Abstract
Accurate multiple alignments of 86 domains that occur in signaling proteins have been constructed and used to provide a Web-based tool (SMART: simple modular architecture research tool) that allows rapid identification and annotation of signaling domain sequences. The majority of signaling proteins are multidomain in character with a considerable variety of domain combinations known. Comparison with established databases showed that 25% of our domain set could not be deduced from SwissProt and 41% could not be annotated by Pfam. SMART is able to determine the modular architectures of single sequences or genomes; application to the entire yeast genome revealed that at least 6.7% of its genes contain one or more signaling domains, approximately 350 greater than previously annotated. The process of constructing SMART predicted (i) novel domain homologues in unexpected locations such as band 4.1-homologous domains in focal adhesion kinases; (ii) previously unknown domain families, including a citron-homology domain; (iii) putative functions of domain families after identification of additional family members, for example, a ubiquitin-binding role for ubiquitin-associated domains (UBA); (iv) cellular roles for proteins, such predicted DEATH domains in netrin receptors further implicating these molecules in axonal guidance; (v) signaling domains in known disease genes such as SPRY domains in both marenostrin/pyrin and Midline 1; (vi) domains in unexpected phylogenetic contexts such as diacylglycerol kinase homologues in yeast and bacteria; and (vii) likely protein misclassifications exemplified by a predicted pleckstrin homology domain in a Candida albicans protein, previously described as an integrin.
Keywords: computer analysis, diacylglycerol kinases, DEATH domain, disease genes, automatic sequence annotation
The functions of only a small fraction of known proteins have been determined by experiment. As a result, the use of computational sequence analysis tools is essential for the annotation of novel genes or genomes, and the prediction of protein structure and function. Currently, the most informative of these techniques are database search tools such as blast (1) and fasta (2) that identify similar sequences with associated statistical significance estimates. Current limitations of the use of these programs concern less the aspects of search sensitivity and more the functional annotation of identified homologues. Annotation terms such as “hypothetical protein” or “suppressor of spt3 mutations” are helpful neither to the user’s prediction of structure and function, nor to computational procedures attempting to automatically predict function from sequence.
An additional aspect concerns the annotation of complete genomes. Existing eubacterial and archaeal genomes have been analyzed with little regard to the existence of domains, because multidomain proteins in these organisms are relatively few in number. The domain as a functional and structural unit in eukaryotic proteins, however, is pre-eminent. For example, the majority of human extracellular proteins are multidomain in character (for reviews see refs. 3 and 4) and many complex eukaryotic signaling networks involve proteins containing multiple domains with catalytic, adaptor, effector, and/or stimulator functions (5). Several dozen of such “signaling domains” are known (for a review see ref. 6). The importance of modular proteins in disease is emphasized by the recent observation that the majority of positionally cloned human disease genes encode multidomain proteins, many of which are, in fact, signaling proteins (7). On the other hand, the view of the domain as a fundamental unit of structure and function is not universally accepted: not a single noncatalytic signaling domain is annotated in the widely distributed Saccharomyces cerevisiae genome directory that catalogs the genes of this complete genome (8).
Thus, there is a need to coordinate knowledge stored in the literature with that stored in sequence databases to facilitate the research of those in the scientific community who require the annotation of genes and genomes. It is our goal to provide an extensively annotated collection of cytoplasmic signaling domain alignments that enables rapid and sensitive detection of additional domain homologues as a Web-based tool.
Because it is difficult to distinguish those domains that perform cytoplasmic signaling roles from those that primarily function in transport, protein sorting, or cell cycle regulation, and for reasons of brevity, we shall discuss those domains that fall under two categories. (i) Cytoplasmic domains that possess kinase, phosphatase, ubiquitin ligase, or phospholipase enzymatic activities or those that stimulate GTPase-activation or guanine nucleotide exchange; these activities are known to mediate transduction of an extracellular signal toward the nucleus resulting in the initiation of a cellular response. (ii) Cytoplasmic domains that occur in at least two proteins with different domain organizations, of which one also contains a domain that is categorized under 1) (for a complete list of such domains see Table 1).
Table 1.
Numbers of domains detected by SMART in the yeast genome, and in the yeast and human fractions of the Swiss Prot database
| Domain | Full name or function | Number of
domains annotated
|
||||||
|---|---|---|---|---|---|---|---|---|
| Yeast genome
|
SwissProt:
S. cerevisiae
|
SwissProt: Homo sapiens
|
||||||
| SMART | SMART | SPa | Pfamb | SMART | SPa | Pfamb | ||
| 14-3-3 | 14-3-3 proteins | 2 | 2 | 2 | 2 | 6 | 6 | 6 |
| ADF | Actin depolymerization factor | 4 | 3 | 2 | 3 | 4 | 2 | 2 |
| ARF | Arf subfamily of small GTPases | 5 | 4 | 4 | 3 | 12 | 12 | 12 |
| ArfGap | GAP for Arf-like GTPases | 6 | 5 | 0 | – | 2 | 0 | – |
| ARM | Armadillo repeat | 14 | 14 | 14 | 1 | 40 | 21 | 13 |
| B41 | Band 4.1 homology | 0 | 0 | 0 | 0 | 9 | 9 | 9 |
| BTK | BTK-like zinc finger | 0 | 0 | 0 | – | 4 | 0 | – |
| C1 | PKC conserved region 1 | 2 | 2 | 2 | 2 | 38 | 32 | 30 |
| C2 | PKC conserved region 2 | 18 | 14 | 11 | 10 | 21 | 19 | 17 |
| CARD | Caspase recruitment domain | 0 | 0 | 0 | 0 | 9 | 0 | – |
| CBS | Cystathionine β-synthase-like domain | 17 | 17 | 0 | – | 23 | 0 | – |
| CH | Calponin homology | 6 | 4 | 4 | 2 | 35 | 26 | 18 |
| CNH | Citron homology | 3 | 2 | 0 | – | 0 | 0 | – |
| cNMP | Cyclic nucleotide monophosphate-binding domain | 4 | 4 | 2 | 3 | 12 | 12 | 14 |
| CYCc | Adenylyl/guanylyl cyclase | 1 | 1 | 1 | 0 | 15 | 15 | 15 |
| DAGKa | DAG kinase (accessory) | 0 | 0 | 0 | – | 6 | 0 | – |
| DAGKc | DAG kinase (catalytic) | 2 | 0 | 0 | – | 6 | 6 | – |
| DAX | In Dsh and axin | 0 | 0 | 0 | – | 1 | 0 | – |
| DEATH | Regulator of cell death | 0 | 0 | 0 | – | 14 | 10 | – |
| DEP | In Dsh. egl-10 and pleckstrin | 5 | 5 | 0 | – | 2 | 0 | – |
| DYNc | Dynamin | 3 | 3 | 3 | 3 | 4 | 4 | 4 |
| EFh | EF-hand | 24 | 24 | 17 | 7 | 124 | 124 | 62 |
| FCH | Fes/CIP4 homology domain | 4 | 4 | 0 | – | 3 | 0 | – |
| FHA | Forkhead associated domain | 14 | 9 | 8 | 9 | 1 | 1 | 1 |
| FYVE | In Fab1, YOTB, Vac1, and EEA1 | 6 | 5 | 2 | – | 1 | 0 | – |
| GAF | In cGMP-PDEs, adenylyl cyclases, and E. coli fh1A | 0 | 0 | 0 | – | 4 | 0 | – |
| HECTc | Homologous to E6-AP carboxyl terminus; ubiquitin ligases | 5 | 4 | 3 | – | 4 | 4 | – |
| HR1 | PRK kinase homology region 1 | 2 | 2 | 0 | – | 0 | 0 | – |
| IPPc | Inositol polyphosphate phosphatase | 4 | 2 | 2 | 2 | 2 | 2 | 2 |
| IQ | Calmodulin-binding motif | 6 | 6 | 3 | – | 10 | 2 | – |
| KISc | Kinesin, catalytic domain | 6 | 6 | 6 | 6 | 6 | 6 | 6 |
| LIM | Zinc finger in Lin-11, Is1-1, Mec-3 | 9 | 7 | 5 | 3 | 50 | 50 | 30 |
| MYSc | Myosin, catalytic domain | 5 | 5 | 5 | 5 | 6 | 6 | 6 |
| OPR | Octicosapeptide repeat | 1 | 1 | 0 | – | 4 | 0 | – |
| PAC | Motif, C-terminal to PAS | 0 | 0 | 0 | – | 3 | 0 | – |
| PAS | Domain in Per, ARNT, Sim | 1 | 1 | 0 | – | 6 | 6 | – |
| PBD | p21 Rho-binding domain | 3 | 3 | 0 | – | 3 | 2 | – |
| PDZ | In PSD-95, D1g, ZO-1 | 0 | 0 | 0 | – | 31 | 15 | – |
| PH | Pleckstrin homology domain | 27 | 19 | 12 | 13 | 36 | 30 | 27 |
| PI3K_PI3Kb | PI3K: p85-binding domain | 0 | 0 | 0 | 0 | 2 | 0 | – |
| PI3K_rbd | P13K: Ras-binding domain | 0 | 0 | 0 | 0 | 3 | 0 | – |
| P13Ka | P13K accessory domain | 2 | 2 | 0 | – | 4 | 0 | – |
| P13Kc | Phosphoinositide kinase | 8 | 8 | 8 | 7 | 5 | 5 | 5 |
| PLAc | Phospholipase A2, catalytic domain | 4 | 4 | 4 | – | 1 | 1 | – |
| PLCXc | Phospholipase C, domain X | 1 | 1 | 1 | 1 | 5 | 5 | 5 |
| PLCYc | Phospholipase C, domain Y | 1 | 1 | 1 | 1 | 5 | 5 | 5 |
| PLDc | Phospholipase D, conserved motif | 2 | 2 | 0 | – | 2 | 2 | – |
| PP2Ac | Protein phosphatases 2A | 12 | 12 | 12 | 12 | 10 | 10 | 10 |
| PP2Cc | Protein phosphatases 2C | 7 | 6 | 5 | 2 | 2 | 2 | 2 |
| Protein kinases | TyrKc: Tyr-specific | 0 | 0 | 0 | – | 70 | 69 | |
| S_TKc: Ser/Thr-specific | 61 | 58 | 58 | – | 62 | 62 | – | |
| STYKc: Ser/Thr/Tyr-specific | 56 | 49 | 46 | 104(all) | 51 | 49 | 184(all) | |
| Protein phosphatases | PTPc: Tyr-specific | 3 | 3 | 3 | 3 | 35 | 34 | 35 |
| DSPc: Dual-specific | 5 | 5 | 5 | – | 7 | 7 | – | |
| PTPc_DSPc: Tyr-,/Ser-/Thr- | 3 | 3 | 2 | – | 2 | 1 | – | |
| PTB | Phosphotyrosine-binding domain | 0 | 0 | 0 | 0 | 4 | 4 | – |
| PX | Phox proteins’ domain | 14 | 10 | 1 | – | 3 | 0 | – |
| RA | In RalGDS, AF-6 (Ras-associating) | 1 | 1 | 0 | – | 4 | 0 | – |
| RAS-like small | RAB | 9 | 9 | 9 | – | 19 | 19 | – |
| GTPases | RAN | 2 | 2 | 2 | – | 1 | 1 | – |
| RAS | 3 | 3 | 3 | – | 11 | 11 | – | |
| RHO | 6 | 6 | 6 | – | 13 | 13 | – | |
| SAR | 1 | 1 | 1 | – | 0 | 0 | – | |
| Others | 11 | 10 | 7 | 24(all) | 5 | 3 | 48(all) | |
| RanBD | Ran-binding domain | 3 | 3 | 1 | – | 5 | 5 | – |
| RasGAP | GAP for Ras-like GTPases | 4 | 3 | 3 | – | 3 | 3 | – |
| RasGEF | GEF for Ras-like GTPases | 5 | 5 | 4 | – | 0 | 0 | – |
| RasGEFN | In some RasGEFs | 4 | 4 | 0 | – | 0 | 0 | – |
| RGS | Regulator of G-protein signaling | 3 | 1 | 1 | – | 11 | 6 | – |
| RhoGAP | GAP for Rho-like GTPases | 9 | 6 | 3 | – | 8 | 4 | – |
| RhoGEF | GEF for Rho-like GTPases | 4 | 4 | 3 | – | 7 | 6 | – |
| SAM | Sterile alpha motif | 6 | 3 | 0 | – | 11 | 1 | – |
| SH2 | Src homology 2 | 1 | 1 | 1 | 1 | 51 | 51 | 51 |
| SH3 | Src homology 3 | 28 | 25 | 25 | 25 | 65 | 63 | 57 |
| SPRY | In sp1A and Ryanodine receptors | 3 | 3 | 0 | – | 7 | 0 | – |
| TBC | In Tre-2, BUB2p, and Cdc16p | 10 | 7 | 0 | – | 1 | 0 | – |
| TPR | Tetratricopeptide repeat | 72 | 69 | 39 | 16 | 40 | 0 | 7 |
| UBA | Ubiquitin-associated domain | 10 | 8 | 0 | – | 12 | 0 | – |
| UBCc | Ubiquitin-conjugating enzyme | 13 | 13 | 13 | 13 | 12 | 12 | 12 |
| UBX | Ubiquitin-related domain | 8 | 4 | 0 | – | 1 | 0 | – |
| VHS | In VPS-27, Hrs and STAM | 4 | 3 | 0 | – | 0 | 0 | – |
| VPS9 | In VPS-9-like proteins | 2 | 1 | 1 | – | 1 | 0 | – |
| WH1 | WASp homology domain 1 | 1 | 1 | 0 | – | 2 | 0 | – |
| WW | Conserved WW motif | 9 | 8 | 7 | 7 | 9 | 9 | 9 |
| ZU5 | In ZO-1 and UNC-5 | 0 | 0 | 0 | – | 4 | 0 | – |
| ZZ | Dystrophin-like zinc finger | 2 | 2 | 0 | – | 4 | 1 | – |
| Totals | 86 | 622 | 544 | 383 | 290 | 1,137 | 886 | 704 |
Numbers of domains detected by SMART in the yeast genome, and in the yeast and human fractions of the SwissProt database are compared with the numbers of domains derived from HMMer analysis and Pfam HMMs scanned against these database fractions, and the numbers of annotations in SwissProt. Many of these domains are reviewed elsewhere (5, 6), and additional references may be found via the SMART Web site (http://www.bork.embl-heidelberg.de/Modules/sinput.shtml).
Annotations in SwissProt.
Annotations using the hmmfs program of the HMMer package with Pfam-derived HMMs (“–” indicates where no Pfam HMM was available).
Domain collections that cover a wide spectrum of cellular functions do exist in the forms of motif, alignment block, or profile databases such as prosite (9), blocks (10), prints (11), or Pfam (12) and provide a guide for the annotation of new proteins. However, there is a necessary trade-off in these collections between exhaustive coverage of domains and optimal sensitivity, specificity, and annotation quality. We have chosen to initiate the collection of gapped alignments of signaling domains because these are imperfectly covered in large collections and often include homologues with extremely divergent sequences. This collection is designed to be updated easily and is provided with a Worldwide Web interface enabling automatic sequence annotation with evolutionary, functional, and structural information. The resulting SMART procedure, a simple modular architecture research tool, offers a high level of sensitivity and specificity coupled with ease of use.
METHODS
Construction of Multiple Sequence Alignments and Choice of the Search Program.
Of the 86 domain families, multiple alignments of 83 had been published previously (for references, see the annotation that accompanies the SMART Web site). These alignments were refined according to constraints described elsewhere (13) that included minimization of insertions/deletions in conserved alignment blocks, optimization of amino acid property conservation within these blocks, and closing of unnecessary gaps within insertion/deletion regions. Gapped alignments were constructed in preference to ungapped ones to allow the prediction of domain limits and as a result of their greater information content. Care was taken to build alignments that encompassed all secondary structures of domains whose tertiary structures are known. For remaining domains, investigations of sequence similarities beyond previously published domain limits were undertaken; this resulted in N-terminal extension of the previously described PX domain alignment by a single predicted β-strand, and identification of a conserved N-terminal motif in guanine nucleotide exchange factors for Ras-like GTPases. Prediction of domain limits also was aided by close proximities of domains to others with well-known limits, and to bona fide N- and C-terminal residues.
Alignments were updated to include additional predicted homologues. Because no single database searching algorithm currently is able to detect all putative homologues that are detectable by the combination of all searching methods (13), three iterative methods—HMMer, MoST, and WiseTools (14–16)—were used to detect candidate homologues (HMMer and MoST thresholds: 25 bits and E < 0.01). Before their addition to multiple alignments, candidate homologue sequences were subjected to analyses using blast (1), Ssearch (2), and/or macaw (17) to estimate the statistical significance of sequence similarities (psi-blast, blast, and Ssearch thresholds: E < 0.01). Those sequences that were considered homologues based on statistical significance estimates, and to a lesser extent on experimentally determined biological context, were used to construct alignments, profiles, and Hidden Markov models (HMMs).
As described above, care was taken to establish alignments representing entire structural domains. However, the termini were found to be the least conserved regions of alignments, and several profiles represent incomplete portions of domains. In two cases, phospholipase D and protein tyrosine phosphatase homologues, only short conserved “motifs” (conservation patterns representing an incomplete domain structure) are detectable across the domain family (18–20). For these examples, profiles/HMMs were calculated only from these short motifs to maximize the amino acid similarity signal-to-noise ratio (13).
Assignment and Calibration of Thresholds for Automatic Runs.
Score thresholds are required to provide automatic assignment of true positives and true negatives. There is no current method, including those that provide E- or p-value representations of score significances, that may be relied on to provide reliable values for these thresholds in all cases. As a result, manual intervention was necessary to estimate threshold values on the basis of published homology arguments and, for example, on the results of individual blast or Ssearch queries. SWise (16) was chosen as an established algorithm able to provide similarity scores for query sequences when compared with the alignment database; however, the SMART database method can be applied to any algorithm that provides similarity scores.
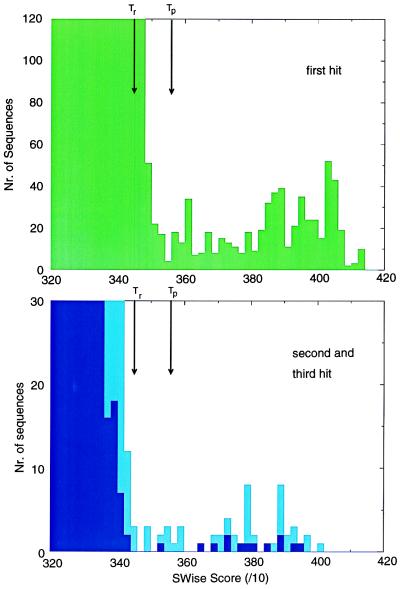
For each alignment an SWise (16) threshold (Tp) was established that represents the lowest score allowable for sequences to be considered as “true positives” or homologues. As such, this single step procedure detects many true positives but does not detect few previously proposed homologues (“false negatives”) that score at levels just below that of the top “true negative.” A proportion of false negatives could not be assigned as homologues without further statistical evidence. However, consideration that domains such as ARM, C2, CBS, IQ, LIM, PDZ, SH2, SH3, and WW (Table 1) frequently are found as repeats, enabled several false negatives to be detected by using estimations of an additional threshold value, Tr (Tr < Tp). Tr represents a repeats’ threshold for a protein where at least one of the repeats scores above Tp (Fig. 1). Two or more repeats scoring above the average of Tp and Tr [(Tp + Tr)/2] also were considered false negatives. Some domains that appear to be found only as tandem repeats (for example, EF-hands, tetratricopeptide repeats, and armadillo repeats) are reported only if two or more copies are found that score above a low threshold Tr. To predict the subfamily of a particular domain (for example, whether a tyrosine or a serine/threonine kinase, or whether a tyrosine-specificity or a dual-specificity phosphatase) further thresholds Ts (Ts > Tp) also were estimated; no subfamily predictions are made for those domain homologues that score above Tp but below Ts.
Figure 1.
Calibration of thresholds. Selection of thresholds from the distributions of SH3 domain scores. (Upper) A histogram of SWise scores for the best match (optimal alignment; in green) of proteins with a SH3 domain profile. (Lower) Similar histograms for the second- and third-best matches (suboptimal alignments; in light blue and dark blue, respectively). Optimal alignment scores less than threshold Tp are mostly derived from sequences considered unlikely to contain SH3 domain homologues. Threshold Tp was selected as the lowest scoring true positive. Domains that are repeated twice or more in the same protein that each score above a lower threshold (Tr) are considered to be true negatives.
Subset alignments of a given domain family were constructed not only to improve the specificity of functional predictions, but also for divergent families for which a single descriptor (profile/HMM) was found to be unable to detect the entire set of known homologues (e.g., C2 and pleckstrin homology (PH) domains; refs. 21 and 22). Construction of multiple profiles each representing different regions of the domain phylogenetic tree resulted in “overlapping” profiles that, when used in combination, found the maximal number of homologues. Sensitivity and specificity is guaranteed with combinations of Ts and Tp. Overlapping hits from nonhomologous profiles, which can occur because of inserted domains (23), all are reported.
Seeding and Updating Procedure.
To reduce redundancy and subfamily bias within sequence families, seed alignments were calculated by using an iterative semiautomatic procedure. In a first step all database sequences considered homologous, given the threshold procedures described above, are subjected to a clustalw phylogenetic tree construction (24). Only a single sequence from every branch of the tree that is shorter than a defined threshold (the default distance is 0.2, which corresponds approximately to 80% identity, ref. 24) is retained in the alignment. From this seed alignment, a profile is derived leading to reiteration of the database search procedure until convergence. For example, four iterations were required to build a Src homology 2 (SH2) seed alignment containing 95 sequences, of a total of 548 SH2 domains identified in the translated EMBL sequence database.
With new sequences entering databases daily, seed alignments and derived profiles need to be updated accordingly. SMART incorporates a facility whereby database daily updates are screened for the presence of signaling domains. Those that represent a new branch of the domain family phylogenetic tree (i.e., with a distance of greater than 0.2) are recorded for inclusion in future SMART domain set updates. The alignments are accessible via the SMART Web server.
Implementation into a Web Server.
SMART has been provided with a user interface (http://www.bork.embl-heidelberg.de/Modules/sinput.shtml) that allows rapid and automatic annotation of the signaling domain composition of any query protein sequence. A graphical display is provided showing domain positions within the query sequence. The SMART set of signaling domains is annotated extensively via hyperlinks to Medline and the Molecular Modeling Database via Entrez (25), thus providing easy access to information relating sequence, homology, structure, and function. As the set of signaling sequences is necessarily incomplete and as there may be other domains represented in the query sequence, direct access also is provided to Pfam (12), a domain database that includes a variety of different domain types, yet provides a lower representation of signaling domains and with lower sensitivity (see Discussion). Intrinsic features of the query such as coiled coil regions (26), low complexity regions (27), and transmembrane regions (28) also are displayed. Annotated or unannotated regions of the query sequence are able to be subjected individually to gapped blast searches (1), thus allowing the advantage of a reduced search space enabling higher sensitivity in searches.
Benchmarking Protocol.
To assess the sensitivity and selectivity of SMART, results were compared with annotations held by SwissProt, because this represents the best-annotated protein sequence database extant, (and includes all those annotations covered by the prosite database) as well as with the Pfam domain collection, because this represents the most comprehensive set of gapped alignments available (12). Our intention here was not to provide justifications for the inclusion or exclusion of particular sequences in domain alignments, but to compare literature information as represented by the SMART database, with the same information as represented by SwissProt and Pfam databases. All S. cerevisiae and human sequences were extracted from SwissProt and annotated by using the SMART protocol. Because these organisms are well-studied and their proteins relatively well-annotated they represent a stringent test for annotation procedures. The SMART domain annotations were compared manually with those derived from HMMer (14) analysis, and those contained in SwissProt (Table 1); the hmmfs program and a 25-bits threshold was used for the HMMer analysis. As the SwissProt release 34 does not contain all yeast sequences, the complete set of S. cerevisiae ORFs also was subjected to SMART analysis (Table 1).
RESULTS
Comparison with SwissProt and Pfam.
Of all protein sequence databases, SwissProt is the most extensively annotated, making use of literature- and sequence-derived (9) data as source material. As a result the SwissProt database is a valuable resource for investigators searching for hints of the structure and function of their sequences of interest. Consequently, it is appropriate to compare SMART-derived annotations with those contained in SwissProt.
SMART detected 548 and 1,137 domains in the yeast and human subsets of SwissProt, respectively (Table 1). Of these, 165 and 251 domains (30% and 22%, respectively) are not annotated in SwissProt. Many of these belong to the 29 domain families that are contained in SMART and yet are not annotated in SwissProt. By contrast, all SwissProt annotations relating to our domain set were detected by SMART, with the exception of a small set of domain fragments. Only 23 of the SMART domain families are represented by Prosite motifs or patterns. Moreover, because Prosite motifs commonly represent active site regions, it is apparent that these do not detect the several homologues of kinases, phosphatases, or ubiquitin-conjugating enzymes that have dispensed with their active site residues.
The current set of Pfam HMMs, when compared with the yeast and human SwissProt subsets, detected 290 and 704 domains. Forty-six of the 86 SMART domain types are not represented currently in Pfam. Moreover, the Pfam set does not yet allow subfamily annotation for domain families such as small GTPases, protein kinases, or protein phosphatases. Pfam and HMMer were able to identify several incomplete domain sequences that SMART could not. SMART was not designed to detect domain fragments because it was considered valuable to detect complete domains, thereby allowing assignment of putative domain boundaries. Consequently, the HMMer (hmmfs) option of SMART has been provided to allow detection of incomplete domain sequences.
Identification of Signaling Domains in Yeast.
Annotation of the complete yeast genome (6218 ORFs) revealed that 420 yeast proteins (6.7%) contain at least one of the domains included in SMART. This is larger than a previous estimate that 2% of yeast proteins are involved in signaling (8), which approximates to the percentage of S. cerevisiae proteins known to be kinase homologues. SMART identifies a total of 622 domains (Table 1); two or more domains occur in 96 of the 420 signaling proteins. Results of the SMART annotation of yeast proteins identified are summarized in a Web page (http://www.bork.embl-heidelberg.de/Modules/syeast.html), which was generated by using SMART’s graphical output features.
These results imply an improvement by SMART on other tools and current best-annotated databases in the particular field of signaling. An additional feature of SMART is its ability to facilitate predictions of the structures and/or functions of proteins when a hit is recorded. The following examples illustrate several such instances that arise from a domain hit.
Domain Annotation and Deduction of Functional Features.
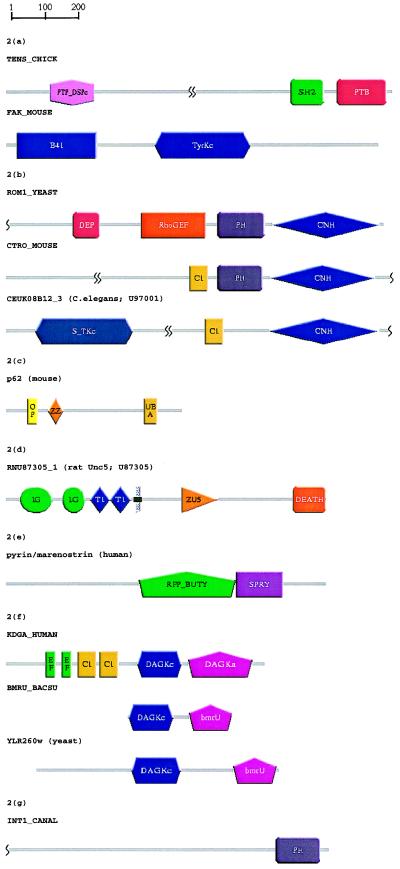
During construction of the SMART database, tensin and focal adhesion kinase (pp125FAK), which both are localized to focal contacts, were found to contain previously unrecognized domains. Fig. 2a shows the modular architecture of tensin, an actin filament capping protein that is known to contain large coiled coil regions, an SH2 (29) and an N-terminal domain homologous to protein tyrosine phosphatases (PTPs) (20). SMART predicts a phosphotyrosine binding domain (PTB; also called phosphotyrosine interaction [PI] domain) (Table 1) in tensin’s most C-terminal region, which has not previously been ascribed a domain homology. Each of tensin’s three globular domains—PTP, SH2, and PTB/PI—have been implicated in phosphotyrosine-mediated signaling. This is consistent with previous findings that tensin is a substrate of the tyrosine kinase pp125FAK (30), which is also highly tyrosine-phosphorylated when activated (reviewed in ref. 31).
Figure 2.
Schematic representations, produced using SMART, of the domain architectures of proteins discussed in the text. See Table 1 for the identified domains; gray lines (no SMART match) might contain other known domains not included in SMART. Putative homologues were identified during SWise (16) searches and/or psi-blast (1) searches (E < 0.01). (a) Domain recognition: A novel PTB domain was identified in tensin, resulting in completion of its modular architecture assignment. A psi-blast search with a previously predicted PTB domain in C. elegans F56D2.1 (53) yields the tensin PTB after four passes. Prediction of molecular function via domain hit: Identification of a domain homologous to band 4.1 protein in focal adhesion kinase (FAK) isoforms. FAKs are predicted to bind cytoplasmic portions of integrins in a similar manner to that of talin, another band 4.1 domain-containing protein. A psi-blast search with a band 4.1-like domain (41 HUMAN, residues 206–401) revealed band 4.1-like domains in human, bovine, and Xenopus FAK isoforms by pass 3. (b) Detection of new domains because of search space reduction: Putative DEP domains in ROM1 and ROM2 were identified by using SWise (16) and HMMer (14), but could not be detected by using psi-blast. Analysis of the regions surrounding identified domains revealed the presence of a novel domain in the C-terminal regions of ROM1 and ROM2 that occurs also in several Ste20-like protein kinases, and mouse citron (CNH, citron homology). A gapped blast search of the region of citron C-terminal to its PH domain (CTRO MOUSE, residues 1134–1457) reveals significant similarity with yeast ROM2 (E = 1 × 10−5). (c) Functional predictions for an entire domain family: A region of p62 known to bind ubiquitin (40), and its homologous sequence in the Drosophila protein ref(2)P, scored as the highest putative true negatives in a SWise search. We predict ubiquitin-binding functions for UBA domains. psi-blast searches were unable to corroborate this prediction. (d) Prediction of cellular functions: Although not indicated in the primary sources (43, 44), a DEATH domain was found in rcm and other UNC5 homologues, in agreement with a previous claim (41). At the molecular level, this domain in UNC5 is predicted to form a heterotypic dimer with an homologous domain in UNC44 implying a cellular role in axon guidance. A gapped blast search with the known DEATH domain of death-associated protein kinase (DAPK HUMAN, residues 1304–1396) predicts a DEATH domain in rat UNC5H1 with E = 9 × 10−3). (e) Signaling domains in “disease genes”: Pyrin or marenostrin, a protein that is mutated in patients with Mediterranean fever and is similar to butyrophilin, contains a SPRY domain. psi-blast with the SPRY domain of human DDX1 (EMBL:X70649, residues 124–240) yields a butyrophilin homologue by pass 5 and pyrin/marenostrin (residues 663–759) by pass 7. (f) Homologues of domains involved in eukaryotic signaling may not be eukaryotic-specific: DAG kinases have been found previously in mammals, invertebrates, plants, and slime mold. However, it is apparent that DAG kinase homologues of unknown function are present in yeasts and in eubacteria (see Fig. 3). A gapped blast search with Bacillus subtilis bmrU (BMRU BACSU) yields significant similarities with Arabidopsis thaliana DAG kinase (KDG1 ARATH; E = 4 × 10−4) and a Schizosaccharomyces pombe ORF (SPAC4A8.07c; E = 1 × 10−7). (g) Identification of potential misclassifications: A PH domain and the lack of an obvious transmembrane sequence indicates a cytoplasmic and signaling role for a protein (INT1 CANAL) previously thought to be a yeast integrin. A psi-blast search with the N-terminal PH domain of pleckstrin yielded INT1 CANAL in pass 3.
Application of SMART procedures to pp125FAK homologues predicts band 4.1-homologous domains in their N-terminal regions that bind the cytoplasmic regions of integrins (32) (Fig. 2a). Although one has to be cautious when inferring functional information simply from domain identification, on this occasion the band 4.1 domains are likely to perform similar molecular functions because talin, another band 4.1 domain-containing protein, is known also to bind integrin cytoplasmic domains (33).
Reducing the Search Space Enables Identification of Novel Domains.
S. cerevisiae ROM1 and ROM2 are sequence-similar proteins that each contain a PH domain and a RhoGEF domain that stimulates exchange of Rho1GDP with Rho1GTP (34). Construction of the SMART databases led to the identification of a putative DEP domain (35) in both ROM1 and ROM2 (Fig. 2b). Comparison of the ROM1 and ROM2 sequences showed a further region of similarity C-terminal to their PH domains. This region [“citron-homology” (CNH) domain] was identified as being homologous to the mouse RhoGTP/RacGTP-binding protein, citron (36) and to the C-terminal regions of several Ste20-like protein kinases (Fig. 2b). A novel domain family (VHS) of unknown function(s) also has been detected in Vps27, Hrs, and STAM, and other proteins.
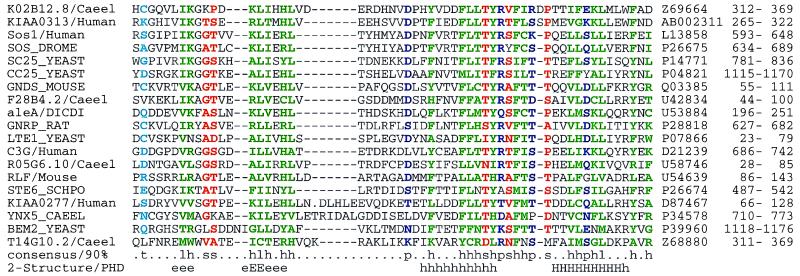
A conserved domain in Cdc25p-like proteins mediates their activities as guanine nucleotide exchange factors for Ras or Ral (37). Each of these molecules contain N-terminal extensions. We find additional amino acid similarities in these regions, and these represent a novel domain family (Fig. 3). Surprisingly, this domain (which we call RasGEFN) can be contiguous to, or far from, the catalytic domain. A construct of p140 Ras-GRF that lacks this region is constitutively active (38), so it is likely that the RasGEFN domain performs a suppressor function.
Figure 3.
Multiple alignments of selected RasGEFN domains. A conserved region was found in the N-terminal regions of several proteins with RasGEF (Cdc25-like) domains (37). Surprisingly, this N-terminal domain may be present in the sequence either close to, of far from, the RasGEF domain. A psi-blast search using a region (residues 898–946) of C. albicans Cdc25 (CC25 CANAL) and E < 0.01, identified each of the sequences in Fig. 3 within nine passes before convergence. Predicted (54) secondary structure and 90% consensus sequences are shown beneath the alignments; SwissProt/PIR/EMBL accession codes and residue limits are given after the alignments. Residues are colored according to the consensus sequence [green: hydrophobic (h), ACFGHIKLMRTVWY; blue: polar (p), CDEHKNQRST; red: small (s), ACDGNPSTV; red: tiny (u), AGS; cyan: turn-like (t), ACDEGHKNQRST; green: aliphatic (l), ILV; and, magenta: alcohol (o), ST). The SwissProt sequence KMHC DICDI has been altered to account for probable frameshifts.
Deducing Functional Features of a Domain Family Via a Protein Hit.
Although rare, we have identified additional members of a domain family in regions of proteins that already have been shown to perform particular functions. Such findings often suggest comparable functions for all other members of the domain family. The ubiquitin-associated (UBA) domain (Table 1) has been shown to be contained in several enzymes implicated in ubiquitination (39). We have identified a UBA domain in a region of p62, a phosphotyrosine-independent ligand of the p56lck SH2 domain (40) that is known to bind ubiquitin (Fig. 2c). Ubiquitin-binding functions are predicted for other UBA domains.
Prediction of Cellular Function.
Particular domains have been implicated in certain cellular events. For example, DEATH domains (Table 1) are present in proteins associated with apoptosis and/or axonal guidance (41, 42). Recent reports (43, 44) identify the rostral cerebellar malformation gene product (rcm) and similar homologues as putative netrin receptors. These reports do not indicate the presence of a DEATH domain in rcm or its homologues, even though the domain’s presence may be readily demonstrated by sequence analysis (Fig. 2d) or from its identification in the rcm Caenorhabditis elegans orthologue, UNC-5 (41). As the DEATH domain of UNC-5 is not annotated in databases, this is one of many instances where the potential of domain identification to predict cellular function has been unfulfilled. DEATH domains often form homotypic or heterotypic dimers (42). Because DEATH domain-containing proteins UNC-44 (45) and the putative netrin-receptor UNC-5 are known to be involved in axonal guidance, we predict that transduction of the netrin-initiated signal involves heterodimerization of UNC-5 and UNC-44 DEATH domains.
Identification of Signaling Domains in Genes That Are Involved in Diseases.
A recent study of 70 positionally cloned human genes mutated in diseases found that a significantly high proportion of these “disease genes” possess roles in cell signaling (7). In accordance with this, the SMART alignment database contains several novel signaling domains in these genes (including the DEATH domain in rcm-like netrin receptors, see above). Fig. 2e shows the modular architecture of pyrin (46) (also called marenostrin; ref. 47). Mutations in the pyrin gene result in Mediterranean fever syndromes that are inherited inflammatory disorders. In addition to its ret-like zinc finger, pyrin/marenostrin and other butyrophilin-like homologues contain a SPRY domain, a domain of unknown function found triplicated in ryanodine receptors and singly in other proteins (48) (Table 1). Midline 1, a pyrin-homologue that also contains a SPRY domain, is mutated in patients with Opitz G/BBB syndrome (49).
Identification of Domains in Different Phyla.
The range of species in which a particular domain type is found can correlate with the evolution of specific signaling pathways; many of the known cascades are expected only in animals or eukaryotes (3). Thus, identification of DAG kinase homologues in yeast and eubacteria (Fig. 2f) is clearly a surprise. Although further experimentation is required to infer functional features, the presence of conserved, presumably catalytic, residues in the alignment (data not shown) and the occurrence of DAG kinase activities in prokaryotes (50) suggests that the yeast and bacterial DAG kinase homologues possess similar molecular, but perhaps not cellular, roles to those of their animal and plant homologues.
Significance of Domain Detection and Functional Prediction.
Annotation of molecular function in sequence databases and even in the literature is difficult to interpret given that the term function may describe phenomena occurring at distinct levels, such as those of amino acids, domains, proteins, molecular complexes, cells, or organisms. Nevertheless, the examples shown above demonstrate that annotation of a certain domain can provide useful hints toward experimental characterization of function at different levels. Domain identification also might provide a counter-argument to a previously proposed molecular function. For example, identification of a PH domain and the absence of a detectable transmembrane region in a supposed integrin from C. albicans (Fig. 2g) argues strongly against its proposed role in cell adhesion (51). Integrins are transmembrane proteins that link the extracellular matrix with the cytoskeleton and normally contain, except for the B-4 subunit, short cytoplasmic sequences. The finding of a PH domain and high sequence similarity to S. cerevisiae BUD4 argues for its signaling role in bud site selection.
DISCUSSION
Many proteins are multidomain in character and possess multiple functions that often are performed by one or more component domains. A Web-based tool (SMART) has been designed that makes use of mainly public domain information to allow easy and rapid annotation of signaling multidomain proteins. The tool contains several unique aspects, including automatic seed alignment generation, automatic detection of repeated motifs or domains, and a protocol for combining domain predictions from homologous subfamilies. The ability of SMART to annotate single sequences or large datasets is exemplified by the cases described in Results, including annotation of the complete set of yeast ORFs.
Currently, large-scale or genome analysis is commonly performed by annotating ORFs with a single “best hit” from similarity searches. Ambiguities whether hits represent orthologs (i.e., homologues in different organisms that arose from speciation rather than intragenome duplication and are likely to have a corresponding function; ref. 52) or else paralogs (other members of multigene families) are not solved and omission of domain annotation also leads to misprediction of function. As most signaling proteins are multidomain in character, only annotation at the domain level avoids ambiguities in assigning homologies and functions to sequences, which may propagate further on additional findings of homology. Furthermore, deduction of the modular architecture is essential for the understanding of the complexities of multidomain eukaryotic signaling molecules; current annotation, however, does not adequately provide this information (Table 1). As examples of this, the existence of noncatalytic signaling domains cannot be deduced from the current yeast genome directory (8) and no human RasGEF domains currently are annotated in SwissProt. Graphical representation of the complement of modular proteins in a completed genome (e.g., the 622 signaling domains in 420 yeast proteins: http://www.bork.embl-heidelberg.de/Modules/syeast.html) might provide the basis for relating experimentally derived information concerning domains and multidomain proteins, to cellular events such as signaling.
Although other collections, such as prosite, Pfam, blocks, and prints, contain many more distinct domains or motifs, the focus of SMART on signaling allows significantly enhanced detection sensitivity, the inclusion of many families that are not represented in other collections, and offers a high level of specificity (i.e., a low rate of false positives that is essential for large-scale analysis). The SMART database shall be continually updated; alignment updates shall be semiautomated to avoid misalignments. Thus, forthcoming SMART database versions shall be hand-checked to provide datasets of high quality. In future, experimental findings that advance the understanding of domain structure and function also shall be provided via updates. As SMART is designed to obtain biologically relevant results without dependency on a single database search technique, there is potential to modify underlying methods to improve performance.
Acknowledgments
We thank colleagues at the European Molecular Biology Laboratory and Ewan Birney for many helpful discussions. We also thank Bernhard Sulzer for computational assistance. C.P.P. is a Wellcome Trust Career Development Fellow and a member of the Oxford Centre for Molecular Sciences, and was supported in part by a European Molecular Biology Organization Short-Term Fellowship. J.S. and P.B were supported by the European Union, Bundesministerium für Bildung und Forschung (Germany), and the Deutsche Forschungsgemeinschaft.
ABBREVIATIONS
- SMART
simple modular architecture research tool
- DAG
diacylglycerol
- PH
pleckstrin homology
- PTB
phosphotyrosine binding
- SH
Src homology
- rcm
rostral cerebellar malformation gene product
- HMM
Hidden Markov model
Note Added in Proof
Recent improvements to the SMART system include implementation of SWise-derived E-values and addition of more than 80 extracellular domains. A ProfileScan Server (http://ulrec3.unil.ch/software/PFSCAN_form.html) has appeared recently that includes facilities that are similar or complementary to those of SMART.
References
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearson W R. Genomics. 1991;11:635–650. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- 3.Doolittle R F. Annu Rev Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- 4.Bork P, Downing A K, Kieffer B, Campbell I D. Q Rev Biophys. 1996;29:119–167. doi: 10.1017/s0033583500005783. [DOI] [PubMed] [Google Scholar]
- 5.Bork P, Schultz J, Ponting C P. Trends Biochem Sci. 1997;22:296–298. doi: 10.1016/s0968-0004(97)01084-0. [DOI] [PubMed] [Google Scholar]
- 6.Ponting, C. P., Schultz, J. & Bork, P. (1997) Trends Biochem. Sci. 22, Poster Suppl. C04. [DOI] [PubMed]
- 7.Mushegian A R, Bassett D E, Jr, Borguski M, Bork P, Koonin E V. Proc Natl Acad Sci USA. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mewes, H. W., Albermann, K., Bahr, M., Frishman, D., Gleissmer, A., Hami, J., Heumann, K., Kleine, K., Muier, A., Oliver, S. G., et al. (1997) Nature (London) 387, Suppl., 7–65. [DOI] [PubMed]
- 9.Bairoch A, Bucher P, Hofmann K. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henikoff J G, Pietrokovski S, Henikoff S. Nucleic Acids Res. 1997;25:222–225. doi: 10.1093/nar/25.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attwood T K, Beck M E, Bleasby A J, Degtyarenko K, Michie A D, Parry-Smith D J. Nucleic Acids Res. 1997;25:212–217. doi: 10.1093/nar/25.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonnhammer E L, Eddy S R, Durbin R. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 13.Bork P, Gibson T J. Methods Enzymol. 1996;266:162–184. doi: 10.1016/s0076-6879(96)66013-3. [DOI] [PubMed] [Google Scholar]
- 14.Eddy S R, Mitchison G, Durbin R J. Comput Biol. 1995;2:9–23. doi: 10.1089/cmb.1995.2.9. [DOI] [PubMed] [Google Scholar]
- 15.Tatusov R L, Altschul S F, Koonin E V. Proc Natl Acad Sci USA. 1994;91:12091–12095. doi: 10.1073/pnas.91.25.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birney E, Thompson J, Gibson T. Nucleic Acids Res. 1996;24:2730–2739. doi: 10.1093/nar/24.14.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuler G D, Altschul S F, Lipman D J. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 18.Ponting C P, Kerr I D. Protein Sci. 1996;5:914–922. doi: 10.1002/pro.5560050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin E V. Trends Biochem Sci. 1996;21:242–243. [PubMed] [Google Scholar]
- 20.Haynie D T, Ponting C P. Protein Sci. 1996;5:2643–2646. doi: 10.1002/pro.5560051227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponting C P, Parker P J. Protein Sci. 1996;5:162–166. doi: 10.1002/pro.5560050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson T J, Hyvonen M, Musacchio A, Saraste M, Birney E. Trends Biochem Sci. 1994;19:349–353. doi: 10.1016/0968-0004(94)90108-2. [DOI] [PubMed] [Google Scholar]
- 23.Russell R B. Protein Eng. 1994;7:1407–1410. doi: 10.1093/protein/7.12.1407. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogue C W V, Ohkawa H, Bryant S H. Trends Biochem Sci. 1996;21:226–229. [PubMed] [Google Scholar]
- 26.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 27.Wootton J C, Federhen S. Methods Enzymol. 1996;266:554–573. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 28.Fasman G D, Gilberts W A. Trends Biochem Sci. 1990;15:89–92. doi: 10.1016/0968-0004(90)90187-g. [DOI] [PubMed] [Google Scholar]
- 29.Davis S, Lu M L, Lo S H, Lin S, Butler J A, Druker B J, Roberts T M, An Q, Chen L B. Science. 1991;252:712–715. doi: 10.1126/science.1708917. [DOI] [PubMed] [Google Scholar]
- 30.Richardson A, Parsons J T. Nature (London) 1996;380:538–540. doi: 10.1038/380538a0. [DOI] [PubMed] [Google Scholar]
- 31.Ilic D, Damsky C H, Yamamoto T. J Cell Sci. 1997;110:401–407. doi: 10.1242/jcs.110.4.401. [DOI] [PubMed] [Google Scholar]
- 32.Schaller M D, Otey C A, Hildebrand J D, Parsons J T. J Cell Biol. 1995;130:1181–1187. doi: 10.1083/jcb.130.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knezevic I, Leisner T M, Lam S C T. J Biol Chem. 1996;271:16416–16421. doi: 10.1074/jbc.271.27.16416. [DOI] [PubMed] [Google Scholar]
- 34.Ozaki K, Tanaka K, Imamura H, Hihara T, Kameyama T, Nonaka H, Hirano H, Matsuura Y, Takai Y. EMBO J. 1996;15:2196–2207. [PMC free article] [PubMed] [Google Scholar]
- 35.Ponting C P, Bork P. Trends Biochem Sci. 1996;21:245–246. [PubMed] [Google Scholar]
- 36.Madaule P, Furuyashiki T, Reid T, Ishizaki T, Watanabe G, Morii N, Narumiya S. FEBS Lett. 1995;377:243–248. doi: 10.1016/0014-5793(95)01351-2. [DOI] [PubMed] [Google Scholar]
- 37.Boguski M S, McCormick F. Nature (London) 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 38.Buchsbaum R, Telliez J-B, Goonesekera S, Feig L A. Mol Cell Biol. 1996;16:4888–4896. doi: 10.1128/mcb.16.9.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofmann K, Bucher P. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 40.Vadlamudi R K, Joung I, Strominger J L, Shin J. J Biol Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 41.Hofmann K, Tschopp J. FEBS Lett. 1995;371:321–323. doi: 10.1016/0014-5793(95)00931-x. [DOI] [PubMed] [Google Scholar]
- 42.Feinstein E, Kimchi A, Wallach D, Boldin M, Varfolomeev E. Trends Biochem Sci. 1995;20:342–344. doi: 10.1016/s0968-0004(00)89070-2. [DOI] [PubMed] [Google Scholar]
- 43.Leonardo E D, Hinck L, Masu M, Keino-Masu K, Ackerman S L, Tessier-Lavigne M. Nature (London) 1997;386:833–838. doi: 10.1038/386833a0. [DOI] [PubMed] [Google Scholar]
- 44.Ackerman S L, Kozak L P, Przyborski S A, Rund L A, Boyer B B, Knowles B B. Nature (London) 1997;386:838–842. doi: 10.1038/386838a0. [DOI] [PubMed] [Google Scholar]
- 45.Otsuka A J, Franco R, Yang B, Shim K H, Tang L Z, Zhang Y Y, Boontrakulpoontawee P, Jeyaprakash A, Hedgecock E, Wheaton V I, et al. J Cell Biol. 1995;129:1081–1092. doi: 10.1083/jcb.129.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.The International FMF Consortium. Cell. 1997;90:797–807. doi: 10.1016/s0092-8674(00)80539-5. [DOI] [PubMed] [Google Scholar]
- 47.The French FMF Consortium. Nat Genet. 1997;17:25–31. doi: 10.1038/ng0997-25. [DOI] [PubMed] [Google Scholar]
- 48.Ponting C P, Schultz J, Bork P. Trends Biochem Sci. 1997;22:193–194. doi: 10.1016/s0968-0004(97)01049-9. [DOI] [PubMed] [Google Scholar]
- 49.Quaderi N A, Schweiger S, Gaudenz K, Franco B, Rugarli E I, Berger W, Feldman G J, Volta M, Andolfi G, Gilgenkrantz S, et al. Nat Genet. 1997;17:285–291. doi: 10.1038/ng1197-285. [DOI] [PubMed] [Google Scholar]
- 50.Loomis C R, Walsh J P, Bell R M. J Biol Chem. 1985;260:4091–4097. [PubMed] [Google Scholar]
- 51.Gale C, Finkel D, Tao N, Meinke M, McClellan M, Olson J, Kendrick K, Hostetter M. Proc Natl Acad Sci USA. 1996;93:357–361. doi: 10.1073/pnas.93.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fitch W M. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 53.Bork P, Margolis B. Cell. 1995;80:693–694. doi: 10.1016/0092-8674(95)90347-x. [DOI] [PubMed] [Google Scholar]
- 54.Rost B, Sander C, Schneider R. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]