Abstract
S100B is a member of the S100 subfamily of EF-hand proteins that has been implicated in malignant melanoma and neurodegenerative conditions such as Alzheimer's and Parkinson's disease. Calcium-induced conformational changes expose a hydrophobic binding cleft, facilitating interactions with a wide variety of nuclear, cytoplasmic, and extracellular target proteins. Previously, peptides derived from CapZ, p53, NDR, HDM2 and HDM4 have been shown to interact with S100B in a calcium-dependent manner. However, the thermodynamic and kinetic basis of these interactions remains largely unknown. To gain further insight, these peptides were screened against the S100B protein using isothermal titration calorimetry and nuclear magnetic resonance. All peptides were found to have binding affinities in the low micromolar to nanomolar range. Binding-induced changes in the line shapes of S100B backbone 1H and 15N were monitored to obtain the dissociation constants and the kinetic binding parameters. The large microscopic Kon rate constants observed in this study, Kon ≥1×107 M-1s-1, suggest that S100B utilizes a “fly casting mechanism” in the recognition of these peptide targets.
The S100 protein family consists of small (9-13 kDa), dimeric, calcium-binding proteins that are unique to vertebrates (1). These proteins play a role in a variety of cell functions, including phosphorylation (2, 3), enzyme regulation (4, 5), and cytoskeleton dynamics (6), and have also been implicated in a variety of cancers (7-10), inflammatory conditions (11-13), and neurological diseases (14-17). Unlike calmodulin, a ubiquitous calcium-binding protein, expression patterns of S100 protein family members appear to be cell specific, with individual expression levels depending on local environmental factors (18). Each monomer consists of four alpha helices and contains two calcium-binding domains: a high affinity, canonical EF-hand in the C-terminus and a low affinity ‘psuedo’ EF-hand in the N-terminus (19). The disparity in calcium-binding affinity between the two sites is much greater than that seen in the different isoforms of parvalbumins, and originates in local sequence differences rather than long range contacts (20-22). Cation binding induces a large conformational change, during which helix 3 reorients itself ~90° relative to helix 4 (23-29). This exposes a large patch of hydrophobic surface area that is generally required for protein binding and various S100 functions (30).
S100B is the best studied member of the S100 family and was the first to be discovered (31). To date, over 29 structures of the protein have been deposited in the Protein Data Bank (PDB) (32). These include both crystallographic and solution NMR studies that span all functionally relevant states (apo, cation bound, and peptide bound) and orthologs from three species (Homo sapiens, Rattus norveggicus, Bos taurus). S100B is known to interact with over 20 targets, including nuclear, cytoplasmic, and extracellular ligands (30). It has established roles in Parkinson's, Alzheimer's, Down Syndrome, and it is currently the best prognostic marker for malignant melanoma (15-17, 33). Previously, it has been shown to interact with peptides derived from CapZ (34, 35), p53 (36-38), NDR (5), MDM2/HDM2 (39, 40) and MDM4/HDM4 (39). Despite the large amount of data concerning the identification of the binding targets, there remains ambiguity about the structural, thermodynamic, and kinetic basis of these interactions.
The TRTK12 peptide is derived from C-terminal region the actin-capping protein CapZ (residues 265-276), but was originally identified as a S100B binding motif using a random phage display library. The consensus sequence (K/R)(L/I)XWXXIL was shown to compete with known S100B binding partners, including CapZ and glial fibrillary acidic protein, in a calcium-dependent manner (34). Previous studies had identified helix 3, the hinge region, and helix 4 as the most likely regions for target recognition, due to their structural rearrangement during calcium binding and their sequence divergence amongst the S100 proteins (5, 36, 38, 39, 41-45). Mapping of the interaction surface by NMR identified residues involved in binding the TRTK12(265-276) peptide and provided an important step in confirming the target binding site for S100B. TRTK12(265-276) also helped establish the tendency for binding-competent peptides to have Arg or Lys residue(s) in addition to hydrophobic residues (46).
The p53 protein is a transcription activator that up regulates genes critical to cell cycle arrest and apoptosis (47). If mutated or down regulated, cell proliferation can proceed unchecked and otherwise healthy cells proliferate and become cancerous. Conversely, if up regulated, premature aging and cell damage can occur (48). The p53 protein consists of an N-terminal transactivation domain, a DNA-binding domain, a tertamerization domain, and a C-terminal regulation domain. S100B has been shown to bind to individual subdomains with varying affinities in a complex manner, depending on the oligomerization state of p53 and its post-translational modifications (37, 49, 50). S100B preferentially binds to the tetramerization domain of p53 during basal expression, preventing the more active tetrameric form of p53 from entering the nucleus. Binding to the extreme C-terminus prevents protein kinase C-dependent phosphorylation of Ser376 and Ser378 (49). Interestingly, this region is unstructured both as a functional domain in the p53 protein and as a free peptide in solution (51-53). Upon binding S100B, this sequence adopts a partially helical structure (29, 54-56).
Nuclear Dbf2-related kinases (NDR's) are a subgroup of the serine/threonine AGC kinases and are highly conserved amongst eukaryotes (57). NDR activity is critical to mitotic progression, cytokinesis, morphological changes, cell proliferation, and apoptosis (57). S100B binds to a basic/hydrophobic sequence at the junction of the N-terminal regulatory and catalytic domains (5). Mutagenesis studies have shown that Thr74 plays a crucial role in binding and it is likely that S100B binding triggers autophosphorylation of Ser281, activating the kinase (58). Similar to the p53 protein, the S100B binding site of NDR includes an unstructured region that adopts a three-turn helical structure upon complex formation(5).
Human Double Minute 2 (HDM2) and Human Double Minute 4 (HDM4) proteins are both critical regulators of p53 (59). The former acts as a tumor suppressor by binding to p53 and inhibiting N-terminal activation, as well as targeting the protein for ubiquitin-dependent proteosomal degradation (60). The latter acts in a more complex fashion, either inhibiting p53's apoptotic activity or stabilizing the protein and promoting the mitochondrial apoptotic response (61). In addition, HDM4 has been shown to form heterodimers with HDM2, which adds an additional layer of regulation to the activity and effective concentration of the HDM2 protein (62). S100B has been shown to interact with peptides derived from the N-terminal domains of both proteins, as well as the full length HDM2 protein (39, 40). It is still unclear whether interactions with these binding sites are physiologically relevant. The HDM2(25-47) peptide, consisting of residues 25-47 of the full length protein, is derived from a region that is buried in the three-dimensional structure (39). In contrast, the HDM4 protein appears to be intrinsically more flexible, adopting several conformations in the unbound state (63). The HDM2(25-47) and HDM4(25-47) peptides, both consisting of residues 25-47, were identified based on their sequence similarity to TRTK12 (39). Competition experiments using a phage display have demonstrated that the binding site for the full-length HDM2(25-47) protein at least partially overlaps with the binding site for TRTK12(265-276) (5, 39, 40). In addition, no structure exists of S100B bound to either target. Interestingly, the available data suggests these peptides may interact with S100B through a unique and asymmetric mechanism (40).
Here, we performed a full thermodynamic and kinetic characterization of the TRTK12(265-276), p53(367-388); NDR(62-87), HDM2(25-47); and HDM4(25-47) peptides binding to the S100B protein. All peptides were found to have dissociation constants in the low micromolar to nanomolar range, as determined by isothermal titration calorimetry (ITC) and nuclear magnetic resonance (NMR). In addition, experimentally measured changes in the heat capacity upon binding were found to be in excellent agreement with predicted values in structure-based calculations that take into account changes in solvation upon binding. Binding induced changes in the chemical shift of S100B backbone 1H and 15N were monitored to confirm the binding site for these peptides and to obtain the kinetic parameters of binding. The relatively fast microscopic on rate constants observed in this study suggest that S100B utilizes a “fly casting mechanism” or “folding after binding” in the recognition of these peptide targets.
EXPERIMENTAL PROCEDURES
Protein and Peptide Purification
Unlabeled and double-labeled 15N/13C human S100B proteins were overexpressed in Escherichia coli and purified as previously described (64-66). The protein concentration was determined using the molar extinction coefficient at 280 nm (ε280nm) of 1,490 M–1 cm–1. All peptides, with their N-termini acetylated and their C-termini amidated, were synthesized using standard Fmoc chemistry at the Penn State College of Medicine Macromolecular Core Facility. The TRTK12(265-276) peptide derived from the actin-binding protein CapZ (Ac-TRTKIDWNKILS-Am), the nuclear Dbf2-related kinase (NDR(62-87); amino acid sequence: Ac-KRLRRSAHARKETEFLRLKRTRLGLEY-Am), and the C-terminal p53 peptide (p53(367-388); amino acid sequence: Ac-YSHLKSKKGQSTSRHKKLMFKTE-Am) have all previously been used in structural studies (5, 35, 36, 67). The human homologue of marine double minute 2 peptide (HDM2(25-47); amino acid sequence: Ac-ETLVRPKPLLLKLLKSVGAY-Am) and the human homologue of marine double minute 4 peptide (HDM4(25-47); amino acid sequence: Ac-NQVRPKLPLLKILHAAGAQY-Am) are the same sequences used by Weber et al (36, 39). With the exception of TRTK12(265-276), all peptides have an additional Tyr residue on their N-termini or C-termini, to allow for quantification by UV absorbance spectroscopy. For the NDR(62-87) and p53(367-388) peptides, this residue is known to exist outside of the binding site in an unstructured region of the peptide and is expected to have little to no impact on binding (5, 29). Similarly, the sequence alignment data for the HDM2(25-47) and HDM4(25-47) peptides suggest that the extreme C-termini exists outside of the S100B binding motif (39). The peptides were purified as previously described (65), and their masses were confirmed with mass spectrometry. The concentration of peptides was determined using the molar extinction coefficients at 280 nm (ε280nm) of 5,500 M–1 cm–1 for TRTK12(265-276) and 1,490 M–1 cm–1 for p53(367-388), NDR(62-87), HDM2(25-47), and HDM4(25-47), respectively (68).
Isothermal Titration Calorimetry (ITC)
Isothermal titration calorimetry measurements were performed using a VP-ITC instrument (MicroCal, Inc., Northhampton, MA) as previously described (65, 69, 70). Prior to all experiments, protein and peptide were dialyzed with two buffer changes of 20 mM Tris base, 0.2 mM disodium EDTA, 1 mM TCEP, pH 7.5 and 5 mM calcium chloride at room temperature. For the p53(367-388) peptide, the “high salt” buffer included an additional 120mM NaCl. In general, 0.4-1.6 mM peptide was injected in 2-7 μL increments into the sample cell (1.5 mL) containing 20-50 μM protein. For the titration of Ca2+-S100B and HDM4(25-47), the protein solution was injected into the cell containing the peptide solution. Experiments were performed in the temperature range from 5°C to 35°C, in 5°C increments, except in cases where the heat effects were close to zero and thus there was insufficient signal. The heat of dilution was measured prior to each experiment by performing four peptide injections into buffer, using the experimental concentrations and volumes. It was found to be negligible for all peptides over the temperature and volume ranges reported. The resulting titration curves were analyzed using the Origin for ITC software supplied by MicroCal (Northampton, MA).
It is important to note that ITC data analysis is model dependent. In the absence of additional information, analysis is performed by examining models in the order of increasing complexity. In accordance with Occam's razor, the simplest model that fits all the available data is assumed to be correct. The following binding models were considered:
Single binding site model - one peptide per S100B dimer (71):
| 1 |
where Q is the integral heat at each injection, , n = 1 is the stoichiometry of the protein-peptide complex, [syringe]t is the total concentration of the peptide/protein solution in the ITC syringe, [cell]t is the total concentration of the protein/peptide solution in the ITC cell, and Ka is the association constant.
Two-identical binding sites model - two peptides per S100B dimer (71):
| 2 |
where Q is the integral heat at each injection, , and n = 2 is the stoichiometry of the protein-peptide complex.
Two-sequential binding sites model - two peptides per S100B dimer (71):
| 3 |
where Q is the integral heat at each injection, K1 and K2 are the association constants for binding to sites 1 and 2, respectively, and ΔHcal1 and ΔHcal2 are the calorimetric enthalpies for binding to sites 1 and 2, respectively.
Far-UV Circular Dichroism
The far-UV CD spectra (260-195 nm) of the TRTK12(265-276), p53(367-388), NDR(62-87), HDM2(25-47), and HDM4(25-47) peptides were measured on a Jasco-715 spectropolarimeter in a 1 mm light path length cuvette. Measurements were performed using 50 μM peptide in the presence of 20 mM Tris, 1 mM TCEP, 0.2 mM EDTA, pH 7.5, with or without the presence of 5 mM calcium and with or without the presence of 30% TFE. Ellipticity values (Θ) for the peptides were corrected by subtracting the corresponding values for the buffer and converting to mean residue ellipticity, [Θ] using the following equation:
| 4 |
where MR is the mean molecular mass of the amino acids in each peptide, l is the optical path length in centimeters and c is the peptide concentration in mg/ml. The fraction helicity, fH, was calculated as:
| 5 |
where [Θ]222 is the experimentally determined ellipticity at 222 nm, [Θ]C is the ellipticity of the fully coiled state, and [Θ]H is the ellipticity of the fully helical state.
The ellipticity of the fully coiled state [Θ]C has a temperature dependence described as (72):
| 6 |
where T is the temperature in degrees Celsius. The ellipticity of the fully helical state, [Θ]H, for a protein or peptide consisting of Nr residues, has a temperature dependence described by (72):
| 7 |
where Nr is the number of residues.
Structure based calculation of ΔCp
Structure-based calculations allow for cross-validation of thermodynamic parameters obtained from protein-ligand binding (65, 73). Most often, these calculations of the heat change upon binding, ΔCp, are based on changes in the accessible surface area upon binding, ΔASAtot, of the ligand to the protein. Using the available three-dimensional structures of S100B in complex with the studied peptides, ΔASAtot was modeled as previously described (74). In cases where no structure was available, homology models of the peptide-bound state were generated using Modeler (75) and PDB structure 1DT7 (29). Changes in the accessible surface area upon binding were calculated as follows:
| 8 |
where ΔASAtot is the total change in accessible surface upon peptide binding, ASAdim+pep is the ASA of the protein in complex with the peptide, ASAdim – pep is the ASA of protein in the absence of peptide, and ASAunf pep is the ASA of unfolded peptide. As described previously (65, 73, 76), changes in ΔASA were subdivided into 4 categories: aliphatic surface area, aromatic surface area, peptide backbone surface area, and polar surface area and converted into ΔCp using the following empirical relationship:
| 9 |
where ΔASAalp are the changes in ASA for aliphatic amino acids, ΔASAarm are the changes in ASA for aromatic amino acids, ΔASApol are the changes in ASA for polar amino acids, and ΔASAbb are the changes in ASA for the polypeptide backbone.
NMR Spectroscopy
NMR spectra were collected at 35°C on a Bruker AVANCE II 600MHz spectrometer equipped with a triple-resonance cryoprobe with z-axis gradients and processed using the Bruker software suite TopSpin (version 2.1). Backbone resonance assignments were obtained for S100B in the peptide-free state using the auto assignment program MONTE with the connectivities detected in the spectra of the HNCO, HN(CA)CO, HNCACB, and HN(CO)CACB triple-resonance experiments as input and validated against previously published assignments (77). Reassignment of the amide groups in the peptide-bound state were performed by monitoring changes in chemical shifts in a series of 2D 1H-15N HSQC spectra acquired throughout the peptide titrations and were verified against published assignments when applicable (5, 24, 27, 29, 78). Prior to all experiments, the protein and peptide were simultaneously dialyzed into buffer containing 5 mM Tris, 15 mM NaCl, 0.34 mM NaN3, pH 7.5. After dialysis, 5% D2O and 10 mM CaCl2 were added to the samples. Chemical shift mapping experiments were performed by titrating 0-4 mM peptide into 0.1-0.45 mM calcium- bound S100B and monitoring changes in backbone 15N and 1H. All titrations were continued until there were no longer observable changes in either the 15N or 1H chemical shifts. The combined changes in the proton and nitrogen chemical shifts were quantified according to the following weighted distance metric (79, 80):
| 10 |
where δN and δH are the changes in chemical shift (in ppm) in the nitrogen and proton dimensions, respectively. Spectra were analyzed using the program Sparky (81).
LineShapeKin Analysis
Line shape analysis was performed using the BiophysicsLab Matlab package (82). The 1D slices of the 15N and 1H dimensions from 2D HSQCs were extracted in Sparky (81) and the intensities were normalized by the area under each peak to account for exchange broadening in the other dimension (82). For the TRTK12(265-276) and p53(367-388) peptides, titration data was fit to a model where there are two identical binding sites per S100B dimer, or one peptide interacting with each S100B monomer. The sequential binding model cannot be explicitly modeled using LineShapeKin, so the titration data for the NDR(62-87) peptide was fit to a model where there are two identical binding sites per S100B dimer and compared to the simulated data using a sequential binding model (83) to estimate the upper and lower limits on the kinetic rate constants. For the HDM2(25-47) and HDM4(25-47) peptides, binding was best fit to a single binding site per S100B dimer. The software analytically solves for both the apparent binding constant, KdApp, and off-rate, Koff, simultaneously, which allows the on-rate, Kon, to be readily calculated. The x2 values were examined in a 2D matrix as a function of binding affinity and Koff rates to ensure the data was fitting to a true global minimum (see supplementary materials). In order for data to qualify for line shape analysis, the amide peak must have been determined to have undergone a significant chemical shift change, not overlap with any other amide peak, and be sufficiently resolved in all spectra of the titration series. For each peptide, six to ten residues were found to meet these criteria. As part of the analysis, all residue specific data were assumed to be reporting on the same binding event. As a result, fitted parameters including the rate constants for binding kinetics could be calculated with greater certainty in global fits using all data. To validate this assumption, individual residues were independently fit and these results were compared to each other, as well as the global analysis, and found to be in good agreement (see supplementary materials). All reported values are the result of global fits, which include both 15N and 1H dimensions of all relevant amide groups across all ligand concentrations.
RESULTS
Thermodynamics of Peptide Binding
ITC is a powerful, quantitative method for measuring the thermodynamics of protein-ligand interactions. It is unique in its ability to simultaneously determine the enthalpy (ΔHcal), affinity (Kd) and stoichiometry (n) of an interaction from a single experiment at a given temperature (70, 84). In addition, if experiments are performed over a range of temperatures, one can derive the heat capacity change upon binding (ΔCp) from the temperature dependence of the enthalpy of binding. An important caveat is that all data analysis is model-dependent and the simplest model that accurately fits the data is assumed to be correct.
Below we present the results of analysis of five different peptides binding to calcium-saturated S100B, i.e., Ca2+-S100B. For readability, it will henceforth be referred to simply as ‘S100B’.
TRTK12
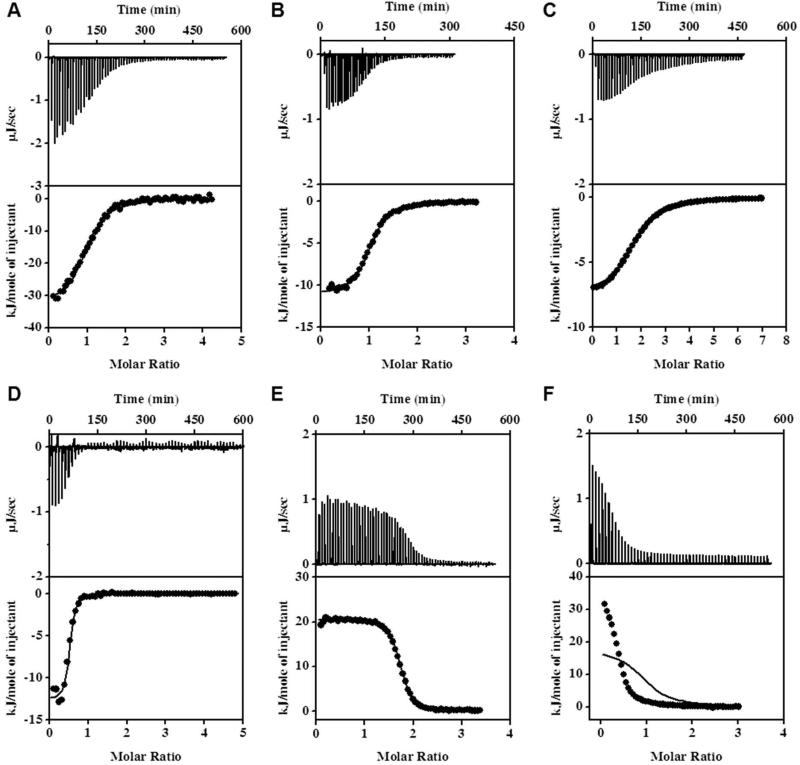
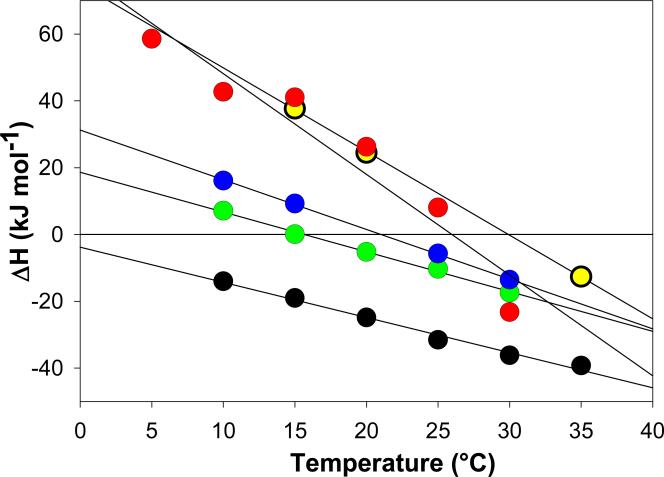
Figure 1A shows representative ITC data from experiments in which the TRTK12(265-276) peptide was titrated into S100B. The titration data are best fit to the simplest model, where there are two identical binding sites per S100B dimer (Equation 2). This stoichiometry is consistent with previous structural studies that show each S100B monomer has an identical binding site (29, 37, 46, 54, 85). The binding affinity per site is determined to be 2 ± 1 μM at 25°C, which is in good agreement with fluorescence- and the weaker values from the conflicting reports from NMR-based assays (54, 86). Figure 2 shows the dependence of the enthalpy of binding, ΔHcal, on temperature. This dependence is a linear function of temperature and the slope of the dependence is the heat capacity change upon binding, ΔCp. It has previously been shown for a variety of protein-protein and protein-peptide interactions that ΔCp can be accurately predicted using structure-based calculations (73, 74). The heat capacity change for the S100B TRTK12(265-276) interaction is not an exception, as the experimentally obtained ΔCp, -1.3 ± 0.2 1 kJ mol-1 K-1, is in excellent agreement with the calculated value based on the bound complex (PDB: 1MQ1) , -1.2 ± 0.1 1 kJ mol-1 K-1 (Equation 9).
Figure 1.
Examples of ITC experiments showing the binding of p53, TRTK12, NDR, HDM2, and HDM4 peptides to S100B in the presence of calcium. Plots represent the raw heat effects (μJ/sec) as a function of time, the cumulative heat effects (kJ/mol, represented by closed circles) as a function of the molar ratio of peptide to protein, and the fits to the experimental data (solid lines) for: p53 binding to S100B at 25 °C (Panel A; data fitted to equation 2), TRTK12 binding to S100B at 25 °C (Panel B; data fitted to equation 2), NDR binding to S100B at 30 °C (Panel C; data fitted to equation 3), HDM2 binding to S100B at 35 °C (Panel D; data fitted to equation 2), HDM4 binding to S100B at 10 °C (Panel E; data fitted to equation 2), HDM2 binding to S100B at 15 °C (Panel E; data fitted to equation 1).
Figure 2.
Temperature dependence of the enthalpies, ΔHcal, of the TRTK12 (green circles), p53 (black circles), NDR (blue circles), HDM2 (yellow circles), and HDM4 (red circles) peptides binding to the S100B protein in the presence of 5 mM calcium. Solid lines represent linear fits of the ΔHcal temperature dependencies for each S100-peptide interaction. The slopes of these lines represent the experimental change in heat capacity, which are summarized in Table 1.
p53
Figure 1B shows the typical ITC titration profile for the p53(376-388) peptide binding to the S100B protein. The titration data fits well to the simplest model, where there are two identical and independent binding sites per S100B dimer. The dissociation constant, Kd, for each site in the presence of low salt was determined to be 2 ± 1 μM, which is in agreement with kinetic phosphorylation assays (3). The dissociation constant in the presence of high salt is an order of magnitude weaker, 20 ± 5 μM. Increasing ioninc strength has been shown to significantly reduce the affinity of the p53 peptide for S100B (39, 49), although the absolute affinity reported here is somewhat tighter than previously reported values measured using competitive binding assays (39) and ultracentrifugation experiments. Figure 2 shows the linear dependence of the enthalpy of binding, ΔHcal, on temperature for the low salt conditions, although it should be noted that the experimentally determined ΔCp values did not change significantly in the presence or absence of salt, -1.2 ± 0.1 kJ mol-1 K-1 and -1.0 ± 0.1 kJ mol-1 K-1, respectively. The calculated ΔCp, -1.0 ± 0.1 kJ mol-1 K-1 (PDB: 1DT7), is well within error of these values (Table 1).
Table 1.
Summary of the thermodynamic properties of S100B binding to p53, TRTK12, NDR, HDM2 and HDM4 and determined using ITC experiments as well as the comparison with structure-based calculations.
| Peptide | Kd μM | ΔCp, exp kJ·mol-1 K-1 | ΔCp, calc kJ·mol-1 K-1 |
|---|---|---|---|
| TRTK12 | 2 ± 1 | -1.3 ± 0.2 | -1.1 ± 0.1 |
| p53, low salt | 2 ± 1 | -1.0 ± 0.1 | -1.0 ± 0.1 |
| p53, high salt | 20 ± 5 | -1.2 ± 0.3 | -1.0 ± 0.1 |
| NDR | 2 ± 1, 0.4 ± 0.2a | -1.5 ± 0.3 | -1.9 ± 0.2 |
| HDM2 | 0.5 ± 0.2 | -1.3 ± 0.2 | -1.0 ± 0.1b |
| HDM4 | 0.07 ± 0.03 | -1.7 ± 0.2 | -0.7 ± 0.1b |
The interaction between S100B and NDR was fit to a sequential binding model, where each S100B monomer had a unique binding affinity.
These values were calculated using homology models of the S100B dimer (PDB id: 1DT7) binding to two peptides.
NDR
Figure 1C shows results from representative ITC experiments in which the NDR(62-88) peptide was titrated into Ca2+-S100B. The titration data could not be accurately fit to the simplest binding model, where there are two identical binding sites per S100B dimer (Equation 2). The data appears to fit well to the more complex sequential binding site model, where there are two non-identical, interacting sites per S100B dimer (Equation 3). This model is consistent with the previously reported stoichiometry (5), i.e., two peptides per S100B dimer. However, it delineates a significantly different mechanism of binding for the NDR peptide as compared to the TRTK12 or p53 peptides. This model suggests that binding of the first NDR peptide either transduces a structural change across the S100B dimer that changes the affinity for the second NDR peptide. The higher affinity site has a Kd1 of 0.4 ± 0.2 μM, while the lower affinity site has a Kd2 of 2 ± 1 μM. The sequential binding model has been observed before for other S100 proteins interacting with target peptides. The negative cooperativity (Kd1 < Kd2) observed here is similar to that observed for the MDM2 N-terminal domain binding to S100B (40), but in contrast to the positive cooperatively (Kd1 > Kd2) observed for the binding of Annexin II to S100A10 (65). Consistent with the sequential binding model, the enthalpies of binding for each site are also different. The temperature dependence of the sum of the individual enthalpies of binding is listed in Table 1. The change in heat capacity for the S100B-NDR interaction is shown in Figure 2. It is important to note that the structure-based calculation for ΔCp can only be calculated for the final bound complex (two peptides per S100B dimer) and not any intermediate states. It is this value that is then compared with the temperature dependence of the total enthalpy of binding, or the sum of the individual enthalpies of binding. In this case, the calculated and experimentally determined ΔCp agree within the error, -1.5 ± 0.3 kJ mol-1 K-1 (PDB: 1PSB) and - 1.9 ± 0.2 kJ mol-K-1, providing internal consistency for the data analysis.
HDM2 and HDM4
Figures 1D and 1E show typical ITC profiles for the HDM2 and HDM4 peptides binding to S100B. The simplest model, where there are two identical binding sites per S100B dimer, was insufficient to fit the binding isotherms (Figure 1E). Subsequently, binding data for the HDM2(25-47) and HDM4(25-47) peptides were found to fit well to a single site binding model (Equation 1), where an individual peptide binds to the S100B dimer. The presence of only one HDM2 and HDM4 peptide binding site per S100B dimer is a distinct stoichiometry compared that of the TRTK12, p53, and NDR peptides. However, the stoichiometry of one peptide per S100B dimer has been previously observed (49, 87). Peptides derived from the tetramerization domains of p53 (distinct sequences from the peptide studied here) and p63 have been reported to bind to S100B with the same ‘one peptide per dimer’ stoichiometry (49, 87). The binding affinity for the HDM2(25-47) peptide was determined here by ITC to be 0.5 ± 0.1 μM, while the affinity for the HDM4(25-47) peptide was almost an order of magnitude tighter, 0.07 ± 0.02 μM. An order of magnitude difference in affinities between the two peptides has been previously reported (39). Furthermore, it has been reported that the affinity for HDM2(25-47) peptide is approximately an order of magnitude tighter than those of the TRTK12(265-276) and p53(367-388) peptides, in agreement with our observations (Table 1). Figure 2 shows the change in heat capacity for both the S100B-HDM2 and S100B-HDM4 interactions, which are significantly larger than the other peptides examined in this study. In addition, the experimentally determined ΔCp for the HDM4(25-47) peptide, -1.7 ± 0.2 kJ mol-1 K-1, is significantly different from that obtained using homology models of the bound complex, -0.7 ± 0.1 kJ mol-1 K-1 (see Methods), where the stoichiometry was modeled as one peptide per S100B dimer (Table 1).
Chemical Shift Perturbations due to Peptide Binding
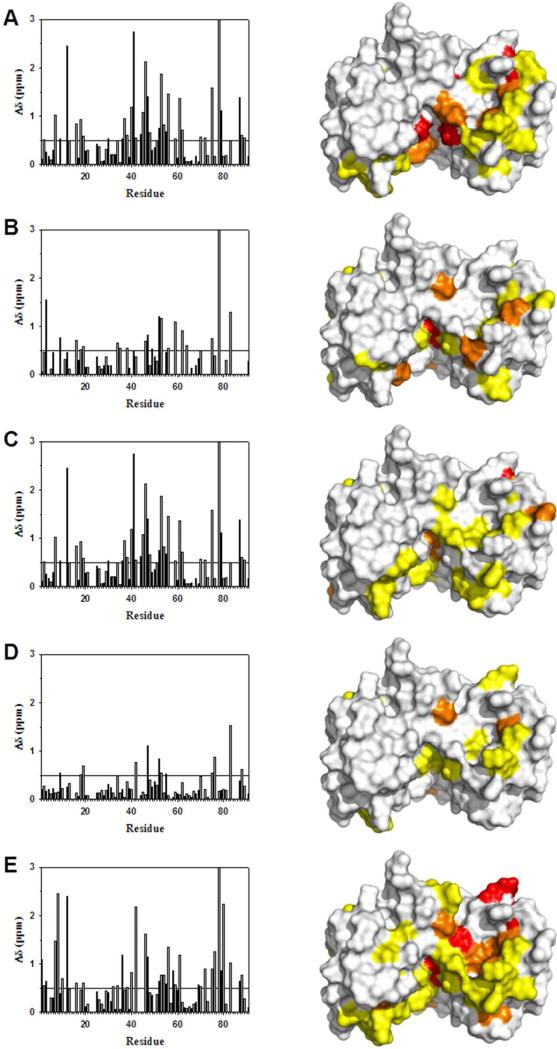
Interactions between S100B and various peptide targets were examined by monitoring binding-induced changes in the chemical shifts of backbone 15N and 1H atoms between the peptide-free state and the peptide-bound state (Figure 3). While resonances undergoing binding-induced chemical shift perturbations are generally limited to those atoms within the interaction surface, it should be noted that chemical shift changes can be detected in residues neighboring those that make direct contacts or in cases where structural changes involving remote sites is coupled with binding events. However, the results suggest that binding-induced chemical shift perturbations are localized to discreet interaction surfaces and that peptide binding does not induce an allosteric effect. The protein concentrations used in this study preclude a full assignment of S100B in the peptide free and peptide bound states. However, the majority of the amino acid residues in S100B that underwent significant chemical shift perturbations were observable and structurally cluster to a region that is in good agreement with the residues reported to be directly involved with TRTK12(265-276), p53(367-388), and NDR(62-87) peptide binding in previous structural studies (see Figure 3) (5, 39, 54). Specifically, residues from loop 2 (S41, H42, F43, L44, E45), helix 3 (V52, V53, V56, T59), and the C-terminus of helix 4 (S78–F88)were found to interact with the p53(367-388), TRTK12(265-276), and NDR(62-87) peptides. There are no structures available for S100B in complex with either HDM2(25-47) or HDM4(25-47). However, we find that the residues involved in binding the HDM2(25-47) and HDM4(25-47) peptides are very similar to those involved in binding the above targets, suggesting that all binding sites at least partially overlap (Figure 3). This is consistent with previous competition assays which show that the S100B-HDM2 interaction can be disrupted by both TRTK12(265-276) and wild type p53 (39). In addition, significant chemical shifts are observed for residues at the dimer interface, including S78, M79, V80, T81, T82, A83, for the HDM2(25-47) and HDM4(25-47) peptide interactions.
Figure 3.
Changes in backbone 1H and 15N chemical shifts for Ca2+-bound S100B upon binding various peptide targets. Similar residues in S100B undergo chemical shift perturbations upon binding TRTK12(265-276) (Panel A), p53(367–388) (Panel B), NDR(62-87) (Panel C), HDM2(25-47) (Panel D), or HDM4(25-47) (Panel E) peptides. The bar graphs show the combined backbone 1H and 15N chemical shift perturbations upon peptide binding to Ca2+–S100B. Residues without Δδ values underwent exchange broadening upon binding, and/or could not be assigned in both the peptide-free and peptide bound states.
Line Shape Analysis
The chemical-shift changes for S100B 1H and 15N resonances were quantified using a series of 2D HSQC experiments with progressive increases in the peptide concentration until there were no longer observable changes in chemical shifts. These experimental data were further analyzed here in order to characterize the thermodynamics and kinetics of the interaction (88, 89). More specifically, line shape analysis was performed on one-dimensional line shapes extracted from the series of 2D HSQC spectra and used to obtain fits for the affinity and kinetic rate constants of binding (88). This method is most accurate when exchange between the bound and free states is not in the extreme ‘fast’ or ‘slow’ exchange regimes, where Kex is approximately two orders of magnitude greater than or lower than < Δω, respectively. The apparent affinities and kinetic constants from the global fit analysis are listed in Tables 2-4.
Table 2.
Summary of the kinetic properties of S100B binding to p53, TRTK12, NDR, HDM2 and HDM4 and determined using NMR and ITC experiments and LineShapeKin analysis.
| Peptide | Kd, App (μM)a | Koff (s-1) | Kon (1×107 M-1 s-1) |
|---|---|---|---|
| TRTK12 | 2 ± 0.5 | 110 ± 20 | 5 ± 2 |
| p53, low salt | 2 ± 0.5 | 760 ± 150 | 40 ± 16 |
| p53, high salt | 30 ± 6 | 900 ± 180 | 3 ± 1 |
| NDR | 0.4 ± 0.2, 2 ± 1b | 50-250, 20-200 b | 13-63, 1-10b |
| HDM2 | 0.5 ± 0.2c | 140 ± 30 | 30 ± 11 |
| HDM4 | 0.07 ± 0.03c | 30 ± 10 | 4 ± 2 |
Peptide binding was fit to a single site model for all peptides during the LineShapeKin analysis. All reported values are for the apparent Kd.
Kinetic rates could not be solved for analytically. See Materials and Methods.
Binding model used was one peptide per S100B dimer. Kd value was taken from ITC and Koff was floated.
Table 4.
Summary of the kinetic properties of S100B binding to p53, TRTK12, NDR, HDM2 and HDM4 and determined using NMR experiments and LineShapeKin analysis.
| Peptide | Kd, App (μM)a | Koff (s-1) | Kon (1×107 M-1 s-1) |
|---|---|---|---|
| TRTK12 | 1.0 ± 0.5 | 100 ± 20 | 10 ± 4 |
| p53, low salt | 1.0 ± 0.5 | 1000 ± 200 | 100 ± 40 |
| p53, high salt | 10 ± 2 | 1000 ± 200 | 10 ± 2 |
| NDR | n.a.b | n.a.b | n.a.b |
| HDM2 | 0.1 ± 0.02c | 100 ± 20 | 100 ± 40 |
| HDM4 | 1.0 ± 0.5c | 10 ± 2 | 1.0 ± 0.4 |
Peptide binding was fit to a single site model for all peptides during the LineShapeKin analysis. All reported values are for the apparent Kd.
Kd values and kinetic rates could not be solved for analytically. See Methods and Table 1.
Binding model used was one peptide per S100B dimer. Kd and Koff is taken directly from 2D matrix.
TRTK12
For the TRTK12(265-276) titration, we established that binding occurs according to the simplest model, where there are two identical sites per S100B dimer. This is consistent with the NMR results, where a single set of chemical shifts were observed for each amino acid residue, suggesting each monomer underwent an identical binding event. In addition, few resonances were observed to have significant line broadening, allowing the population averaged chemical shift values of the peptide-bound and peptide-free states to be tracked throughout the titration. Binding therefore occurs in the intermediate to fast exchange regime relative to the ‘NMR timescale’ (Koff ≥ Δω). The dissociation constant obtained directly from the line shape analysis is in excellent agreement with the Kd from ITC. The obtained kinetic parameters for binding are: Kon = 1×108 ± 4×107 M-1 s-1 and Koff = 100 ± 20 s-1 (Table 3). Similar rates are obtained regardless of whether the Kd is allowed to be fitted in the line shape analysis, or fixed to values obtained by ITC and indicate the robustness of our analysis (Table 2). Previously, Barber et al. examined the chemical shift changes of the Trp7 indole ring of the TRTK12(265-276) peptide throughout a titration with S100B and estimated a Kon of 1.4×108 M-1 s-1 and a Koff of 125 s–1 (86). These results are in excellent agreement with the kinetic rate constants observed in this study.
Table 3.
Summary of the kinetic properties of S100B binding to p53, TRTK12, NDR, HDM2 and HDM4 and determined using NMR experiments and LineShapeKin analysis.
| Peptide | Kd, App (μM)a | Koff (s-1) | Kon (1×107 M-1 s-1) |
|---|---|---|---|
| TRTK12 | 1.0 ± 0.5 | 100 ± 20 | 10 ± 4 |
| p53, low salt | 1.0 ± 0.5 | 800 ± 160 | 80 ± 30 |
| p53, high salt | 10 ± 2 | 750 ± 150 | 7.5 ± 1.5 |
| NDR | n.a.b | n.a.b | n.a.b |
| HDM2 | 0.1 ± 0.02c | 160 ± 30 | 160 ± 70 |
| HDM4 | 1.0 ± 0.5c | 30 ± 10 | 3 ± 1 |
Peptide binding was fit to a single site model for all peptides during the LineshapeKin analysis. All reported values are for the apparent Kd.
Kd values and kinetic rates could not be solved for analytically. See Methods and Table 1.
Binding model used was one peptide per S100B dimer. Kd is taken directly from 2D matrix. Koff is then floated with that Kd held constant.
p53
The chemical shift changes observed for p53(367-388) binding to S100B are similar to those of the TRTK12(265-276) peptide, i.e. fast on the NMR timescale and consistent with the two identical binding sites per dimer model, where one peptide binds each S100B monomer. The dissociation constant calculated from line shape analysis is similar to that of ITC and results in a Kon of 8×108 ± 3.2×107 M-1 s-1 and a Koff of 800 ± 160 s-1 (Table 3). The Kon for the p53(367-388) peptide is the greatest of any of the peptides examined in this study. Such fast Kon rate constants are not frequently observed for protein-peptide interactions but have been reported. Examples include peptides derived from nitric oxide synthase binding to calmodulin: 6.6×108 M–1 s–1 (90) and the phosphorylated kinase inducible domain (pKID) to the structured KIX domain: 6.3×106 M–1 s–1 (91). Interestingly, peptides derived from the N-terminus of p53(17-26) have been observed to bind to the MDM2 protein with a Kon of 2.2×107 M–1 s–1 (60), and p53(38-61) has been observed to bind to CBP with a Kon of 1.7×1010 M–1 s–1 (92). Specifically, the binding kinetics reported for the p53 peptides in the latter study approach the theoretical limit of protein-ligand binding, even when factoring in electrostatic interactions (93-95). Taken together, these studies indicate that fast binding kinetics may be a general property of p53-target protein interactions and that it may be important for the biological functions of p53.
NDR
A single amide peak was observed for equivalent residues of S100B dimer with chemical shift perturbations exhibiting fast exchange behavior in binding studies with the NDR(62-87) peptide. While the results are consistent with earlier observations (5, 29, 35), ITC analysis indicates that a sequential recognition model is employed, with two, non-identical binding sites per S100B dimer. As a result, distinct chemical shifts for amide resonances from each monomeric unit could be anticipated due to loss of symmetry within the dimer complex. However, the rates of exchange between free and bound states are detected here to be sufficiently rapid to result in the time-averaged chemical shifts that are being observed. Indeed, the affinities of each site differ by less than an order of magnitude according to ITC and the exchange between the free and fully bound state is fast on the NMR time-scale. Because there is no analytical solution available for the sequential binding model, one cannot explicitly calculate the kinetic rate constants using LineShapeKin (82). Instead, the titration data was simulated (83) using the dissociation constants obtained with ITC to estimate upper and lower bounds for the kinetic parameters. The Koff rate for the first peptide binding event is estimated to be between 50 s-1 and 250 s-1, while the Koff rate for the second peptide binding event is estimated to be between 10 s-1 and 200 s-1. This results in a Kon between 1×108 M-1 s-1 and 6×108 M-1 s-1 for the first peptide binding event, and a Kon between 1×107 M-1 s-1 and 1×108 M-1 s-1 for the second peptide binding event (Table 2). If the titration data is directly fit to the two identical binding sites model (one peptide per S100B monomer), and the dissociation constant is restrained by the values obtained by ITC, then the Koff rate is estimated to be 180 ± 40 s-1. This results in Kon values between 9×107 M-1 s-1 and 5×108 M-1 s-1, which are in good agreement with the simulated Kon rate constants.
HDM2 and HDM4
Binding of the HDM2(25-47) and HDM4(25-47) peptides occurs in the intermediate and slow time scales, respectively (Kex ≤ Δω). The stoichiometry obtained for both peptides by the ITC and NMR titrations is inconsistent with a two identical binding sites stoichiometry (one peptide per monomer), and fits best to a single binding site model (one peptide per S100B dimer). Furthermore, the concentrations used during the shape
NMR titrations were several orders of magnitude higher than the dissociation constants (Kd < 1μM), where the obtained stoichiometry is the most accurate. The simplest explanation consistent with all the data is that the HDM2(25-47) and HDM4(25-47) peptides interact with S100B through a novel binding mechanism, with the interaction surface of a single peptide possibly utilizing the prototypical binding site of both monomeric units and a portion of the dimer interface. The affinities obtained by ITC and LineShapeKin for the HDM2(25-47) peptide are in good agreement and result in apparent Kon and Koff rates of 2.8×108 ± 1.1×108 M-1 s-1and 140 ± 30 s-1, respectively. The HDM4(25-47) peptide is in a slow exchange regime with S100B, making accurate fitting of the dissociation constant using line shape analysis difficult, so the Kd value, 0.07 ± 0.03 μM, was taken directly from ITC. The obtained kinetic parameters: Kon = 4×107 ± 2×107 M-1 s-1 and Koff = 30 ± 10 s-1, are the slowest of any of the peptides examined here.
DISCUSSION
Thermodynamic Mechanisms of Binding
In this study, we present detailed thermodynamic analysis for five different peptides binding to calcium-saturated S100B. Even though all five peptides bind to a similar region on S100B, they interact with the protein through three unique binding mechanisms (Equations 1-3). It is well established that both the TRTK12 and p53 peptides interact with S100B according to the simplest model, where there are two identical binding sites per S100B dimer (29, 35-37, 39, 49, 78, 96). Here, we validate this model and present novel thermodynamic data for these interactions. There was significantly less information available for the S100B-NDR interaction. Our work suggests the NDR peptide interacts with S100B protein through a sequential binding model with negative cooperativity. Although a highly symmetric structure for the S100B-NDR complex has been reported, it involves the full NDR N-terminal domain in complex with bovine S100B, which differs from the sequence of human S100B at three residue positions (Val7Met, Ser62Asn, and Ile80Val) (5). Interestingly, these few changes may actually result in a significantly altered bound state for the NDR(62-88) peptide. Additional precedence for the differences in the conformation of the bound state is established by the available structures for TRTK12 interacting with rat and human S100B, which only differ at two positions (Glu62Asn and Ser78Ala), yet show the peptide adopting a different orientation and structure in each case (5, 35, 85). When bound to human S100B, it remains in an extended conformation, forming multiple contacts with helix 3, the loop region, and helix 4 of the protein. When bound to rat S100B, it forms a short helix and the N-terminal region of the peptide is aligned more closely with helix 4 of S100B. The interactions between S100B and the HDM2(25-47) and HDM4(25-47) peptides are even less well understood than the other targets examined in this study, with no structural information available. This study suggests both the HDM2(25-47) and HDM4(25-47) peptides interact with the S100B protein with a stoichiometry of one peptide per S100B dimer. Considering that a single protein is able to bind multiple targets with unique mechanisms, this suggests a certain degree of promiscuity in the S100B binding site, similar to that of Calmodulin (97). It appears that this is a general property of the S100 proteins. For example, the interactions between Annexin I peptide and S100A11 are best described by the two identical binding sites model, while the interactions of the Annexin II peptide with the same protein are best described by the two sequential binding sites model (65).
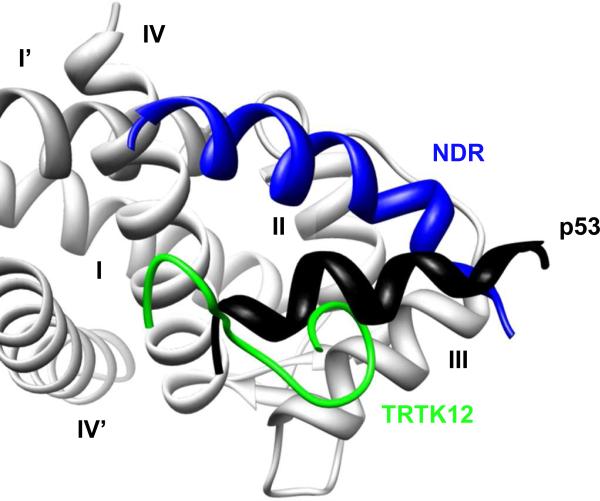
The promiscuity of the S100 binding site is also evident at the structural level. The difference in the orientation of TRTK12 peptide in complex with human and bovine S100B proteins has been discussed above. The binding of the non-muscle myosin IIA peptide fragment to S100A4 (98) is another stark example. It shows a stoichiometry of one peptide per S100A4 dimer, with the peptide occupying binding sites on both monomers. In general, the peptide binding site on the S100 proteins is defined by residues from helix 3, the loop region, and helix 4, but the orientation of different peptides bound to this site is very distinct (Figure 4).
Figure 4.
S100 Proteins Interacts with Peptide Targets through a Promiscuous Binding Site. A ribbon cartoon representing the converged NMR ensemble for the S100B protein interacting with the TRTK12 (green, PDB: 1MQ1), p53 (black, PDB: 1DT7), and NDR (blue, PDB: 1PBS) peptides in the presence of calcium. Only the binding site of one S100B monomer (light gray, Helices I-IV) is shown. Although all three peptides interact with similar amino acid residues in the binding pocket of S100B and with the same stoichiometry, each peptide binds in unique orientation with a distinct structure.
This difference in peptide orientation in the binding site and dissociation constants partially originates from the variations in peptide sequences. It has been proposed that the S100B binding motif consists of a core of hydrophobic residues flanked by positively charged Arg or Lys residues on one or both sides (39). The importance of hydrophobic residues for binding is evident by the large negative heat capacity change upon binding for all peptides studied both here (Table 1) and in previous work (65, 69, 70, 73). Of these, the HDM2(25-47) and HDM4(25-47) peptides have the largest absolute values of ΔCp and also have the tightest binding affinities to S100B (Table 1). Importantly, both peptides contain the largest fraction of non-polar residues in their sequences. This suggests that the percent hydrophobicity of a peptide is an important parameter when defining binding affinity for S100B. The aliphatic index for HDM2(25-47) and HDM4(25-47) is several times higher than similarly-sized ligands, including the NDR(62-87), p53(367-388), and RAGE(42-59) peptides (99). Interestingly, one of the only other peptides known to bind S100B with sub micromolar affinity, ROS-GC1(962-981), shares these same features. The majority of the residues in this peptide are also non-polar and the aliphatic index is similar to that of the HDM peptides, thus supporting the importance of hydrophobicity for the dissociation constant for S100B.
Kinetic Mechanism of Binding
Interactions between S100B and its peptide targets reveal complex and coupled folding and binding events. Previous studies have observed that binding of p53 induces helical formation in the peptide (38). In contrast, TRTK12(265-276) in complex with rat S100B was observed to have a single helical turn, but to remain completely unstructured in complex with human S100B (5, 54). For NDR(62-87), over half of the peptide adopts a helical structure in the bound state (5). There are no available structures for S100B-HDM2 or S100B-HDM4 complexes.
This study is the first to experimentally examine the kinetics of peptide binding to S100B and its implication for the coupled binding and folding mechanism. While the exact time constants of helix formation are still disputed, it is generally accepted that the coil-helix transition takes place in less than 1 μs, with residues first slowly overcoming the high energy barrier for nucleation, and then elongating with a time constant proportional to the number of residues in the final helix (100, 101). All five peptides were predicted to be unstructured in the unbound state using the Agadir algorithm and the lack of significant secondary structure content was subsequently confirmed by CD spectroscopy (see supplementary materials). This strongly suggests these peptides can be considered to be intrinsically disordered (IDP's) (92), i.e. that the folded state for the peptide found in the final complex with S100B only exists transiently in the unbound state. We find a “fly-casting” or “folding after binding” mechanism (102, 103) to be the most likely explanation for the large observed bimolecular rates for these peptides, as opposed to conformational selection (104, 105). The distinction between these two mechanisms for peptides which become structured in the bound state can be made from the analysis of the Kon rates (106). It is generally considered that the fly-casting mechanism for such systems has a Kon rate on the order of 107-1010 M-1 s-1, while the conformational selection mechanism has been proposed to have a Kon rate constant slower than 107 M-1 s-1 (106). For the p53(367-388) and NDR(62-87) peptides, electrostatic steering between the positively charged Arg and Lys residues of the ligands and the negatively charged Glu residues in the S100B binding pocket may also contribute significantly to the large Kon rates. This is directly supported by experimental evidence. For the p53(367-388) interaction with S100B, the Kon is reduced by an order of magnitude in the presence of 120 mM NaCl, resulting in a similar reduction in the dissociation constant (Table 1; Table 2). Several computational simulations that have examined the S100B-p53 interaction specifically (52, 107), and the binding of unstructured ligands in general (52, 102, 106, 108-111), have arrived at a similar conclusion: namely, that “folding after binding” is the principal mechanism for these species. Unfortunately, only a limited number of experimental studies have examined this hypothesis (91, 92, 112-114). The kinetic results reported here represent an important contribution to the increasing body of evidence for this “fly casting” mechanism.
Supplementary Material
ACKNOWLDGEMENTS
Instrumentation at the Core Facilities at the Center of Biotechnology and Interdisciplinary Studies at RPI were used for some of the experiments reported in this paper. We would like to thank Anthony Chiarella for his contributions to protein expression and purification, and Dr. Evgenii Kovrigin and for providing the LineShapeKin software advice on its usage.
Funding Statement
This work was supported by a grant from NIH/NIGMSRO1-GM054537.
Abbreviations
- CD
circular dichroism
- DNA
deoxyribonucleic acid
- EDTA
ethylenediaminetetraacetic acid
- EF hand
calcium-binding domain consisting of a helix-loop-helix structure
- Fmoc
Fluorenylmethyloxycarbonyl chloride
- HDM2
human homologue of murine double minute 2 protein
- HDM4
human homologue of murine double minute 4 protein
- HSQC
heteronuclear single quantum coherence
- IPTG
isopropyl β-D-thiogalactopyranoside
- ITC
isothermal titration calorimetry
- MDM2
murine double minute 2
- MDM4
murine double minute 4
- NDR
Nuclear Dbf2-related kinase
- NMR
nuclear magnetic resonance
- TCEP
tris(2-carboxyethyl)phosphine
- TFE
trifluoroethanol
- UV
ultraviolet
Footnotes
Supporting Information
Supplementary methods and Figures 1–3. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Schafer BW, Heizmann CW. The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem Sci. 1996;21:134–140. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- 2.Pritchard K, Marston SB. Ca(2+)-dependent regulation of vascular smooth-muscle caldesmon by S.100 and related smooth-muscle proteins. Biochem J. 1991;277(Pt 3):819–824. doi: 10.1042/bj2770819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilder PT, Rustandi RR, Drohat AC, Weber DJ. S100B(betabeta) inhibits the protein kinase C-dependent phosphorylation of a peptide derived from p53 in a Ca2+-dependent manner. Protein Sci. 1998;7:794–798. doi: 10.1002/pro.5560070330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heierhorst J, Kobe B, Feil SC, Parker MW, Benian GM, Weiss KR, Kemp BE. Ca2+/S100 regulation of giant protein kinases. Nature. 1996;380:636–639. doi: 10.1038/380636a0. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya S, Large E, Heizmann CW, Hemmings B, Chazin WJ. Structure of the Ca2+/S100B/NDR kinase peptide complex: insights into S100 target specificity and activation of the kinase. Biochemistry. 2003;42:14416–14426. doi: 10.1021/bi035089a. [DOI] [PubMed] [Google Scholar]
- 6.Sorci G, Agneletti AL, Donato R. Effects of S100A1 and S100B on microtubule stability. An in vitro study using triton-cytoskeletons from astrocyte and myoblast cell lines. Neuroscience. 2000;99:773–783. doi: 10.1016/s0306-4522(00)00238-4. [DOI] [PubMed] [Google Scholar]
- 7.Tarabykina S, Scott DJ, Herzyk P, Hill TJ, Tame JR, Kriajevska M, Lafitte D, Derrick PJ, Dodson GG, Maitland NJ, Lukanidin EM, Bronstein IB. The dimerization interface of the metastasis-associated protein S100A4 (Mts1): in vivo and in vitro studies. J Biol Chem. 2001;276:24212–24222. doi: 10.1074/jbc.M009477200. [DOI] [PubMed] [Google Scholar]
- 8.Gibadulinova A, Tothova V, Pastorek J, Pastorekova S. Transcriptional regulation and functional implication of S100P in cancer. Amino Acids. 2011 doi: 10.1007/s00726-010-0495-5. [DOI] [PubMed] [Google Scholar]
- 9.Nakazato Y, Ishizeki J, Takahashi K, Yamaguchi H. Immunohistochemical localization of S-100 protein in granular cell myoblastoma. Cancer. 1982;49:1624–1628. doi: 10.1002/1097-0142(19820415)49:8<1624::aid-cncr2820490816>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 10.Salama I, Malone PS, Mihaimeed F, Jones JL. A review of the S100 proteins in cancer. Eur J Surg Oncol. 2008;34:357–364. doi: 10.1016/j.ejso.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Goyette J, Geczy CL. Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids. 2010;41:821–842. doi: 10.1007/s00726-010-0528-0. [DOI] [PubMed] [Google Scholar]
- 12.Leclerc E, Fritz G, Vetter SW, Heizmann CW. Binding of S100 proteins to RAGE: an update. Biochim Biophys Acta. 2009;1793:993–1007. doi: 10.1016/j.bbamcr.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 13.Moroz OV, Burkitt W, Wittkowski H, He W, Ianoul A, Novitskaya V, Xie J, Polyakova O, Lednev IK, Shekhtman A, Derrick PJ, Bjoerk P, Foell D, Bronstein IB. Both Ca2+ and Zn2+ are essential for S100A12 protein oligomerization and function. BMC Biochem. 2009;10:11. doi: 10.1186/1471-2091-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Wang H, Zhang L, Xu Y, Deng W, Zhu H, Qin C. S100B transgenic mice develop features of Parkinson's disease. Arch Med Res. 2011;42:1–7. doi: 10.1016/j.arcmed.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Esposito G, Imitola J, Lu J, De Filippis D, Scuderi C, Ganesh VS, Folkerth R, Hecht J, Shin S, Iuvone T, Chesnut J, Steardo L, Sheen V. Genomic and functional profiling of human Down syndrome neural progenitors implicates S100B and aquaporin 4 in cell injury. Hum Mol Genet. 2008;17:440–457. doi: 10.1093/hmg/ddm322. [DOI] [PubMed] [Google Scholar]
- 16.Chaves ML, Camozzato AL, Ferreira ED, Piazenski I, Kochhann R, Dall'igna O, Mazzini GS, Souza DO, Portela LV. Serum levels of S100B and NSE proteins in Alzheimer's disease patients. J Neuroinflammation. 2010;7:6. doi: 10.1186/1742-2094-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leclerc E, Sturchler E, Vetter SW. The S100B/RAGE Axis in Alzheimer's Disease. Cardiovasc Psychiatry Neurol. 2010;2010:539581. doi: 10.1155/2010/539581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech. 2003;60:540–551. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- 19.Fritz G, Heizmann CW. Encyclopedia of Inorganic and Bioinorganic Chemistry. John Wiley & Sons, Ltd.; 2011. 3D Structures of the Calcium and Zinc Binding S100. Proteins. [Google Scholar]
- 20.Henzl MT, Davis ME, Tan A. Leucine 85 is an important determinant of divalent ion affinity in rat beta-parvalbumin (Oncomodulin) Biochemistry. 2008;47:13635–13646. doi: 10.1021/bi8014899. [DOI] [PubMed] [Google Scholar]
- 21.Henzl MT, Tanner JJ. Solution structure of Ca2+-free rat beta-parvalbumin (oncomodulin) Protein Sci. 2007;16:1914–1926. doi: 10.1110/ps.072837307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isobe T, Okuyama T. The amino-acid sequence of S-100 protein (PAP I-b protein) and its relation to the calcium-binding proteins. Eur J Biochem. 1978;89:379–388. doi: 10.1111/j.1432-1033.1978.tb12539.x. [DOI] [PubMed] [Google Scholar]
- 23.Potts BC, Smith J, Akke M, Macke TJ, Okazaki K, Hidaka H, Case DA, Chazin WJ. The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins. Nat Struct Biol. 1995;2:790–796. doi: 10.1038/nsb0995-790. [DOI] [PubMed] [Google Scholar]
- 24.Drohat AC, Baldisseri DM, Rustandi RR, Weber DJ. Solution structure of calcium-bound rat S100B(betabeta) as determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1998;37:2729–2740. doi: 10.1021/bi972635p. [DOI] [PubMed] [Google Scholar]
- 25.Kilby PM, Van Eldik LJ, Roberts GC. The solution structure of the bovine S100B protein dimer in the calcium-free state. Structure. 1996;4:1041–1052. doi: 10.1016/s0969-2126(96)00111-6. [DOI] [PubMed] [Google Scholar]
- 26.Matsumura H, Shiba T, Inoue T, Harada S, Kai Y. A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain. Structure. 1998;6:233–241. doi: 10.1016/s0969-2126(98)00024-0. [DOI] [PubMed] [Google Scholar]
- 27.Smith SP, Shaw GS. A novel calcium-sensitive switch revealed by the structure of human S100B in the calcium-bound form. Structure. 1998;6:211–222. doi: 10.1016/s0969-2126(98)00022-7. [DOI] [PubMed] [Google Scholar]
- 28.Maler L, Potts BC, Chazin WJ. High resolution solution structure of apo calcyclin and structural variations in the S100 family of calcium-binding proteins. J Biomol NMR. 1999;13:233–247. doi: 10.1023/a:1008315517955. [DOI] [PubMed] [Google Scholar]
- 29.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(betabeta) Nat Struct Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 30.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396:201–214. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore B. A soluble protein characteristic of the nervous system. Biochem Biophys Res Commun. 1965:739–744. doi: 10.1016/0006-291x(65)90320-7. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr., Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank. A computer-based archival file for macromolecular structures. Eur J Biochem. 1977;80:319–324. doi: 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- 33.Nonaka D, Chiriboga L, Rubin BP. Differential expression of S100 protein subtypes in malignant melanoma, and benign and malignant peripheral nerve sheath tumors. J Cutan Pathol. 2008;35:1014–1019. doi: 10.1111/j.1600-0560.2007.00953.x. [DOI] [PubMed] [Google Scholar]
- 34.Rezvanpour A, Phillips JM, Shaw GS. Design of high-affinity S100-target hybrid proteins. Protein Sci. 2009;18:2528–2536. doi: 10.1002/pro.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inman KG, Yang R, Rustandi RR, Miller KE, Baldisseri DM, Weber DJ. Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. Journal of molecular biology. 2002;324:1003–1014. doi: 10.1016/s0022-2836(02)01152-x. [DOI] [PubMed] [Google Scholar]
- 36.Rustandi RR, Drohat AC, Baldisseri DM, Wilder PT, Weber DJ. The Ca(2+)-dependent interaction of S100B(beta beta) with a peptide derived from p53. Biochemistry. 1998;37:1951–1960. doi: 10.1021/bi972701n. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez-Fernandez MR, Rutherford TJ, Fersht AR. Members of the S100 family bind p53 in two distinct ways. Protein Sci. 2008;17:1663–1670. doi: 10.1110/ps.035527.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rustandi RR, Baldisseri DM, Drohat AC, Weber DJ. Structural changes in the C-terminus of Ca2+-bound rat S100B (beta beta) upon binding to a peptide derived from the C-terminal regulatory domain of p53. Protein Sci. 1999;8:1743–1751. doi: 10.1110/ps.8.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilder PT, Lin J, Bair CL, Charpentier TH, Yang D, Liriano M, Varney KM, Lee A, Oppenheim AB, Adhya S, Carrier F, Weber DJ. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim Biophys Acta. 2006;1763:1284–1297. doi: 10.1016/j.bbamcr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 40.van Dieck J, Lum JK, Teufel DP, Fersht AR. S100 proteins interact with the N-terminal domain of MDM2. FEBS Lett. 2010;584:3269–3274. doi: 10.1016/j.febslet.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 41.Rety S, Sopkova J, Renouard M, Osterloh D, Gerke V, Tabaries S, Russo-Marie F, Lewit-Bentley A. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat Struct Biol. 1999;6:89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- 42.Rety S, Osterloh D, Arie JP, Tabaries S, Seeman J, Russo-Marie F, Gerke V, Lewit-Bentley A. Structural basis of the Ca(2+)-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Structure. 2000;8:175–184. doi: 10.1016/s0969-2126(00)00093-9. [DOI] [PubMed] [Google Scholar]
- 43.Gribenko AV, Makhatadze GI. Oligomerization and divalent ion binding properties of the S100P protein: a Ca2+/Mg2+-switch model. Journal of molecular biology. 1998;283:679–694. doi: 10.1006/jmbi.1998.2116. [DOI] [PubMed] [Google Scholar]
- 44.Pozdnyakov N, Margulis A, Sitaramayya A. Identification of effector binding sites on S100 beta: studies with guanylate cyclase and p80, a retinal phosphoprotein. Biochemistry. 1998;37:10701–10708. doi: 10.1021/bi9802115. [DOI] [PubMed] [Google Scholar]
- 45.Garbuglia M, Verzini M, Rustandi RR, Osterloh D, Weber DJ, Gerke V, Donato R. Role of the C-terminal extension in the interaction of S100A1 with GFAP, tubulin, the S100A1- and S100B-inhibitory peptide, TRTK-12, and a peptide derived from p53, and the S100A1 inhibitory effect on GFAP polymerization. Biochem Biophys Res Commun. 1999;254:36–41. doi: 10.1006/bbrc.1998.9881. [DOI] [PubMed] [Google Scholar]
- 46.Ivanenkov VV, Jamieson GA, Jr., Gruenstein E, Dimlich RV. Characterization of S-100b binding epitopes. Identification of a novel target, the actin capping protein, CapZ. J Biol Chem. 1995;270:14651–14658. doi: 10.1074/jbc.270.24.14651. [DOI] [PubMed] [Google Scholar]
- 47.Ashcroft M, Kubbutat MH, Vousden KH. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, Hee Park S, Thompson T, Karsenty G, Bradley A, Donehower LA. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 49.Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc Natl Acad Sci U S A. 2005;102:4735–4740. doi: 10.1073/pnas.0501459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Dieck J, Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. J Biol Chem. 2009;284:13804–13811. doi: 10.1074/jbc.M901351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bell S, Klein C, Muller L, Hansen S, Buchner J. p53 contains large unstructured regions in its native state. Journal of molecular biology. 2002;322:917–927. doi: 10.1016/s0022-2836(02)00848-3. [DOI] [PubMed] [Google Scholar]
- 52.Chen J. Intrinsically disordered p53 extreme C-terminus binds to S100B(betabeta) through “fly-casting”. J Am Chem Soc. 2009;131:2088–2089. doi: 10.1021/ja809547p. [DOI] [PubMed] [Google Scholar]
- 53.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 9 Suppl. 2008;1:S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charpentier TH, Thompson LE, Liriano MA, Varney KM, Wilder PT, Pozharski E, Toth EA, Weber DJ. The effects of CapZ peptide (TRTK-12) binding to S100B-Ca2+ as examined by NMR and X-ray crystallography. Journal of molecular biology. 2010;396:1227–1243. doi: 10.1016/j.jmb.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liriano M, Varney K, Wright N, Hoffman C, Toth E, Ishima R, Weber D. Target Binding to S100B Reduces Dynamic Properties and Increases Ca(2+)-Binding Affinity for Wild Type and EF-Hand Mutant Proteins. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.07.011. PMID:22824086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Markowitz J, Rustandi RR, Varney KM, Wilder PT, Udan R, Wu SL, Horrocks WD, Weber DJ. Calcium-binding properties of wild-type and EF-hand mutants of S100B in the presence and absence of a peptide derived from the C-terminal negative regulatory domain of p53. Biochemistry. 2005;44:7305–7314. doi: 10.1021/bi050321t. [DOI] [PubMed] [Google Scholar]
- 57.Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- 58.Tamaskovic R, Bichsel SJ, Rogniaux H, Stegert MR, Hemmings BA. Mechanism of Ca2+-mediated regulation of NDR protein kinase through autophosphorylation and phosphorylation by an upstream kinase. J Biol Chem. 2003;278:6710–6718. doi: 10.1074/jbc.M210590200. [DOI] [PubMed] [Google Scholar]
- 59.Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, Bellefroid E, Marine JC. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A. 2006;103:3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schon O, Friedler A, Bycroft M, Freund SM, Fersht AR. Molecular mechanism of the interaction between MDM2 and p53. Journal of molecular biology. 2002;323:491–501. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 61.Mancini F, Di Conza G, Monti O, Macchiarulo A, Pellicciari R, Pontecorvi A, Moretti F. Puzzling over MDM4-p53 network. Int J Biochem Cell Biol. 2010;42:1080–1083. doi: 10.1016/j.biocel.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 62.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 63.Sanchez MC, Renshaw JG, Davies G, Barlow PN, Vogtherr M. MDM4 binds ligands via a mechanism in which disordered regions become structured. FEBS Lett. 2010;584:3035–3041. doi: 10.1016/j.febslet.2010.05.058. [DOI] [PubMed] [Google Scholar]
- 64.Gribenko A, Lopez MM, Richardson JM, 3rd, Makhatadze GI. Cloning, overexpression, purification, and spectroscopic characterization of human S100P. Protein Sci. 1998;7:211–215. doi: 10.1002/pro.5560070123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Streicher WW, Lopez MM, Makhatadze GI. Annexin I and annexin II N-terminal peptides binding to S100 protein family members: specificity and thermodynamic characterization. Biochemistry. 2009;48:2788–2798. doi: 10.1021/bi8019959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gribenko AV, Hopper JE, Makhatadze GI. Molecular characterization and tissue distribution of a novel member of the S100 family of EF-hand proteins. Biochemistry. 2001;40:15538–15548. doi: 10.1021/bi0114731. [DOI] [PubMed] [Google Scholar]
- 67.Wright NT, Cannon BR, Wilder PT, Morgan MT, Varney KM, Zimmer DB, Weber DJ. Solution structure of S100A1 bound to the CapZ peptide (TRTK12) Journal of molecular biology. 2009;386:1265–1277. doi: 10.1016/j.jmb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 69.Gribenko AV, Guzman-Casado M, Lopez MM, Makhatadze GI. Conformational and thermodynamic properties of peptide binding to the human S100P protein. Protein Sci. 2002;11:1367–1375. doi: 10.1110/ps.0202202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lopez MM, Makhatadze GI. Isothermal titration calorimetry. Methods Mol Biol. 2002;173:121–126. doi: 10.1385/1-59259-184-1:121. [DOI] [PubMed] [Google Scholar]
- 71.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 72.Rohl CA, Baldwin RL. Deciphering rules of helix stability in peptides. Methods Enzymol. 1998;295:1–26. doi: 10.1016/s0076-6879(98)95032-7. [DOI] [PubMed] [Google Scholar]
- 73.Brokx RD, Lopez MM, Vogel HJ, Makhatadze GI. Energetics of target peptide binding by calmodulin reveals different modes of binding. J Biol Chem. 2001;276:14083–14091. doi: 10.1074/jbc.M011026200. [DOI] [PubMed] [Google Scholar]
- 74.Makhatadze GI, Privalov PL. Energetics of protein structure. Adv Protein Chem. 1995;47:307–425. doi: 10.1016/s0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- 75.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007 doi: 10.1002/0471140864.ps0209s50. Chapter 2, Unit 2 9. [DOI] [PubMed] [Google Scholar]
- 76.Wafer LN, Streicher WW, Makhatadze GI. Thermodynamics of the Trp-cage miniprotein unfolding in urea. Proteins. 2010;78:1376–1381. doi: 10.1002/prot.22681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hitchens TK, Lukin JA, Zhan Y, McCallum SA, Rule GS. MONTE: An automated Monte Carlo based approach to nuclear magnetic resonance assignment of proteins. J Biomol NMR. 2003;25:1–9. doi: 10.1023/a:1021975923026. [DOI] [PubMed] [Google Scholar]
- 78.McClintock K,A, Shaw, Gary S. Letter to the Editor: Assignments of 1H, 13C and 15N resonances of human Ca2+-S100B in complex with the TRTK-12 peptide. J. Biomol. NMR. 2002;23:255–256. doi: 10.1023/a:1019886414743. [DOI] [PubMed] [Google Scholar]
- 79.Rule GS, T. K. H. Fundamentals of Protein NMR Spectroscopy. Springer; New York: 2005. [Google Scholar]
- 80.Farmer BT, 2nd, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr., Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat Struct Biol. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- 81.Goddard K. Sparky. 2008 ( www.cgl.ucsf.edu/home/sparky/, Ed.)
- 82.Kovrigin EL, Loria JP. Enzyme dynamics along the reaction coordinate: critical role of a conserved residue. Biochemistry. 2006;45:2636–2647. doi: 10.1021/bi0525066. [DOI] [PubMed] [Google Scholar]
- 83.Kovriguine E. LineShapeKin Simulation. (4.1 ed.) 2009 www.lineshapekin.net.
- 84.Garcia-Fuentes L, Baron C, Mayorga OL. Influence of dynamic power compensation in an isothermal titration microcalorimeter. Anal Chem. 1998;70:4615–4623. doi: 10.1021/ac980203u. [DOI] [PubMed] [Google Scholar]
- 85.McClintock KA, Shaw GS. A novel S100 target conformation is revealed by the solution structure of the Ca2+-S100B-TRTK-12 complex. J Biol Chem. 2003;278:6251–6257. doi: 10.1074/jbc.M210622200. [DOI] [PubMed] [Google Scholar]
- 86.Barber KR, McClintock KA, Jamieson GA, Jr., Dimlich RV, Shaw GS. Specificity and Zn2+ enhancement of the S100B binding epitope TRTK-12. J Biol Chem. 1999;274:1502–1508. doi: 10.1074/jbc.274.3.1502. [DOI] [PubMed] [Google Scholar]
- 87.van Dieck J, Brandt T, Teufel DP, Veprintsev DB, Joerger AC, Fersht AR. Molecular basis of S100 proteins interacting with the p53 homologs p63 and p73. Oncogene. 2010;29:2024–2035. doi: 10.1038/onc.2009.490. [DOI] [PubMed] [Google Scholar]
- 88.McCallum SA, Hitchens TK, Torborg C, Rule GS. Ligand-induced changes in the structure and dynamics of a human class Mu glutathione S-transferase. Biochemistry. 2000;39:7343–7356. doi: 10.1021/bi992767d. [DOI] [PubMed] [Google Scholar]
- 89.Lian L-Y, Roberts GCK. NMR of Macromolecules. IRL Press; Oxford, U.K: 1993. [Google Scholar]
- 90.Wu G, Berka V, Tsai AL. Binding kinetics of calmodulin with target peptides of three nitric oxide synthase isozymes. J Inorg Biochem. 2011;105:1226–1237. doi: 10.1016/j.jinorgbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 92.Arai M, Ferreon JC, Wright PE. Quantitative Analysis of Multisite Protein-Ligand Interactions by NMR: Binding of Intrinsically Disordered p53 Transactivation Subdomains with the TAZ2 Domain of CBP. J Am Chem Soc. 2012;134:3792–3803. doi: 10.1021/ja209936u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biophys Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 94.Zhou G, Wong MT, Zhou GQ. Diffusion-controlled reactions of enzymes. An approximate analytic solution of Chou's model. Biophys Chem. 1983;18:125–132. doi: 10.1016/0301-4622(83)85006-6. [DOI] [PubMed] [Google Scholar]
- 95.Zhou GQ, Zhong WZ. Diffusion-controlled reactions of enzymes. A comparison between Chou's model and Alberty-Hammes-Eigen's model. Eur J Biochem. 1982;128:383–387. [PubMed] [Google Scholar]
- 96.Wright NT, Varney KM, Ellis KC, Markowitz J, Gitti RK, Zimmer DB, Weber DJ. The three-dimensional solution structure of Ca(2+)-bound S100A1 as determined by NMR spectroscopy. Journal of molecular biology. 2005;353:410–426. doi: 10.1016/j.jmb.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 97.Yamniuk AP, Vogel HJ. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 98.Kiss B, Duelli A, Radnai L, Kekesi KA, Katona G, Nyitray L. Crystal structure of the S100A4-nonmuscle myosin IIA tail fragment complex reveals an asymmetric target binding mechanism. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1114732109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ikai A. Thermostability and aliphatic index of globular proteins. J Biochem. 1980;88:1895–1898. [PubMed] [Google Scholar]
- 100.De Sancho D, Best RB. What is the time scale for alpha-helix nucleation? J Am Chem Soc. 2011;133:6809–6816. doi: 10.1021/ja200834s. [DOI] [PubMed] [Google Scholar]
- 101.Serrano AL, Tucker MJ, Gai F. Direct assessment of the alpha-helix nucleation time. J Phys Chem B. 2011;115:7472–7478. doi: 10.1021/jp200628b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci U S A. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Koshland DE. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc Natl Acad Sci U S A. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Berger C, Weber-Bornhauser S, Eggenberger J, Hanes J, Pluckthun A, Bosshard HR. Antigen recognition by conformational selection. FEBS Lett. 1999;450:149–153. doi: 10.1016/s0014-5793(99)00458-5. [DOI] [PubMed] [Google Scholar]
- 105.Foote J, Milstein C. Conformational isomerism and the diversity of antibodies. Proc Natl Acad Sci U S A. 1994;91:10370–10374. doi: 10.1073/pnas.91.22.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kiefhaber T, Bachmann A, Jensen KS. Dynamics and mechanisms of coupled protein folding and binding reactions. Curr Opin Struct Biol. 2011;22:21–29. doi: 10.1016/j.sbi.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 107.Ganguly D, Chen J. Topology-based modeling of intrinsically disordered proteins: balancing intrinsic folding and intermolecular interactions. Proteins. 2011;79:1251–1266. doi: 10.1002/prot.22960. [DOI] [PubMed] [Google Scholar]
- 108.Trizac E, Levy Y, Wolynes PG. Capillarity theory for the fly-casting mechanism. Proc Natl Acad Sci U S A. 2010;107:2746–2750. doi: 10.1073/pnas.0914727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang Y, Liu Z. Kinetic advantage of intrinsically disordered proteins in coupled folding-binding process: a critical assessment of the “fly-casting” mechanism. Journal of molecular biology. 2009;393:1143–1159. doi: 10.1016/j.jmb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 110.Turjanski AG, Gutkind JS, Best RB, Hummer G. Binding-induced folding of a natively unstructured transcription factor. PLoS Comput Biol. 2008;4:e1000060. doi: 10.1371/journal.pcbi.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Freer ST, Rose PW. Simulating disorder-order transitions in molecular recognition of unstructured proteins: where folding meets binding. Proc Natl Acad Sci U S A. 2003;100:5148–5153. doi: 10.1073/pnas.0531373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Narayanan R, Ganesh OK, Edison AS, Hagen SJ. Kinetics of folding and binding of an intrinsically disordered protein: the inhibitor of yeast aspartic proteinase YPrA. J Am Chem Soc. 2008;130:11477–11485. doi: 10.1021/ja803221c. [DOI] [PubMed] [Google Scholar]
- 113.Onitsuka M, Kamikubo H, Yamazaki Y, Kataoka M. Mechanism of induced folding: Both folding before binding and binding before folding can be realized in staphylococcal nuclease mutants. Proteins. 2008;72:837–847. doi: 10.1002/prot.21978. [DOI] [PubMed] [Google Scholar]
- 114.Bachmann A, Wildemann D, Praetorius F, Fischer G, Kiefhaber T. Mapping backbone and side-chain interactions in the transition state of a coupled protein folding and binding reaction. Proc Natl Acad Sci U S A. 2011;108:3952–3957. doi: 10.1073/pnas.1012668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.