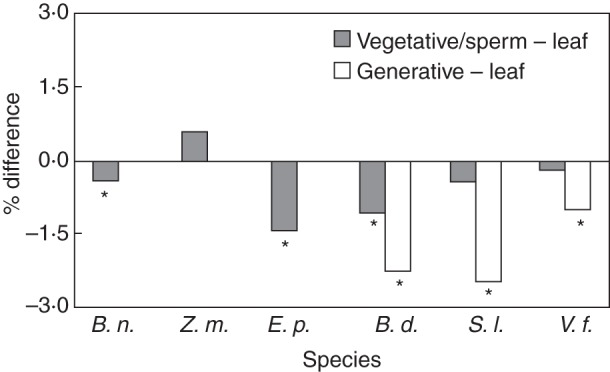
Percentage difference between genome size estimates obtained from pollen nuclei and estimates from leaf nuclei [(pollen – leaf estimate)/average]. Negative values indicate lower estimates from pollen. Asterisks indicate significant differences between leaf and pollen estimates (α = 0·05). B.n. = Brassica napus; Z. m. = Zea mays; E. p = Eupatorium perfoliatum; B. d. = Buddleia davidii; S. l. = Solanum lycopersicum; V. f. = Vicia faba.
The leaf genome size estimates averaged higher than the pollen in all species except Z. mays (pollen 0·6 % higher, P = 0·18, Fig. 3). In comparisons between fluorescence peaks from single species samples and mixed samples (inhibition tests), no clear overall pattern was detected, with both upwards and downwards shifts in fluorescence when species were mixed. However, we observed two trends that would contribute to higher leaf DNA content estimates. In three species (Br. napus, E. perfoliatum and S. lycopersicum) the standard peak showed reduced fluorescence in the presence of leaf, more so than any similar reduction in the presence of pollen. In three species (Br. napus, E. perfoliatum and V. faba) the pollen peak shifted downward in the presence of the standard, more so than any similar shift in leaf tissue. These shifts in fluorescence averaged 1·6–4·5 %, depending on the species and tissue.
DISCUSSION
Effectiveness of filter bursting across species
The range of species we tested (80 species, 64 genera and 33 families) exceeds the combined total for all previous studies using flow cytometry to measure relative DNA content of pollen nuclei (46 species, 14 genera and 12 families, with multiple hybrids and varieties of Petunia and Lilium; Table 1). Filter bursting success rates were high when measured against commonly applied genome size and cell cycle analysis quality standards: 81–90 % for genome size quality and 51–71 % for cell cycle quality (Table 2). Furthermore, cell cycle quality in combination with data from 10 000 pollen grains was achieved in 33 species, within 28 genera and 19 families.
Fifteen species failed to meet the minimum genome size criteria (Supplementary Data Table S1). This failure rate is probably conservative because eight of these species were tested only once and, hence, adjustments to protocols (e.g. buffer choice, pollen amount, selecting healthier individuals) would potentially improve results. For four of the 16 failed species (Arabidopsis thaliana, Artemisia biennis, Galium mullugo and Vicia villosa) pollen collection was difficult and low nuclei counts were the main cause of failure. Note, however, that we restricted ourselves to single day collections of single plants, a requirement that may not apply in all studies. In two trinucleate species (Lychnis noctiflora and Symphyotrichum novae-angliae), CVs exceeded 5 % (but were <6 %) due to differential staining of vegetative and sperm nuclei that resulted in a near-double peak. For each of the remaining nine species (Aeschynanthus lobbianus, Alstroemeria cultivar, Crocus sativus, Cucurbita pepo, Hemerocallis fulva, Iris reticulata, Lilium Asiatic cultivars, Strelitzia reginae and Taraxacum officinale), some combination of high debris, high CV and/or low nuclei number was at fault. The reasons for poor quality in these species remain to be determined. Problems are often associated with large pollen size, with most species with maximum pollen diameters over 100 µm failing to meet genome size quality, although this may be confounded with family effects (e.g. Iridaceae, Liliaceae). In addition, some commonly cultivated genera (e.g. Aeschynanthus, Alstroemeria, Crocus, Lilium) may include hybrids or odd-numbered polyploids, and their pollen may have high rates of infertility or aneuploidy. The quality of results may therefore be species- or even cultivar-specific. For example, we had considerable difficulty obtaining good quality histograms with Lilium Asiatic cultivars, testing multiple buffers and pollen amounts before getting one sample with near-genome size quality (>1290 nuclei). In contrast, obtaining genome size quality was not difficult in Lilium longiflorum, and we achieved cell cycle quality or better in some samples for this species (Fig. 1).
Comparisons with other methods
It is difficult to evaluate previously published data against the specific genome size and cell cycle criteria described here, because the required detail is lacking in most cases. It is likely, however, that several studies include examples that meet or approach the minimum requirements for genome size testing, including examples of chopping (e.g. Van Tuyl et al., 1989; Bino et al., 1990; Mishiba et al., 2000, Sugiura et al., 1998, 2000; Pichot and El Maâtaoui, 2000; Błocka-Wandas et al., 2007; Stehlik et al., 2007), sonication (Pan et al., 2004) and osmotic shock combined with germination (Dewitte et al., 2009). However, none of the studies in Table 1 presents data or histograms obviously meeting the standards of cell cycle analysis, with the possible exception of Pan et al. (2004), which does not provide specifics for CV or BAD. This apparent absence of examples of cell cycle quality may simply reflect the fact that such quality was not always demanded by the studies in question. However, it may also reflect the difficulties inherent in obtaining a combination of high nuclei numbers, low CVs and low debris.
In our direct comparisons with chopping, filter bursting produced significantly higher nuclei counts, slightly improved BAD and essentially identical CVs (Fig. 2). Chopping frozen samples produced significantly poorer quality than filter bursting, and our filter-burst samples of R. nivalis produced much higher quality histograms (Fig. 1) than our previous use of freeze-chopping did in this same species (Stehlik et al., 2007). Our nuclei yields for filter-burst B. napus pollen (1067 nuclei/1000 pollen grains) exceeded by 2·5 times the maximum yield reported for sonication (400 nuclei/1000 pollen with 10 min of treatment; Pan et al., 2004). Pan et al. (2004) did not report debris measures and so we cannot directly compare our results with theirs, but we note that they recommended a shorter sonication time (4–5 min, yields of 300 nuclei/1000 pollen) and multiple centrifugation and resuspension steps to reduce debris to acceptable levels. Debris levels following sonication have also been reported as a problem in Begonia pollen flow cytometry (Dewitte et al., 2009). We did not compare filter bursting with osmotic bursting of germinated or enzymatically treated pollen, methods with much longer sample preparation times, nor did we did make direct comparisons with bead-beating, a fast and simple extraction method (Roberts, 2007). Although bead beating is the one nuclei extraction method with a shorter preparation time than filter bursting, extensive experience with this method in our lab indicates that extracting nuclei of high enough quality for flow cytometry is very species-dependent, and debris levels higher than chopping (and by extension, filter bursting) are the norm. Our output for filter burst pollen of Rosa rugosa (Fig. 1) can be compared quite favourably with that for the same species in Roberts (2007).
It is likely that the high nuclei yield and relatively clean samples produced by filter bursting result from the efficient application of pressure on most pollen grains in the confined space provided by the filters, and the subsequent filtering out of the relatively intact pollen walls following bursting. Chopping is probably less efficient with respect to contact with the pollen, and both chopping and sonication produce more fragments of pollen wall that will pass through the filter and contribute to debris. Filter bursting may also result in less damage to nuclei than more energetic methods. For example, Roberts (2007) and Dewitte et al. (2009) reported substantial reductions in expected numbers of vegetative nuclei (relative to generative) using bead beating and sonication. Similar but less pronounced patterns have been reported for chopped samples (e.g. Van Tuyl et al., 1989) and are sometimes seen with filter bursting as well. This effect might arise from deterioration of vegetative nuclei at maturity (Brewbaker, 1967), failure of vegetative nuclei to escape from the opened pollen walls (Van Laere et al., 2009) or counting inaccuracies when debris is high (Van Tuyl et al., 1989), but an alternative hypothesis is that more energetic extraction methods may damage fragile vegetative nuclei.
Use in genome size studies
The exact quality criteria for genome size data may be debated, and not everyone will wish to restrict themselves to CV < 5 % and nuclei numbers >1300. These guidelines do provide a useful test for evaluating the method, however, and the 81–90 % success rates indicate that genome size measurement with pollen is a viable option when filter bursting is used. This option is further strengthened by the observation that leaf and pollen estimates did not differ by more than 2·5 % in the six species we tested (1·5 % if tests are restricted to the 1C vegetative nuclei in binucleate species; Fig. 3). A number of authors have suggested that fluorescence of leaf and pollen nuclei appear to be similar but may not be identical (Van Tuyl et al., 1989; Bino et al., 1990; Dewitte et al., 2006), but only two previous studies have compared such estimates using internal standardization and replication. Roberts (2007) reported pollen DNA content estimates 0·7 % lower than those for leaves in bead-beaten Rosa rugosa samples, while Błocka-Wandas et al. (2007) found that pollen estimates were 4–6 % lower than theoretical expectations based on somatic tissue in Rumex acetosa.
The general trend towards slightly higher DNA content estimates with leaf nuclei raises the question of why these differences occur, especially as there is no a priori reason to assume that leaf estimates are the more accurate of the two. The same question applies in the case of vegetative and generative nuclei in binucleate species, where the two types provide slightly different genome size estimates, with those from vegetative nuclei tending to be most similar to leaf nuclei estimates (Fig. 3). A likely explanation lies in the pronounced structural differences often observed between vegetative and generative nuclei, and presumably leaf and pollen nuclei (Fig. 4). Associated differences, such as levels of chromatin condensation, may lead to differences in stain uptake by the nuclei, a problem recognized in the use of feulgen microdensitometry (Verma and Rees, 1974; Bennett and Leitch, 2005) and previously suggested for staining differences in pollen flow cytometry (Suda et al., 2007a; Błocka-Wandas et al., 2007). We also noted two weak trends in our data consistent with fluorescence inhibition effects (sensu Price et al., 2000) operating differently in pollen and leaf samples. In three species, there was some evidence that leaf tissue affects fluorescence of the standard plant's nuclei more than pollen. Also, in three species, there were signs that the standard might be inhibiting pollen nuclei fluorescence more than it was inhibiting leaf nuclei. These trends produce conflicting viewpoints on the relative merits of leaf and pollen tissue. Although such inhibition effects may contribute to higher estimates for leaf tissue, they are less likely than structural differences to explain differences between vegetative and generative nuclei estimates, because in any given sample, both nuclei types would be equally exposed to secondary compounds from the standard. With these issues unresolved, we would recommend that in binucleate species, the vegetative nuclei should be used for genome size estimates, to facilitate comparison with species for which leaf tissue is used.
Fig. 4.
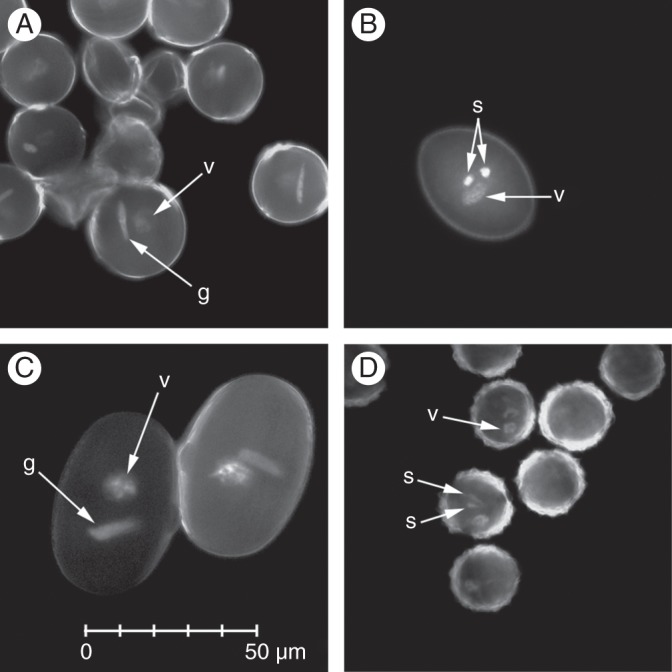
Fluorescence images of binucleate (A, C) and trinucleate (B, D) pollen, showing variation in morphology of pollen nuclei. (A) Solanum lycopersicum; (B) Brassica napus; (C) Vicia faba; (D) Eupatorium perfoliatum. Pollen was stained with DAPI (A, C) or propidium iodide (B, D). The scale in C applies to all four images. Abbreviations: g, generative nucleus; v, vegetative nucleus; s, sperm nucleus.
There are some limitations to using pollen for genome size estimation. Pollen is generally less available and less abundant than leaf tissue. Despite the improvements offered by filter bursting, it may still be more challenging to obtain the required number of nuclei with pollen than with leaf tissue, as our own genome size tests show. Pollen ploidy may not relate to somatic ploidy in the expected manner, for example in rare cases of plants that produce only unreduced pollen (e.g. Cupressus dupreziana, Pichot and El Maâtaoui, 2000). At the same time, pollen use may have strong advantages in certain scenarios. When it is difficult to get high-quality histograms with somatic tissue, for example, due to secondary compounds in the leaves, pollen may be a good alternative. This will not always be the case, as we had greater difficulty acquiring good data from Asiatic Lilium pollen than we did from leaves, and for Malus coronaria, similar quality problems were found with leaf tissue and pollen. Pollen nuclei may be useful for verifying the genome size in endopolyploid species, which sometimes have low proportions of 2n cells and readily overlooked 2C peaks in somatic tissue (Suda et al., 2007a). Where intraspecific variation is suspected, testing the DNA content of different individuals with pollen as well as leaf tissue may be helpful in ruling out staining artefacts and establishing the repeatability of DNA content differences. This may be particularly valuable where the suspected intraspecific differences are so small that the recommended method for verifying them is inconclusive (i.e. co-chopping individuals and checking for double peaks; Suda et al., 2007b). Finally, the high quality of the data from 5-year-old R. nivalis pollen suggests that pollen may have long-term storage advantages, although this may vary by species, and further tests would be needed to determine whether genome size estimates differ between fresh and dried pollen.
Potential for use in unreduced gamete estimation
The potential utility of flow cytometry for quantifying unreduced gamete frequency has been recognized for some time (Van Tuyl et al., 1989; Bino et al., 1990). It has been successfully applied in horticultural and agricultural contexts to identify individuals or experimental treatments associated with increases in unreduced pollen production (Sugiura et al., 2000; Okazaki et al., 2005; Akutsu et al., 2007; Dewitte et al., 2009; Van Laere et al., 2009; Nukui et al., 2011). It would be incorrect, therefore, to suggest that useful information about unreduced gametes can only be obtained with cell cycle quality data. However, errors will be reduced when such quality is achieved, for two main reasons. First, high nuclei counts are essential for good estimates of rare events, because error terms for proportions depend on these counts. Secondly, high debris levels seriously compromise the accurate measurement of event numbers in nuclei peaks. When debris consists of damaged nuclei, it contributes to high CVs and non-Gaussian peaks and can be difficult to gate out in a repeatable manner (and perhaps should not be, as damaged nuclei are themselves potential data points). Other forms of debris (e.g. exine fragments) may be more easily gated out on the basis of fluorescence wavelength or scatter properties, but such gating must still be done cautiously so as not to compromise some methods of histogram analysis (e.g. modelling of debris curves and doublet identification; Verity Software House, Inc., 2000). Estimates of unreduced gamete numbers from histograms with high debris and low nuclei counts should therefore be evaluated cautiously. Filter bursting produced histograms meeting consensus standards for cell cycle analysis in half of all species tested, and in more than 40 % of species, this included data from 10 000 or more pollen grains. For many species, data of this quality would seem to be a reasonable and attainable goal in studies of unreduced gamete frequency as well as in other studies emphasizing proportions of different nuclei types.
SUMMARY
The filter bursting method for extracting pollen nuclei requires no special equipment beyond filters that are required for any extraction method. It is comparable to chopping in terms of the time and effort required. CV and debris measures are as good as or better than other commonly used methods, and nuclei yields are higher than reported for other methods. Quality levels reached or exceeded commonly accepted standards for genome size studies in the great majority of species, and estimates of genome size did not differ from leaf estimates by more than 2·5 % in the six species tested. These results show that using pollen for genome size estimation is a viable option when other considerations (e.g. availability) are taken into account. Furthermore, the high consensus standards for cell cycle analysis were met in roughly half of all species tested. Use of this method will therefore allow for high-quality data in studies emphasizing the measurement of proportions of rare events, particularly studies of unreduced pollen production.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Jillian Bainard for comments on the manuscript and Petr Šmarda for providing seeds. This research was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) and Canada Research Chair (CRC) grants to B.C.H., and by equipment purchased through the Canada Foundation for Innovation (CFI).
LITERATURE CITED
- Akutsu M, Kitamura S, Toda R, Miyajima I, Okazaki K. Production of 2n pollen of Asiatic hybrid lilies by nitrous oxide treatment. Euphytica. 2007;155:143–152. [Google Scholar]
- Arumuganathan K, Earle ED. Estimation of nuclear DNA content of plants by flow cytometry. Plant Molecular Biology Reporter. 1991;9:229–233. [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms. Annals of Botany. 1995;76:113–176. doi: 10.1093/aob/mci003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. Gregory TR. The evolution of the genome. Amsterdam: Elsevier Academic Press; 2005. Genome size evolution in plants; pp. 89–162. [Google Scholar]
- Bino RJ, Van Tuyl JM, De Vries JN. Flow cytometric determination of relative nuclear DNA contents in bicellulate and tricellulate pollen. Annals of Botany. 1990;65:3–8. [Google Scholar]
- Bino RJ, De Vries JN, Kraak HL, Van Pijlen JG. Flow cytometric determination of nuclear replication stages in tomato seeds during priming and germination. Annals of Botany. 1992;69:231–236. [Google Scholar]
- Błocka-Wandas M, Sliwinska E, Grabowska-Joachimiak A, Musial K, Joachimiak AJ. Male gametophyte development and two different DNA classes of pollen grains in Rumex acetosa L., a plant with an XX/XY1Y2 sex chromosome system and a female-biased sex ratio. Sexual Plant Reproduction. 2007;20:171–180. [Google Scholar]
- Brewbaker JL. The distribution and phylogenetic significance of binucleate and trinucleate pollen grains in the angiosperms. American Journal of Botany. 1967;54:1069–1083. [Google Scholar]
- De Laat AMM, Blaas J. Flow-cytometric characterization and sorting of plant chromosomes. Theoretical and Applied Genetics. 1984;67:463–467. doi: 10.1007/BF00263414. [DOI] [PubMed] [Google Scholar]
- Dewitte A, Eeckhaut T, Van Huylenbroeck J, Van Bockstaele E. Flow cytometric detection of unreduced pollen in Begonia. Acta Horticulturae. 2006;714:107–112. [Google Scholar]
- Dewitte A, Eeckhaut T, Van Huylenbroeck J, Van Bockstaele E. Occurrence of viable unreduced pollen in a Begonia collection. Euphytica. 2009;168:81–94. [Google Scholar]
- Doležel J, Binarová P, Lucretti S. Analysis of nuclear DNA content in plant cells by flow cytometry. Biologia plantarum (Praha) 1989;31:113–120. [Google Scholar]
- Doležel J, Sgorbati S, Lucretti S. Comparison of three DNA fluorochromes for flow-cytometric estimation of nuclear DNA content in plants. Physiologia Plantarum. 1992;85:625–631. [Google Scholar]
- Doležel J, Doleželová M, Novák FJ. Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana) Biologia Plantarum. 1994;36:351–357. [Google Scholar]
- Doležel J, Greilhuber J, Lucretti S, et al. Plant genome size estimation by flow cytometry: inter-laboratory comparison. Annals of Botany. 1998;82A:17–26. [Google Scholar]
- Doležel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Greilhuber J, Temsch EM, Loureiro JCM. Nuclear DNA content measurement. In: Doležel J, Greilhuber J, Suda J, editors. Flow cytometry with plant cells: analysis of genes, chromosomes and genomes. Weinheim: Wiley-VCH Verlag; 2007. pp. 67–101. [Google Scholar]
- Hirano T, Hoshino Y. Detection of changes in the nuclear phase and evaluation of male germ units by flow cytometry during in vitro pollen tube growth in Alstroemeria aurea. Journal of Plant Research. 2009;122:225–234. doi: 10.1007/s10265-008-0208-2. [DOI] [PubMed] [Google Scholar]
- Jacob Y, Priol V, Ferrero F, Coudret C, Sallanon H. Fluorescent staining of roses pollen tubes and nuclei by microscopy and flow cytometry analysis. Acta Horticulturae. 2001;547:383–385. [Google Scholar]
- Kron P, Suda J, Husband BC. Applications of flow cytometry to evolutionary and population biology. Annual Review of Ecology, Evolution and Systematics. 2007;38:847–876. [Google Scholar]
- Leitch IJ, Bennett MD. Flow cytometry with plant cells: analysis of genes, chromosomes and genomes. Weinheim: Wiley-VCH Verlag; 2007. Genome size and its uses: the impact of flow cytometry; pp. 153–176. Doležel J, Greilhuber J, Suda J. [Google Scholar]
- Leus L, Van Laere K, Dewitte A, Van Huylenbroeck J. Flow cytometry for plant breeding. Acta Horticulturae. 2009;836:221–226. [Google Scholar]
- Mishiba KI, Ando T, Mii M, et al. Nuclear DNA content as an index character discriminating taxa in the genus Petunia sensu Jussieu (Solanaceae) Annals of Botany. 2000;85:665–673. [Google Scholar]
- Nukui S, Kitamura S, Hioki T, et al. N2O induces mitotic polyploidization in anther somatic cells and restores fertility in sterile interspecific hybrid lilies. Breeding Science. 2011;61:327–337. doi: 10.1270/jsbbs.61.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Kurimoto K, Miyajima I, et al. Induction of 2n pollen in tulips by arresting meiotic process with nitrous oxide gas. Euphytica. 2005;143:101–114. [Google Scholar]
- Ormerod MG, Tribukait B, Giaretti W. Consensus report of the task force on standardisation of DNA flow cytometry in clinical pathology. Analytical Cellular Pathology. 1998;17:103–110. doi: 10.1155/1998/842306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Zhou Y, Fowke LC, Wang H. An efficient method for flow cytometric analysis of pollen and detection of 2n nuclei in Brassica napus pollen. Plant Cell Reports. 2004;23:196–202. doi: 10.1007/s00299-004-0830-y. [DOI] [PubMed] [Google Scholar]
- Pichot C, El Maâtaoui M. Unreduced diploid nuclei in Cupressus dupreziana A. Camus pollen. Theoretical and Applied Genetics. 2000;101:574–579. [Google Scholar]
- Price HJ, Hodnett G, Johnston JS. Sunflower (Helianthus annuus) leaves contain compounds that reduce nuclear propidium iodide fluorescence. Annals of Botany. 2000;86:929–934. [Google Scholar]
- Roberts AV. The use of bead beating to prepare suspensions of nuclei for flow cytometry from fresh leaves, herbarium leaves, petals and pollen. Cytometry Part A71A. 2007:1039–1044. doi: 10.1002/cyto.a.20486. [DOI] [PubMed] [Google Scholar]
- Shankey TV, Rabinovitch PS, Bagwell B, et al. Guidelines for implementation of clinical DNA cytometry. Cytometry. 1993;14:472–477. doi: 10.1002/cyto.990140503. [DOI] [PubMed] [Google Scholar]
- Stehlik I, Kron P, Barrett SCH, Husband BC. Sexing pollen reveals female bias in a dioecious plant. New Phytologist. 2007;175:185–194. doi: 10.1111/j.1469-8137.2007.02093.x. [DOI] [PubMed] [Google Scholar]
- Suda J, Kron P, Husband BC, Trávnícek P. Flow cytometry with plant cells: analysis of genes, chromosomes and genomes. Weinheim: Wiley-VCH Verlag; 2007a. Flow cytometry and ploidy: applications in plant systematics, ecology and evolutionary biology; pp. 103–130. In: Doležel J, Greilhuber J, Suda J. [Google Scholar]
- Suda J, Weiss-Schneeweiss H, Tribsch A, Schneeweiss GM, Trávníček P, Schönswetter P. Complex distribution patterns of di-, tetra-, and hexaploid cytotypes in the European high mountain plant Senecio carniolicus (Asteraceae) American Journal of Botany. 2007b;94:1391–1401. doi: 10.3732/ajb.94.8.1391. [DOI] [PubMed] [Google Scholar]
- Sugiura A, Tao R, Ohkuma T, Tamura M. Pollen nuclear number in four Diospyros species. HortScience. 1998;33:149–150. [Google Scholar]
- Sugiura A, Ohkuma T, Choi YA, Tao R, Tamura M. Production of nonaploid (2n=9x) Japanese persimmons (Diospyros kaki) by pollination with unreduced (2n=6x) pollen and embryo rescue culture. Journal of the American Society for Horticultural Science. 2000;125:609–614. [Google Scholar]
- Van Laere K, DeWitte A, Van Huylenbroeck J, Van Bockstaele E. Evidence for the occurrence of unreduced gametes in interspecific hybrids of Hibiscus. Journal of Horticultural Science & Biotechnology. 2009;84:240–247. [Google Scholar]
- Van Tuyl JM, De Vries JN, Bino RJ, Kwakkenbos TAM. Identification of 2n-pollen producing interspecific hybrids of Lilium using flow cytometry. Cytologia. 1989;54:737–745. [Google Scholar]
- Vergne P, Delvallee I, Dumas C. Rapid assessment of microspore and pollen development stage in wheat and maize using DAPI and membrane permeabilization. Stain Technology. 1987;62:299–304. doi: 10.3109/10520298709108014. [DOI] [PubMed] [Google Scholar]
- Verity Software House, Inc. ModFit LT User Guide. Topsham, ME: Verity Software House, Inc; 2000. [Google Scholar]
- Verma SC, Rees H. Nuclear DNA and the evolution of allotetraploid Brassicae. Heredity. 1974;33:61–68. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.