Abstract
The protein nanopore Mycobacteria smegmatis porin A (MspA), can be used to sense individual nucleotides within DNA, potentially enabling a technique known as nanopore sequencing. In this technique, single stranded DNA electrophoretically moves through the nanopore and results in an ionic current that is nucleotide-specific. However, with a high transport velocity of the DNA within the nanopore, the ionic current cannot be used to distinguish signals within noise. Through extensive (~100 μs in total) all-atom molecular dynamics simulations, we examine the effect of positively charged residues on DNA translocation rate and the ionic current blockades in MspA. Simulation of several arginine mutations show a ~10–30 fold reduction of DNA translocation speed without eliminating the nucleotide induced current blockages. Comparison of our results with similar engineering efforts on a different nanopore (alpha-hemolysin) reveals a non-trivial effect of nanopore geometry on the ionic current blockades in mutant nanopores.
Keywords: Nanopore, sequencing, membrane channel, DNA-protein interactions, stochastic sensor, protein engineering
Nanopores have emerged as versatile research tools for single molecule detection and analysis.1 One of the most prominent applications of nanopores is rapid, label-free, and real-time sequencing of DNA.2 In the original concept of nanopore sequencing,3 single stranded DNA (ssDNA) is electrophoretically driven through a nanopore. Inside the nanopore, DNA nucleotides uniquely modify the co-passing ionic current, yielding an ionic current trace that contains information about the DNA sequence. With a number of recent advances in both biological and solid-state nanopores, nanopore sequencing of DNA is becoming more of a reality.2
The most common nanopores used for DNA transport measurements are naturally occurring or genetically engineered proteins alpha-hemolysin (αHL)4 and Mycobacteria smegmatis porin A (MspA)5 and solid-state nanopores made in thin silicon nitride6–8 or graphene9–11 membranes. In the case of MspA, experimental studies using immobilized DNA strands have already demonstrated the possibility of distinguishing all four DNA nucleotides via ionic current measurement12–14 and detection of single nucleotides in a random sequence background.14 Nanopores have also been used to detect RNA,15–17 epigenetic modifications such as methylated cytosine and 8-oxoguanidine14,18, 19 and DNA damage.19
A common problem with using nanopores for single molecule analysis of DNA is the speed of the analyte through a nanopore. The average speed of DNA transport in free translocation experiments typically exceeds 1 nucleotide per microsecond, which is too high to detect the local structure or sequence of the transported DNA via ionic current measurement. Consequently, a number of techniques have been proposed to reduce and control the speed of DNA transport through nanopores.20, 21 Simple techniques, such as increasing the buffer viscosity or decreasing temperature can reduce the translocation speed22–25 but also reduce the ionic current signal as the ion mobility also decreases. More complex techniques such as coupling the nanopore with processive enzymes that sequentially displace DNA26, 27 through a nanopore have shown to be successful, but require specific conditions to keep the DNA processing enzymes operational. Yet another approach is to modify the nanopore surface by attaching complementary DNA sequences28, modifying the DNA analyte,13, 29 changing the charge state of the nanopore30 or using the steric confinement of a nanopore to arrest DNA motion.31
In this article, we report extensive all-atom molecular dynamics (MD) simulations that assessed the feasibility of slowing DNA transport through modified MspA nanopores and using such nanopores for analysis of nucleic acids. We chose to focus our efforts on MspA, as this nanopore has features32 sufficiently small to enable detection of individual DNA nucleotides by measuring ionic current.13, 14 Our baseline system is the MspA nanopore, denoted M1-NNN, which was previously used to demonstrate nucleotide-specific ionic current blockades. The constriction of this nanopore already contains asparagine substitutions at positions 90, 91 and 93 in all chains of the octomer. We hypothesize that additional positively charged arginine substitutions near the constriction of MspA can reduce the speed of DNA transport while preserving the nucleotide-type specificity of the ionic current blockades.
Results and discussion
To determine the effect of arginine mutations on the transport rate of ssDNA through MspA, we constructed all-atom models of the MspA nanopore, embedded in a lipid bilayer membrane and submerged in 1 M KCl electrolyte. Figures 1a shows one such system featuring a full-length MspA nanopore and a 58-nucleotide DNA strand threaded through it using the phantom nanopore method.33 Using the all-atom molecular dynamics method,34 we simulated electric-field driven transport of the DNA strand through several variants of the MspA nanopore. Short (<100 ns in duration) exploratory simulations performed under a 1.2 V transmembrane bias35 revealed transient arrests of ssDNA transport as multiple salt-bridges formed between the phosphate groups of the DNA backbone and the amine groups of arginine substitutions of leucine 88 residues of the L88R MspA nanopore, see Supporting Information Figure S1. However, the high bias used in these simulations prevented the effect of arginine substitutions to fully develop as the energy gain from transporting one nucleotide through the constriction of MspA (~1.2 eV or 46 kBT) is considerably greater than a typical energy of an ionic salt bridge in water (~ 5 kBT). Furthermore, applying a 1.2 V bias in experiment is known to render the bilayer unstable on the experimental time scale.
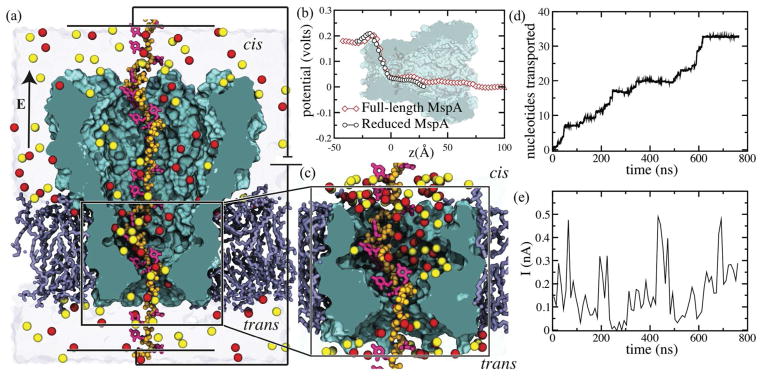
Figure 1.
MD simulations of ssDNA transport through MspA. (a) Cut-away view of a full-length MspA system. The MspA nanopore is shown as a teal molecular surface, the POPC lipid bilayer is shown as purple lines, the backbone of a DNA strand threaded through the nanopore is shown as ochre spheres, the DNA bases are shown as thick magenta lines. K+ and Cl− ions within a representative 1-nm-thick cross section of the system are shown as red and lemon yellow spheres, respectively. Water is not shown. The locations of the cis and trans sides are specified in the image. (b) Average electrostatic potentials along the symmetry axis of the MspA nanopores at a transmembrane bias of 180 mV. The electrostatic profiles were computed from all-atm MD simulation of open (no DNA) MspA nanopores. In panels a and c, the z axis points normal to the plane of the lipid bilayer. The background image of the nanopore faithfully aligns with the profiles of the electrostatic potential. (c) Cut-away view of a reduced MspA system. In the reduced-MspA system, the DNA strand is covalently joined to itself across the periodic boundary, forming an effectively infinite DNA strand. The coloring scheme is the same as in panel (a). (d) The number of cytosine nucleotides translocated through the constriction of the M1 MspA in a typical MD trajectory performed at 180 mV bias. (e) The ionic current trace for the MD trajectory featured in panel (d), sampled at 10 ns.
To observe multiple, microsecond duration trajectories of ssDNA transport through MspA nanopores at the experimental bias of 180 mV, we used an ensemble of sixteen reduced-length MspA systems, each having different starting conformation of a DNA strand in the nanopore. Each simulation system contained a single copy of a reduced MspA nanopore (residues 75 to 120), a poly(dC) strand containing from 13 to 21 nucleotides (depending on the system) and connected to itself via periodic boundary conditions, a patch of a lipid bilayer membrane containing about 60 lipid molecules and 1M KCl solution. Figure 1-c shows one such reduced-length MspA system; all sixteen systems are shown in the Supporting Information Figure S2. To prevent structural deterioration of the truncated nanopore, all Ca atoms of the protein were restrained to their crystallographic coordinates. Our 200-ns simulations of full-length and reduced-length MspA systems revealed very similar electrostatic potential distributions in the nanopore constriction at a 180mV bias, a bias typically used in experiment.
Figure 1-d and 1-e illustrate a typical simulation of a reduced-length M1 MspA system. In that particular trajectory, approximately 33 nucleotides passed through the nanopore constriction in 750 ns, which is consistent with the experimental estimates12 of the nucleotide transport rate. The translocation proceeds in bursts, where long pauses follow rapid advancements, similar to transport observed in αHL.36 The ionic current computed for the same trajectory (Figure 1-e) exhibits large fluctuations and no apparent correlation with the DNA transport rate. An animation provided in the Supporting Information illustrates this trajectory.
Figure 2 shows the number of nucleotides transported through the constriction of MspA versus simulation time for the ensemble of 16 simulation systems and four MspA variants: the baseline M1-NNN nanopore, single arginine mutant L88R-NNN, and two triple arginine mutants L88R/T83R/S116R-NNN and L88R/A96R/S116R-NNN. The location of the arginine substitutions is shown in Figures 2-b, c, and d. Due to the octomeric architecture of MspA, the total number of arginine residues is 8 in L88R-NNN and 24 in the triple mutant nanopores. Within ~14 μs of aggregate simulation time, ~250 nucleotides were observed to pass through the constriction of M1-MspA, against 45 nucleotides observed to pass through the L88R-NNN nanopore within ~12 μs. A linear fit to the cumulative permeation traces suggests a 5-fold reduction of the permeation rate through L88R-NNN nanopore in comparison to M1-NNN MspA. A closer inspection of the trajectories reveals that the permeation rates are not constant for the L88R-NNN nanopore: the nucleotides appear to move faster at the beginning of the individual patches than at the end, as formation of DNA-Arg contacts does not occur instantaneously. Furthermore, even after the contacts are formed, DNA can still be displaced by several nucleotides through the constriction of MspA before the effect DNA-Arg contacts becomes apparent.
Figure 2.
Ensemble simulation of ssDNA transport through mutant variants of MspA. The number of nucleotides translocated through the constriction of MspA at a 180 mV bias is plotted as a function of time for 16 independent systems in the case of (a) M1, (e) M1 L88R, (f) M1 L88R/A96R/S116R and (g) M1 L88R/T83R/S116R variants of the MspA nanopore and cytosine homopolymers. Each trace is shown in two colors, red and black, indicating multiple trajectories (differing in their initial DNA conformation) that were added consecutively to produce the total permeation trace. In panel (g), the last 4-μs trace corresponds to a single MD trajectory (blue). Panels (b), (c) and (d) show the locations of the arginine residues (blue vdW spheres). The backbone of the DNA strand is shown as yellow spheres with the DNA bases depicted by thick magenta lines.
In Figure 3, we plot the nucleotide translocation rate averaged over 16 different systems as a function of the time elapsed from the beginning of the simulations. The average transport rate through the M1-NNN MspA nanopore gradually decreases with time and is approximately 20±5 nucleotides/μs. A linear fit to the cumulative trace in Figure 2-a yields a similar value. In contrast, the average permeation rate for the L88R-NNN mutant is clearly higher within the first 200 ns of the trajectories than within the last 450 ns. Excluding the initial 300 ns, the average permeation rate is about 0.7±0.4 nucleotides/μs. Thus, our MD simulations suggest that L88R-NNN mutation can reduce the transport velocity of ssDNA through MspA by at least 20–30-fold, compared to M1–NNN MspA.
Figure 3.
Simulated effect of arginine mutations on DNA transport rate. (a) Averaged over 16 independent trajectories, the transport rate of cytosine homopolymers is plotted versus the simulation time for M1 and M1 L88R nanopores. The error bars represent the standard deviation of the mean. The inset shows the average permeation rates for triple mutants M1 L88R/T83R/S116R and M1 L88R/A96R/S116R. A break marked by ‘//’ indicates a switchover between linear and logarithmic scales. (b) The number of persistent contacts between the DNA and the arginine residues as a function of simulation time, averaged over 16 independent trajectories. To count as a persistent contact, non-hydrogen atoms of the same DNA nucleotide must reside within 4 Å of the same arginine residue for 10 ns at least 80% of the time. The inset shows the corresponding plot for the single 4-microsecond trajectory of the M1 L88R/T83R/S116R system. (c–d) Cut away view of the M1 L88R/T83R/S116R nanopore showing contacts between (c) a phosphate group of the DNA backbone and an arginine residue and (d) a cytosine base and an arginine residue. The DNA backbone and bases are shown as vdW spheres and bonds, respectively; the MspA protein is shows as a semitransparent surface; the arginine residues are shown in all-atom detail.
Our simulations of the triple mutants suggest the possibility of reducing the DNA translocation velocity even further. Although the total number of nucleotides transported through triple mutants in our ensemble simulations (Figures 2g and 2f) appears to be similar to that observed for the L88R-NNN mutant, the permeation rates (Figure 3-a, inset) drop to zero as the simulation progresses. The trajectories of the two triple mutants were shorter than those of the L88R-NNN mutant and the permeation rates did not reach a steady value. Unable to accurately determine the steady-state transport rate from ensemble simulations, we continued one of the L88R/T83R/S116R-NNN trajectories for additional 4 μs using DESRES supercomputer Anton37 The transport rate obtained from this continuous 4-μs trajectory was close to zero (−0.4 ± 0.6 nucleotides/μs), which indicates a reduction of the DNA transport rate in comparison with an ensemble-average rate of the L88R mutant. An animation provided in the supporting information illustrates the 4-μs trajectory.
In Figure 3-b, we plot the ensemble average of the number of persistent contacts between the DNA and the arginine residues versus the simulation time. Here, a persistent contact is defined to exist between a nucleotide and an arginine residue if the non-hydrogen atoms of the nucleotide reside within a cutoff of 4 Å of the non-hydrogen atoms of the same arginine residue for at least 80% time within 10 ns. For all three mutant nanopores, the number of DNA-arginine contacts increases steeply in the initial 150 ns before reaching an approximately steady-state value, which correlates with the observed reduction of the DNA transport rate, Figure 3-a. For the L88R-NNN nanopore, the number of contacts continues to increase throughout the trajectory at a smaller rate. The L88R/T83R/S116R-NNN mutant formed the highest steady-state number of contacts (~7), the L88R/A96R/S116R-NNN nanopore had about 6 while the L88R-NNN mutant had the least number of contacts (~2–3). A scatter diagram of the number of persistent contacts versus the DNA transport rate, Supporting Information Figure S3, indicates that a higher number of persistent contacts correlates with a smaller rate of DNA transport.
In view of the larger number of contacts formed, the L88R/T83/S116R-NNN is likely to be more effective than the other mutants studied in controlling the translocation of the DNA. Figure S4 in the supporting material shows the number of contacts for each of the mutated residues in the two triple mutant nanopores. Residues R116 are seen to be the most effective in both cases, forming between 2 and 3 contacts with the DNA. Residues R83 are the second most effective, forming ~2 contacts. R88 and R96 residues are seen to form fewer than two contacts with the DNA.
The anticipated effect of arginine mutations was electrostatic attraction between positively charged amine groups and negatively charged DNA backbone. Surprisingly, visual inspection of the MD trajectories revealed two types of persistent DNA-arginine contacts. As expected, phosphate groups of the DNA backbone formed salt bridges with the amine groups of arginine residues, as shown in Figure 3-c. The second type of persistent contact involved stacking of the cytosine base with the arginine side chain, as shown in Figure 3-d. This type of contact is similar to the stacking observed in the crystal structure of the human protein U1A bound to RNA.38 Supporting Information Figure S5 displays the number of salt-bridge and stacking-type contacts for the three mutant nanopores. The L88R/A96R/S116R-NNN nanopore is seen to form a greater number of stacking-type contacts (~3) than salt bridges (~2). For the L88R-NNN and M1L88R/T83R/S116R mutants, the stacking and salt bridge contacts are almost equally prevalent (~1 for the L88R-NNN and ~3 for the L88R/T83R/S116R-NNN mutants respectively).
Recent experimental30 and computational39 studies systematically examined the effect of arginine mutations on the DNA transport rate and ionic current blockades in αHL. In the experiments with αHL, the residual current was ~0 pA when the arginine substitutions were in the stem of αHL, rendering the nanopores less suitable for DNA sequence detection. In our simulations of MspA mutants, we find ionic current blockades in the modified MspA to retain sensitivity to the sequence of DNA strands threaded through the nanopore’s constriction.
To investigate the effect of arginine substitutions on ionic current blockades in both MspA and αHL nanopores, we computed the residual ionic current from our ensemble simulations of M1-NNN and arginine mutants of the MspA nanopore. For comparison, we performed two 150-ns simulations of ionic current blockades in wild-type and triple-mutant αHL nanopores (Figure 4), using previously reported computational models.33,36 For MspA we find the residual current for M1 to be 127 ± 8 pA and for L88R to be 120 ± 6 pA. These values are indistinguishable within the statistical uncertainties. Here and in the rest of the manuscript, the standard error was computed using current values block-averaged over 100 ns and the 16 independent trajectories. The average currents through triple mutants were identifiably greater with currents of 164 ± 13 pA for L88R/T83R/S116R-NNN and 205 ±15 pA for L88R/A96R/S116R-NNN. The simulated blockade currents are systematically higher than those measured experimentally using NeutrAvidin anchors12–14 as the outer rim of the MspA nanopore are absent in our reduced-length systems. In terms of percentage of the open nanopore currents, the average simulated blockade current is 13.8±1.2% for the M1-NNN nanopore, 13.2 ±0.9 % for the L88R, 19.2±1.9% for the L88R/T83R/S116R and 23.8±1.8 % for the L88R/A96R/S116R mutations. Figure 4c shows the distributions of the blockade currents for M1 and L88R/A96R/S116R nanopores. Thus, the residual current through triple mutant MspA nanopore is almost twice the value of the M1 nanopore. Figure 4-d shows similar data for our simulations of αHL with three arginine substitutions resulting in 21 additional positive charges. In the case of αHL, triple arginine substitutions completely block the nanopore current. Thus, the arginine substitutions have opposite effect on the residual current in αHL and MspA, which, as we show below, originates from the difference in the shape of the two nanopores.
Figure 4.
The effect of arginine mutations on the ionic current. (a, b) Cut-away view of the MspA (a) and αHL (b) nanopores containing a DNA strand. The locations of arginine substitutions are indicated by horizontal blue arrows. Simulated values of the K+ and Cl− currents at a 180 mV (MspA) or 240 mV (αHL) bias are specified in the figure. (c, d) Histograms of ionic current obtained from ensemble simulations of the MspA nanopores (c) and single trajectories of the αHL systems (d). The overlap of the two histograms is shown in purple. The histograms were constructed using 100 (MspA) and 1 (αHL) ns averages of instantaneous current. (e, f) Local concentration of K+ and Cl− ions along the symmetry axis of the MspA (e) and αHL (f) nanopores. The blue bars indicate the location of the arginine residues in the nanopores; the z coordinate is defined in panels a and b. The local ion concentration is expressed in molals to avoid uncertainties associated with defining the local volume of a nanopore containing a strand of DNA.
For both M1 MspA and WT αHL nanopores, a negatively charged DNA backbone inside the nanopores ensures that the total ionic current is primarily carried by potassium ions (87% of total current in MspA and >90% of total current in αHL). With the additional positive charges in the triple arginine αHL mutant, the current of K+ drops to zero while the current of Cl− remains at the same (negligible) level (Figure 4-b). In contrast, both K+ and Cl− currents increase when positively charged arginine side chains are introduced near the constriction of MspA (Figure 4-a). The change in the ion density in both nanopores (Figures 4-e,f) reveals the microscopic effect of the positive charges. For αHL, the arginine substitutions deplete K+ ions in the stem of the channel and do not considerably affect the distribution of Cl− (Figure 4-e). However, for MspA, the arginine substitutions considerably increase Cl− concentration in the vestibule of MspA without altering the density of K+ near the trans entrance of the nanopore. The difference in the effects arises from the shape of the channels. The stem of αHL is so narrow that the negatively charged backbone of DNA serves as a poly-counterion to the positively charged side chain of the substitutions and vice versa, excluding ions. In contrast, the vestibule of MspA is considerably wider than the Debye length at 1M KCl, which allows arginine side chains at one side of the channel to interact with DNA, while the remaining side chains to attract Cl− ions. The open geometry combined with the arginine substitutions allows an increase in the overall concentration of charge carriers in the channel, thereby increasing the residual current.
To demonstrate that arginine mutations in MspA do not eliminate the nucleotide sensitivity of ionic current blockades, we repeated our ensemble simulations of the M1, L88R and the two triple mutant nanopores using thymine homopolymers. Similar to the poly(dC) systems, we observed reduction of ssDNA translocation velocity in the poly(dT) systems, Figure S6. The average currents recorded for poly(dC) and poly(dT) strands and the four MspA systems are specified in Table 1. In the case of M1 MspA, the ensemble-average current of the poly(dT) systems is considerably greater than that of poly(dC), in agreement with experiment14. Similar currents are recorded in the case of M1 L88R mutant. Rather surprisingly, the blockade current difference between poly(dT) and poly(dC) systems in the triple arginine mutants differs from those in the M1 and M1 L88R systems. In the case of L88R/A83R/S116R, the current blockade difference is within the statistical error, however, in the case of L88R/A96R/S116R, the ensemble-average current of the poly(dC) systems is considerably greater than that of the poly(dT) systems, opposite to what is observed in M1 MspA. Such a reversal of the blockade current difference cannot be directly linked to the increased current of chloride ions, as the fractional currents carried by ionic species are similar for the two DNA sequences.
Table 1.
Averaged over 16 independent simulation systems blockade current for the four MspA variants and poly(dC) and poly(dT) homopolymers.
| poly(dC) | poly(dT) | |||||
|---|---|---|---|---|---|---|
| System | I(K+)(pA) | I(Cl−) (pA) | I(total) | I(K+)(pA) | I(Cl−) (pA) | I(total) |
| M1 | 111±6 | 16±2 | 127±8 | 141±9 | 31±3 | 172±11 |
| M1 L88R | 87±4 | 33±2 | 120±6 | 117±7 | 49±4 | 166±11 |
| M1 L88R/T83R/S116R | 110±8 | 54±5 | 164±13 | 98±12 | 47±8 | 145±20 |
| M1 L88R/A96R/S116R | 140±10 | 65±5 | 205±15 | 114±10 | 53±9 | 167±17 |
In general, the charge state of a nanopore can have a non-trivial effect on the overall transport rate of a charged analyte. Thus, introducing negatively charged side chains in the lumen of alpha-hemolysin was shown to both catalyze the transport of short positively charged peptides 39 and slow the transport of longer positively charged polypeptides.40 Hence our arginine mutations to MspA may both increase the DNA capture12 and slow the DNA transport.
Conclusions
In this study, we performed extensive (~100 μs in total) all-atom MD simulations to investigate electric field-driven transport of single DNA strands and ions through biological nanopore MspA containing arginine mutations. In nanopores decorated with the positive charges above the constriction, persistent contacts between the arginine substitution and DNA backbone, or by base-stacking interactions, could slow the DNA translocation by a factor 10–30. Because of MspA’s geometry, the charge substitutions did not eliminate nucleotide-specific current. These results reveal the importance of nanopore geometry and charge location and may help direct the modification of nanopores such as MspA to improve their utility for nanopore sequencing and other nanopore technologies. Furthermore, our simulations demonstrate that precise placement of arginine mutations can have a non-trivial effect40 on the sequence specificity of the ionic current blockades.
METHODS
General MD Methods
The MD simulations were carried out using program NAMD241 on Cray XT-5 machines or using D.E. Shaw Research’ special purpose Anton supercomputer.37 All simulations employed periodic boundary conditions and multiple timestepping41, 42 wherein local interactions were calculated every 2 fs time step and full electrostatic evaluations were performed every 3 time steps. NAMD simulations employed the particle mesh Ewald method for evaluation of long-range electrostatics.41, 42 Simulations on Anton were performed using the Nose’-Hoover NVT integrator with a k-Gaussian Split Ewald method43 for long-range electrostatics. The CHARMM2744 parameter set, along with TIP3P45 water model were used in all simulations. All covalent bonds involving hydrogen were kept rigid in water and other molecules using SETTLE46 or RATTLE47 algorithms. The cutoff and switching distance for evaluation of van der Waals and short-range electrostatic interactions were set to 8 and 7 Å, respectively.48, 49
After a 2000 step minimization, each systems was equilibrated using NAMD for at least 2 ns in the NPT ensemble using Nose’-Hoover Langevin piston pressure control5 at 295 K with no external electric field applied. Following equilibration, all NAMD simulations were performed in the NVT ensemble using the Lowe Andersen thermostat51 with a voltage bias of 180 mV imposed across the membrane as described in.34, 35 In all simulations, harmonic restraints with a spring constant of 695 pN/nm applied to all Ca atoms of the protein, restraining them to the crystallographic coordinates. Additional simulations were carried out on Anton to obtain a continuous 4-μs long trajectory of ssDNA transport through the truncated L88R-NNN/T83R/S116R nanopore, our trajectories each of 400 ns for the M1 L88R nanopore and nine trajectories for the M1 nanopore varying between 200 and 360 ns. Each Anton simulation was a continuation of the corresponding NAMD trajectory and employed the same force fields and constraints.
Simulation Setup
Full-length MspA
Atomic coordinates of the MspA porin were obtained from the Protein Data Bank (entry 1UUN)5. Eight R96 residues resolved in the crystal structure were changed to alanines in accord with the sequence of WT MspA. 24 aspartate residues in the constriction of the porin were replaced by asparagines to create the D20N/D91N/D93N mutant referred to as M1-NNN MspA. The resulting structure was merged with a 12×12 nm2 patch of POPC lipid bilayer such that the hydrophobic β-barrel of MspA embedded in the bilayer while its hydrophilic cap protruded above the membrane. All lipid molecules overlapping with the nanopore were removed; the remaining bilayer contained 272 lipid molecules. Using the phantom nanopore method,33 a 58 -nucleotide random sequence DNA strand was placed inside the nanopore of MspA with the backbone approximately aligned with the nanopore axis and the 3′ end of the strand emerging at the trans end of the nanopore. The system was immersed in ~35,000 pre-equilibrated TIP3P water molecules. K+ and Cl− ions were added at random positions in the water layers corresponding to a concentration of 1 M. Additional K+ ions were added to compensate the charge of the MspA nanopore and the DNA fragment, bringing the total charge of the final system to zero. The final system, shown in Figure 1-a, consisted of ~167,000 atoms and measured 12 nm × 12 nm × 17 nm.
Reduced-length MspA
The minimal model of the MspA nanopore was constructed by eliminating the vestibule part of MspA that extended away from the lipid membrane on the cis side in our full-length model (Figure 1a–c). The reduced nanopore contained residues 75 to 120. An 8 nm × 8 nm patch POPC bilayer containing 60 lipid molecules was merged with the truncated nanopore. An ensemble of sixteen systems, each differing in the initial conformation of the DNA, was created. The sixteen different DNA configurations were obtained from a ~110-ns trajectory of full-length MspA at a 1.2~V transmembrane bias. Each DNA/protein/lipid system was solvated in a volume of pre-equilibrated TIP3P water containing about 3800 molecules. DNA segments that extended outside the water box were deleted, the nucleotide sequence of the DNA strand was changed to comprise a cytosine (or thymine) homopolymer. K+ and Cl− ions were added at random positions to 1 M concentration and bring the total charge of each system to zero. Following usual minimization and equilibration protocol, the ends of each DNA strand were brought together (across the periodic boundary) in a 240 ps SMD simulation.52 Subsequently, the ends were covalently joined via a custom patch to create an effectively infinite DNA strand joined through periodic boundary conditions. The electrostatic profiles in full-length and reduced MspA systems, Figure 1-b, appear to be very similar, and so are the forces applied to DNA and ions, which validates our reduced nanopore approach.
In addition to M1-NNN MspA nanopore, the following three additional variations were constructed: a single mutant L88R-NNN, with all LEU88 replaced by arginines, and two triple mutants L88R/T83R/S116R-NNN and L88R/A96R/S116R-NNN where all LEU88, THR83, SER 116 or LEU88, ALA96, SER116 were replaced by arginines. Figure 2(b–d) illustrates the location of the mutations. Each reduced system consisted of about 28,000 atoms. Using custom scripts, all sixteen systems of the ensemble could be simulated in parallel on an XT-5 system, producing ~1 μs of aggregate simulation time within 24 hours.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (HG-005115) and the National Science Foundation (DMR-095595). Computer time was provided via Teragrid/XSEDE allocation MCA05S028, Department of Energy INCITE program and the National Resource for Biomedical Supercomputing at the DESRES supercomputer Anton at the Pittsburgh Supercomputing Center (PSCA00052). A. A. would like thank the Department of Bionanosciences at Delft University for hospitality and support from the Netherlands Organisation for Scientific Research (NWO)
Footnotes
Supporting Information Available: MD simulations of full-length MspA and its mutants; molecular graphics images of the sixteen ensemble systems; plots of number of DNA-arginine contacts vs permeation rate; plots of the number of persistent contacts vs time for the two triple mutants; plots of the number of electrostatic and stacking contacts; ensemble permeation rates of the thymine homopolymers; animations illustrating DNA permeation trajectory in M1-NNN and triple mutant MspA nanopores. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dekker C. Solid-State Nanopores. Nat Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 2.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, et al. The Potential Challenges of Nanopore Sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of Individual Polynucleotide Molecules using a Membrane Channel. Proc Natl Acad Sci U S A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song LZ, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of Staphylococcal Alpha-Hemolysin, A Heptameric Transmembrane Pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 5.Faller M, Niederweis M, Schulz GE. The Structure of a Mycobacterial Outer-Membrane Channel. Science. 2004;303:1189–1192. doi: 10.1126/science.1094114. [DOI] [PubMed] [Google Scholar]
- 6.Li HB, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM. Point Mutations Alter The Mechanical Stability Of Immunoglobulin Modules. Nat Struct Biol. 2000;7:1117–1120. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 7.Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Fabrication Of Solid-State Nanopores With Single-Nanometre Precision. Nat Mater. 2003;2:537–540. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 8.Heng JB, Ho C, Kim T, Timp R, Aksimentiev A, Grinkova YV, Sligar S, Schulten K, Timp G. Sizing DNA using a Nanometer-Diameter Pore. Biophys J. 2004;87:2905–2911. doi: 10.1529/biophysj.104.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garaj S, Hubbard W, Reina A, Kong J, Branton D, Golovchenko JA. Graphene as a Subnanometre Trans-Electrode Membrane. Nature. 2010;467:190–U73. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merchant CA, Healy K, Wanunu M, Ray V, Peterman N, Bartel J, Fischbein MD, Venta K, Luo Z, Johnson ATC, Drndic M. DNA Translocation through Graphene Nanopores. Nano Lett. 2010;10:2915–2921. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 11.Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen HW, Vandersypen LMK, Dekker C. DNA Translocation through Graphene Nanopores. Nano Lett. 2010;10:3163–3167. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 12.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-Molecule DNA Detection With An Engineered Mspa Protein Nanopore. Proc Natl Acad Sci U S A. 2008;105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrington IM, Butler TZ, Collins MD, Manrao E, Pavlenok M, Niederweis M, Gundlach JH. Nanopore DNA Sequencing with MspA. Proc Natl Acad Sci U S A. 2010;107:16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manrao EA, Derrington IM, Pavlenok M, Niederweis M, Gundlach JH. Nucleotide Discrimination with DNA Immobilized in the MspA Nanopore. PloS One. 2011:6. doi: 10.1371/journal.pone.0025723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndic M. Rapid Electronic Detection of Probe-Specific MicroRNAs using Thin Nanopore Sensors. Nat Nanotechnol. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 16.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond Time-Scale Discrimination among Polycytidylic Acid, Polyadenylic Acid, and Polyuridylic Acid as Homopolymers or as Segments within Single RNA Molecules. Biophys J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Hout M, Skinner GM, Klijnhout S, Krudde V, Dekker NH. The Passage of Homopolymeric RNA through Small Solid-State Nanopores. Small. 2011;7:2217–2224. doi: 10.1002/smll.201100265. [DOI] [PubMed] [Google Scholar]
- 18.Schibel AEP, An N, Jin Q, Fleming AM, Burrows CJ, White HS. Nanopore Detection of 8-Oxo-7, 8-dihydro-2′-deoxyguanosine in Immobilized Single-Stranded DNA via Adduct Formation to the DNA Damage Site. J Am Chem Soc. 2010;132:17992–17995. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace EVB, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, Bayley H. Identification of Epigenetic DNA Modifications with a Protein Nanopore. Chem Commun. 2010;46:8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keyser UF. Controlling Molecular Transport through Nanopores. J R Soc, Interface. 2011;8:1369–1378. doi: 10.1098/rsif.2011.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luan B, Stolovitzky G, Martyna G. Slowing and Controlling the Translocation of DNA in a Solid-State Nanopore. Nanoscale. 2012;4:1068–1077. doi: 10.1039/c1nr11201e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawano R, Schibel AEP, Cauley C, White HS. Controlling the Translocation of Single-Stranded DNA through alpha-Hemolysin Ion Channels using Viscosity. Langmuir. 2009;25:1233–1237. doi: 10.1021/la803556p. [DOI] [PubMed] [Google Scholar]
- 23.de Zoysa RSS, Jayawardhana DA, Zhao Q, Wang D, Armstrong DW, Guan X. Slowing DNA Translocation through Nanopores Using a Solution Containing Organic Salts. J Phys Chem B. 2009;113:13332–13336. doi: 10.1021/jp9040293. [DOI] [PubMed] [Google Scholar]
- 24.Fologea D, Uplinger J, Thomas B, McNabb DS, Li JL. Slowing DNA Translocation in a Solid-State Nanopore. Nano Lett. 2005;5:1734–1737. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meller A, Branton D. Single Molecule Measurements of DNA Transport through a Nanopore. Electrophoresis. 2002;23:2583–2591. doi: 10.1002/1522-2683(200208)23:16<2583::AID-ELPS2583>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 26.Cockroft SL, Chu J, Amorin M, Ghadiri MR. A Single-Molecule Nanopore Device Detects DNA Polymerase Activity with Single-Nucleotide Resolution. J Am Chem Soc. 2008;130:818. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olasagasti F, Lieberman KR, Benner S, Cherf GM, Dahl JM, Deamer DW, Akeson M. Replication of Individual DNA Molecules under Electronic Control using a Protein Nanopore. Nat Nanotechnol. 2010;5:798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howorka S, Cheley S, Bayley H. Sequence-Specific Detection of Individual DNA Strands Using Engineered Nanopores. Nat Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 29.Soni GV, Meller A. Progress toward Ultrafast DNA Sequencing using Solid-State Nanopores. Clin Chem. 2007;53:1996–2001. doi: 10.1373/clinchem.2007.091231. [DOI] [PubMed] [Google Scholar]
- 30.Rincon-Restrepo M, Milthallova E, Bayley H, Maglia G. Controlled Translocation of Individual DNA Molecules through Protein Nanopores with Engineered Molecular Brakes. Nano Lett. 2011;11:746–750. doi: 10.1021/nl1038874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirsaidov U, Comer J, Dimitrov V, Aksimentiev A, Timp G. Slowing the Translocation of Double-Stranded DNA using a Nanopore smaller than the Double Helix. Nanotechnology. 2010 doi: 10.1088/0957-4484/21/39/395501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niederweis M. Mycobacterial Porins - New Channel Proteins in Unique Outer Membranes. Mol Microbiol. 2003;49:1167–1177. doi: 10.1046/j.1365-2958.2003.03662.x. [DOI] [PubMed] [Google Scholar]
- 33.Mathe J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Orientation Discrimination of Single-Stranded DNA inside the Alpha-Hemolysin Membrane Channel. Proc Natl Acad Sci U S A. 2005;102:12377–12382. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aksimentiev A. Deciphering Ionic Current Signatures of DNA Transport through a Nanopore. Nanoscale. 2010;2:468–483. doi: 10.1039/b9nr00275h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aksimentiev A, Schulten K. Imaging alpha-Hemolysin with Molecular Dynamics: Ionic Conductance, Osmotic Permeability, and the Electrostatic Potential Map. Biophys J. 2005;88:3745–3761. doi: 10.1529/biophysj.104.058727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wells DB, Abramkina V, Aksimentiev A. Exploring Transmembrane Transport through Alpha-Hemolysin with Grid-Steered Molecular Dynamics. J Chem Phys. 2007;127:125101. doi: 10.1063/1.2770738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, Salmon JK, Young C, Batson B, Bowers KJ, Chao JC, et al. Anton a Special-Purpose Machine for Molecular Dynamics Simulation. Comm ACM. 2008;51:91–97. [Google Scholar]
- 38.Guallar V, Borrelli KW. A Binding Mechanism in Protein-Nucleotide Interactions: Implication For U1A RNA Binding. Proc Natl Acad Sci U S A. 2005;102:3954–3959. doi: 10.1073/pnas.0500888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammad MM, Movileanu L. Excursion of a Single Polypeptide into a Protein Pore: Simple Physics, but Complicated Biology. Eur Biophys J. 2008;37:913–925. doi: 10.1007/s00249-008-0309-9. [DOI] [PubMed] [Google Scholar]
- 40.Mohammad MM, Prakash S, Matouschek A, Movileanu L. Controlling a Single Protein in a Nanopore through Electrostatic Traps. J Am Chem Soc. 2008;130:4081–4088. doi: 10.1021/ja710787a. [DOI] [PubMed] [Google Scholar]
- 41.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable Molecular Dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batcho PF, Case DA, Schlick T. Optimized Particle-Mesh Ewald/Multiple-Time Step Integration for Molecular Dynamics Simulations. J Chem Phys. 2001;115:4003–4018. [Google Scholar]
- 43.Shan YB, Klepeis JL, Eastwood MP, Dror RO, Shaw DE. Gaussian split Ewald: A Fast Ewald Mesh Method for Molecular Simulation. J Chem Phys. 2005:122. doi: 10.1063/1.1839571. [DOI] [PubMed] [Google Scholar]
- 44.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-Atom Empirical Potential for Molecular Modeling Dynamics Studies of Proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 45.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison Of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 46.Miyamoto S, Kollman PA. SETTLE - An Analytical Version of the SHAKE and RATTLE Algorithm for Rigid Water Models. J Comput Chem. 1992;13:952–962. [Google Scholar]
- 47.Andersen HC. RATTLE - A Velocity Version of the SHAKE Algorithm for Molecular Dynamics Calculations. J Comput Phys. 1983;52:24–34. [Google Scholar]
- 48.Kowalczyk SW, Wells DB, Aksimentiev A, Dekker C. Slowing down DNA Translocation through a Nanopore in Lithium Chloride. Nano Lett. 2012;12:1038–44. doi: 10.1021/nl204273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wells DB, Bhattacharya S, Carr R, Maffeo C, Ho A, Comer J, Aksimentiev A. Optimization of the molecular dynamics method for simulations of DNA and ion transport through biological nanopores. Methods in molecular biology (Clifton, NJ) 2012;870:165–86. doi: 10.1007/978-1-61779-773-6_10. [DOI] [PubMed] [Google Scholar]
- 50.Martyna GJ, Tobias DJ, Klein ML. Constant-Pressure Molecular Dynamics Algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 51.Koopman EA, Lowe CP. Advantages Of A Lowe-Andersen Thermostat In Molecular Dynamics Simulations. J Chem Phys. 2006:124. doi: 10.1063/1.2198824. [DOI] [PubMed] [Google Scholar]
- 52.Isralewitz B, Izrailev S, Schulten K. Binding Pathway Of Retinal To Bacterio-Opsin; A Prediction By Molecular Dynamics Simulations. Biophys J. 1997;72:TU410–TU410. doi: 10.1016/S0006-3495(97)78326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.