Abstract
Paper-based analytical devices are the subject of growing interest for the development of low-cost point-of-care diagnostics, environmental monitoring technologies and research tools for limited-resource settings. However, there are limited chemistries available for the conjugation of biomolecules to cellulose for use in biomedical applications. Herein, divinyl sulfone (DVS) chemistry was demonstrated to covalently immobilize small molecules, proteins and DNA onto the hydroxyl groups of cellulose membranes through nucleophilic addition. Assays on modified cellulose using protein-carbohydrate and protein-glycoprotein interactions as well as oligonucleotide hybridization showed that the membrane’s bioactivity was specific, dose-dependent, and stable over a long period of time. Use of an inkjet printer to form patterns of biomolecules on DVS-activated cellulose illustrates the adaptability of the DVS functionalization technique to pattern sophisticated designs, with potential applications in cellulose-based lateral flow devices.
Keywords: paper, covalent immobilization, divinyl sulfone, cellulose, inkjet
INTRODUCTION
Lateral flow (LF) has been widely used for the operation of assays in commercial and research applications, particularly as an ideal solution for self-powered, rapid fluid flow for low-cost, distributed diagnostic needs.1–3 For instance, nitrocellulose-based diagnostic strips employ LF for the transport of solutions over immobilized biomolecules (often antibodies) for analyte detection.4 Cellulose, a cheaper and non-hazardous alternative to nitrocellulose, has also recently attracted interest as a LF-based diagnostic platform for low-resource settings.5–8 Advantages of cellulose include low production costs, portability, durability, and excellent wicking ability of aqueous solutions via capillary action. Various cellulose-based diagnostic designs have been introduced, such as microfluidic paper analytical devices (μPADs), 2D-shaped LF strips, and thread-based networks.8–11 These designs vary in sophistication but typically feature isolated channels that wick aqueous solution to a test site; many operate multiple channels simultaneously to demonstrate multiplexing potential. For example, the simple branched design of the μPAD allows a drop of reagent to be transported via capillary action to multiple test sites, yielding colorimetric readouts within minutes. The low-cost and timely convenience demonstrated by the μPAD is a major advantage to operating diagnostics on cellulosic materials. However, further development of diagnostic devices on cellulose is limited by the need for a reliable strategy to immobilize bioactive molecules and impart selective and specific function to cellulose.
Physical adsorption is the simplest strategy for biomolecule immobilization onto solid substrates. In particular, immobilization of proteins onto supports such as polyvinylidene difluoride (PVDF) and nitrocellulose is well documented.12–14 However, substrates such as nitrocellulose are limited in their ability to bind to certain classes of biomolecules, and are generally deficient towards the capture of nucleic acids, carbohydrates, and other small molecules. This restricts the potential scope of nitrocellulose-based LF diagnostics. While proteins do interact with slightly anionic cellulose,15 physical adsorption is unlikely to be effective for biomolecules without cationic character, as they are less firmly bound and easily washed off. This is especially relevant when considering immobilization of non-proteinaceous biomolecules. Therefore, a more versatile material system capable of immobilizing a broader range of biomolecules could be desirable under certain circumstances.
Recently, significant attention has been directed towards the chemical modification of cellulose for biomolecule functionalization. For example, carbonyldiimidazole and 1-cyano-4-dimethylaminopyridinium tetrafluoroborate were used by Stollner et al. to immobilize glucose oxidase and a glycated pentapeptide onto a cellulose dialysis membrane to form part of an immunosensor.16 The Nahar group has developed a UV-based activation strategy using 1-fluoro-2-nitro-4-azidobenzene to generate nitrenes from the azido group and subsequently immobilize unmodified carbohydrates and proteins to the cellulose.17, 18 A chemoenzymatic strategy based on xyloglucan modification was developed by Brumer et al. for the efficient incorporation of chemical functionality onto crystalline cellulose surfaces.19 In a case of industrial interest, several methods for immobilizing invertase on Granocel cellulose carriers (e.g., NH-Granocel generated with pentaethylenehexamine, 1-chloro-2,3-expoxypropane sodium borohydride) have been explored to increase the enzyme’s activity.20 Kong et al. presented an epichlorohydrin method for coupling DNA aptamers to cellulose beads to form an immunoadsorbent that could remove anti-DNA antibodies from the plasma of systemic lupus erythematosus patients.21 Immobilized DNA has also been demonstrated to detect ATP by Su et al., sodium periodate was used to generate aldehyde groups on cellulose that could then bind DNA aptamers.22 Recently, Catarina et al. used the linking agent a 1,4-phenylenediisothiocyanate, to immobilize DNA onto filter paper for target hybridization via capillary action,23 Xu et al. detailed the fabrication of bioactive cellulose via heteropolysaccharide oxidation by galactose 6-oxidase,24 and Filpponen et al. incorporated click chemistry to modify cellulose for immobilization of biomolecules with azide and alkyne groups.25
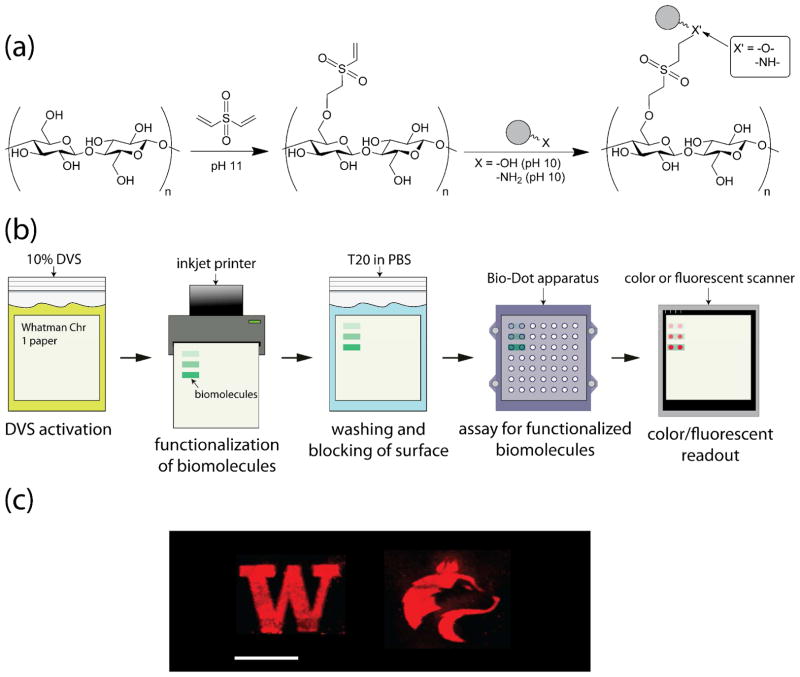
While the aforementioned covalent biomolecular immobilization strategies have successfully demonstrated the potential for chemical modification of cellulose, they are not without their limitations. Most methods require multiple modification steps, involve flammable and hazardous solvents, and are suitable only for specific classes of biomolecules. Therefore, a simple and versatile technique to immobilize a variety of biomolecules onto cellulose is highly desirable. To achieve this aim, we report the use of divinyl sulfone (DVS) in a simple, two-step method for the covalent modification of cellulose with an assortment of biomolecules. The homobifunctional DVS molecule contains two electrophilic vinyl groups which exhibit cross-linking activity with nucleophiles.26 It has been reported to cross-link agarose to increase enzyme-adsorption, and to modify sepharose with mannose to form an adsorbent for a mannose-binding protein.27, 28 In particular, Fornstedt et al. utilized DVS’s cross-linking ability to generate D-mannose-substituted sepharose to capture and purify a Vicia ervilia-derived lectin.28 Recently, our lab used DVS to functionalize hydroxyl-terminated gold surfaces and organophosphonate-coated silicon photonic microring resonators with glycans for biosensing applications.29, 30 Cellulose, which contains many hydroxyl groups on its backbone, presents an ideal surface for functionalization. The chemical strategy involves addition of one of the vinyl groups to the cellulose backbone. Application of nucleophile-bearing biomolecules, either by spotting or inkjet printing, to the remaining unreacted vinyl groups covalently attaches the biomolecules to the cellulose backbone. The derivatization process is illustrated in Scheme 1.
Scheme 1.
(a) Chemical activation and functionalization of cellulose with nucleophile-bearing biomolecules (gray). (b) Schematic of the stepwise process for paper activation via DVS, patterning of biomolecules via inkjet printing, and assaying of bioactivity. (c) Illustration of patterned biofunctional paper, demonstrating the visualization of immobilized carbohydrates via a fluorometric probe, scale bar: 1 cm.
In this article we report the functionalization of DVS-activated cellulose with unmodified and modified carbohydrates, glycoprotein, and DNA. Assays for these biomolecules using carbohydrate-protein interactions and oligonucleotide hybridization indicate that they are specifically immobilized onto the activated membrane. The functionalized membranes also exhibit dose-dependent response to probes, and stability to prolonged storage. Furthermore, we demonstrate that this biofunctional paper is amenable to the patterning of biomolecules with an inkjet printer using an existing bioprinting technique. This biofunctional paper will advance the application of cellulose-based bioassays as low-cost, portable and reliable devices for point-of-care diagnosis, environmental monitoring and biomedical research, particularly in limited-resource settings.
EXPERIMENTAL SECTION
Materials and Reagents
Chr 1 Chromatography paper (Whatman) was purchased from Sigma-Aldrich, St. Louis, MO. 97% DVS stabilized with 0.05% hydroquinone was purchased from Alfa Aesar, Ward Hill, MA. D-(+)-Mannose, D-(+)-Galactose, D-(+)-Maltose, D-(+)Glucose and D-(+)-Lactose, and Sucrose >98% pure powder were purchased from Sigma-Aldrich, St. Louis, MO. Aminopropyl mannoside and aminopropyl galactose were synthesized as previously described.31 Synthesis of the aminated hexafluorobenzamide is described in the supporting information (Scheme S1). Horseradish peroxidase (HRP)-labeled Concanavalin A (Con A-HRP) was purchased from Sigma-Aldrich, St. Louis, MO. HRP-labeled Ricinus communis (RCA120-HRP) was purchased from EY Laboratories, San Mateo, CA. Dylight 649 amine-reactive dye was purchased from Thermo Fisher Scientific, Rockford, IL. RNase B was purchased from New England Biolabs, Ipswich, MA. PBS buffer (2.7 mM KCl, 137 mM NaCl, 10 mM phosphate, pH 7.4) and HEPES buffer (10 mM HEPES, 150 mM NaCl, 1 mM Ca2+ and Mn2+, pH 7.4) were prepared in the lab. Two complementary oligonucleotide sequences were purchased from Invitrogen Corporation, Carlsbad, CA. The sequences (A and A′) are detailed in Table 1:
Table 1.
Oligonucleotide Sequences and Modifications
| 5′ modification | Sequence | 3′ modification | |
|---|---|---|---|
| A | −NH33 | CTGAACGGTAGCATCTTGAC | None |
| A′ | −NH33 | GTCAAGATGCTACCGTTCAG | None |
DVS Activation of Cellulose
A 10% DVS (v/v, 0.1 M sodium carbonate, pH 11) solution was used to chemically activate cellulose membranes; 0.1 M sodium carbonate buffer alone served as a control (referred to in the text as DVS−). 12.0 × 9.0 cm sheets of Chr 1 paper were immersed in either 20 mL 10% DVS solution or in buffer alone, incubated in separate 400 mL-capacity plastic zip-bags, and agitated for 2 h on a rocking shaker. Following the incubation, the DVS-activated (DVS+) and control membranes (DVS−) were removed from the bags and rinsed in a plastic tray with 100 mL Millipore-purified deionized water, three times. These membranes were dried for 2 h in ambient conditions. In cases where the activated membranes were stored, they were either placed in aluminum foil in ambient conditions to protect from light and dust, or stored in a dry, nitrogen-filled plastic box.
Functionalization of DVS-activated Cellulose Membrane
Free reducing sugars (at concentrations ranging from 292 to 1100 mM) and aminopropyl glycosides (at concentrations ranging from 1 to 10 mM) were dissolved in 50 mM sodium carbonate buffer (pH 10). The glycoprotein RNase B was dissolved at 7 mM in PBS. The oligonucleotide sequence A was dissolved at either 0.1 or 1 mM in PBS. 2 μL volumes of these biomolecule solutions were spotted on the DVS+ and DVS- membranes in a 96-well grid format using a custom-made 96-well microplate template. On both membranes one row was not functionalized, to serve as a control for the non-specific binding of probes during detection assays. The spotted sheets were placed in a 75% relative humidity (RH) box at room temperature to react overnight. The sheets were then immersed in 20 mL PBS-T (0.05% w/v Tween® 20 in PBS) for 1 h in plastic zip-bags to block the surface and remove unbound biomolecules.
Inkjet Functionalization of DVS+ Membranes with Galactose
To pattern free galactose on the DVS+ membrane, an Epson R280 printer was modified using a method developed by the Lutz laboratory of the University of Washington (mf20.org). Stock Epson ink cartridges were replaced with empty refill cartridges, and the ink channel of the black ink cartridge was hollowed out via a milling machine to allow a pipette tip to be fitted onto the printhead (in lieu of the cartridge’s ink channel). Bioprinting solution was injected into the pipette tip; this served as the bioprinting solution reservoir. We devised a bioprinting solution composed of 10% w/v galactose, 40% w/v sucrose and 5% w/v TWEEN® 20 in 0.1 M pH 10 sodium carbonate buffer. Prior to printing the solution, 5 mL of a priming solution composed of 50% w/v sucrose and 5% w/v TWEEN® 20 in Millipore water was passed through the printhead via a syringe. This printing preparation process is photographically documented in the Supporting Information. The printed sheet was placed in the 75% RH box to react overnight and blocked as previously described.
Surface Characterization of Modified Cellulose Membrane
X-ray photoelectron spectroscopy (XPS) composition data of the DVS+ and DVS− cellulose membranes were acquired on a Surface Science Instruments S-probe spectrometer equipped with a monochromatic Al-Kα ray source (hv = 1486.6 eV). Data were collected at 55° takeoff angle in the hybrid mode with approximately 5 nm sampling depth, using a pass energy of 150 eV. Three spots for each sample were analyzed. The reported data were averaged over multiple spots. Analysis was performed on the Service Physics ESCA2000 A analysis software (Bend, OR). To facilitate analysis, an aminated F-tag (hexafluorobenzamide, see supporting information) 2.5 mM in 0.1 M pH 10 sodium carbonate buffer was used to mimic the reaction of an amino-bearing sugar with the DVS+ membrane.
To prepare the membranes for scanning electron microscopy (SEM), samples of the DVS+ and DVS− membranes were coated with 5 nm of a gold-palladium alloy with an SPI Module™ sputter coater. SEM micrographs of the membranes were acquired with an FEI Sirion SEM at the University of Washington Nanotechnology User Facility, using a spot size of 3 and a 10 kV beam accelerating voltage.
Tensile Strength Characterization of Modified and Unmodified Cellulose
Untreated and DVS+ membranes were tested for their tensile strength using a Thwing Albert Model EJA II Universal Testing Machine at the University of Washington Paper Science and Engineering Laboratory. 12.0 cm × 3.0 cm sheets of the membranes were affixed onto the two grips of the machine and pulled until the sample experienced necking. The maximum of the stress-strain curve was defined as the tensile strength.
Preparation of Labeled Carbohydrate-Binding Protein Probes
Horseradish peroxidase (HRP)-labeled and Dylight 649-labeled carbohydrate-binding proteins (lectins) were employed to detect unmodified and modified carbohydrates and glycoproteins on the functionalized membrane. Con A-HRP in HEPES-T (0.05% TWEEN® 20 w/v in HEPES) interacts with immobilized mannose, maltose, glucose, aminated mannose, and RNase B.32 RCA120-HRP in PBS-T interacts with immobilized galactose, lactose, and aminopropyl galactose.33 For fluorescent detection, Con A and RCA120 lectins were labeled with Dylight 649 amine-reactive dye. Dylight 649 containing an N-hydroxysuccinimide (NHS) ester was added to the lectins in the appropriate buffer (e.g., Con A in HEPES), incubated in an orbital shaker for 1 h at room temperature, and dialyzed for 16 h against the appropriate buffer (e.g., Dylight 649-labeled Con A against HEPES) to remove unreacted dye.
Preparation of Labeled Nucleotide Probes
Dylight 649-labeled oligonucleotide sequences (A and A′) in hybridization buffer (0.3 M NaCl, 20 mM phosphate, 2 mM EDTA, 0.6 M sodium dodecyl sulfate) were employed to detect immobilized sequences (refer to Scheme S2 for additional detail on the use of the nucleotide probes). Both sequences were labeled with Dylight 649 fluor on their 5′ ends, as described for labeling the carbohydrate-binding proteins.
Colorimetric Detection of Immobilized Biomolecules
Colorimteric assays of the activated and functionalized cellulose membranes were operated on the Bio-Dot apparatus (Bio-Rad laboratories, Hercules, CA). Reagents were added successively to the sheets; each reagent was vacuumed through at 5.0 in Hg pressure before addition of the next reagent (see Figure S6 for more detail on the Bio-Dot setup). 100 μL/well of PBS was first added to rehydrate the sheet. 200 μL/well of HRP-labeled probe solution was added and vacuumed through, and the membrane was incubated for 10 minutes to allow the probes to bind to the immobilized biomolecules. Following incubation, 1200 μL/well of washing buffer (0.05% w/v TWEEN® 20, 0.1% w/v BSA in PBS) was vacuumed through to rinse unbound probes and block the surface. 100 μL/well of tetramethylbenzidine (TMB, Gaithersburg, MD) was added and vacuumed through to obtain a colorimetric readout (the reaction between HRP and TMB produces a blue colored substrate which is apparent on the paper); the color developed in approximately one minute. The sheet was then scanned on a Canon LiDE 600F desktop scanner (Canon Inc., Lake Success, NY).
Fluorometric Detection of Immobilized Biomolecules
Fluorometric assays of the activated and functionalized cellulose membranes were operated on the Bio-Dot apparatus. Reagents were added successively to the sheets; each reagent was vacuumed through at 5.0 in Hg pressure before addition of the next reagent. 100 μL/well of PBS was first added to rehydrate the sheet. 200 μL/well of Dylight 649-labeled probe solution was added and vacuumed through, and the membrane was incubated for 10 minutes to allow the probes to bind to the immobilized biomolecules. Then 1200 μL/well of washing buffer (0.05% w/v TWEEN® 20, 0.1% w/v BSA in PBS) was vacuumed through to wash off unbound probes and block the surface. The sheet was scanned on a Storm 865 system (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) to obtain a fluorometric readout.
Analysis of Assayed Membrane
Scanned colorimetric intensities were grayscaled, inverted, and analyzed with ImageJ (NIH, Bethesda, MD). Circular regions of interest (ROIs) defined by the approximately 0.45 cm-diameter wells of the Bio-Dot apparatus, were analyzed for the Integrated Density (ID) (additional details on ROI selection can be found in the Supporting Information). Scanned fluorometric intensities were analyzed with ImageQuant TL (GE Healthcare) with circular ROIs (also defined by the Bio-Dot wells) for fluorescent intensity. Mean IDs, fluorescent intensities, and standard deviations for spots immobilized with the same biomolecule were calculated in Microsoft Excel. When appropriate, Student’s t-tests were employed at p = 0.05 to determine whether intensities of biomolecule spots on the membranes were statistically higher than the control row, and whether intensities on the DVS+ membrane were higher than the corresponding spots on DVS- membranes. Limit of detection (LOD) was determined via a Student’s t-test (p = 0.05) for a significant difference in intensity between spots tested with a given concentration of probe and those tested with buffer only (absence of probe).
RESULTS AND DISCUSSION
Surface Characterization
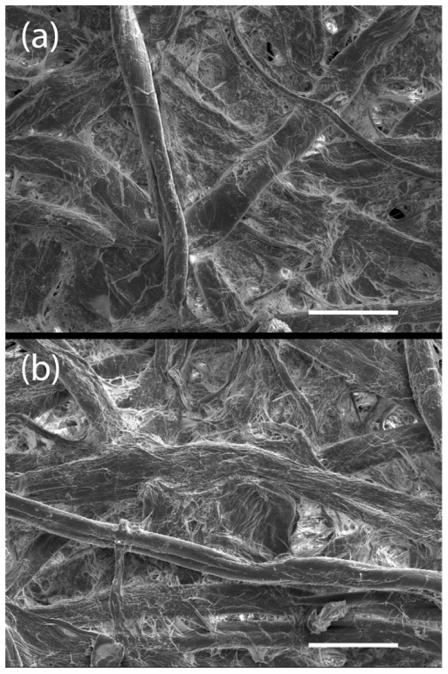
XPS surface analysis was performed to verify DVS-activation and functionalization. As shown in the XPS elemental composition results (Table 2), only carbon and oxygen were detected at the interface of unmodified cellulose membranes. After DVS-activation, the appearance of a sulfur signal in the XPS spectrum indicates that DVS successfully modified the cellulose substrate. Based on the S/O ratio, the DVS activation efficiency is estimated to be ~16%. A fluorine-containing aminated probe (F-tag, Figure S1) was used to mimic the reaction of amino-bearing biomolecules with DVS+ cellulose. After application of the F-tag, fluorine could be observed by XPS on the DVS-activated (DVS+) membrane. Based on the quotient of the observed F/S ratio by the theoretical maximum F/S ratio (each DVS bound to cellulose binds with one F-tag), 19% of the DVS molecules were observed to have reacted with the F-tag. Drawing upon the determined DVS activation and F-tag reaction efficiencies, the XPS measurements suggest that ~3% of the cellulose units are functionalized with F-tag. However, this value represents the total mol% of modified cellulose within roughly 10 nm of the surface of the cellulosic fiber. The percent of F-tag modified cellulose units on the surface of the fiber is predicted to be significantly higher, as the XPS measurement includes an attenuated signal from the bulk carbohydrate substrate. This discrepancy between the percent F-tag modification of the surface versus the bulk cellulose is due to the crystalline nature of cellulose, which restricts chemical modification beyond the surface of the fiber. As illustrated in Figure 1, cellulosic paper is comprised of tightly bundled insoluble fibrils displaying semi-impermeable surfaces available for chemical modification with limited access to the bulk material.
Table 2.
Relative Compositions of Functionalized Cellulose Determined by XPS
| Cellulose | Cellulose + DVS | Cellulose + DVS + F-tag | Cellulose + F-tag | |
|---|---|---|---|---|
| C 1s | 57.7 ± 0.2 | 58.6 ± 0.6 | 56.9 ± 0.1 | 58.0 ± 1.6 |
| O 1s | 42.3 ± 0.2 | 40.0 ± 0.8 | 40.5 ± 0.3 | 42.0 ± 1.6 |
| S 2p | n/d | 1.4 ± 0.2 | 1.2 ± 0.3 | n/d |
| F 1s | n/d | n/d | 1.4 ± 0.2 | n/d |
Note: n/d = not detected
Figure 1.
SEM micrographs at 500x magnification of cellulose membranes: (a) DVS− cellulose membrane (b) DVS+ cellulose membrane. Scale bar: 50 μm (Photo credit, Lauren Cummings).
On the unmodified membrane that was treated only with the buffer (DVS−) and spotted with F-tag, no fluorine was detected, which demonstrates the dependence on DVS for immobilization to the cellulose substrate. This data validated the ability of the DVS conjugation strategy to covalently immobilize amine-bearing molecules on DVS-activated paper.
SEM micrographs illustrate the morphology of DVS+ cellulose compared with the DVS− control membrane. No apparent difference in morphology or fibrosity was observed between the two samples at magnifications ranging from 32x to 500x. A comparison of the micrographs at 500x is depicted in Figure 1. These similarities suggest that DVS modification results in little to no alteration of the gross morphologic characteristics of cellulose fibers as a result of DVS cross-linking. These results suggest that DVS-modification will have minimal impact on the capillarity and LF suitability of the DVS-activated cellulose membranes. Tensile strength measurements of modified and unmodified cellulose showed a modest change in the material properties of the paper, with a decrease in tensile strength from 49±3 N/m2 to 34±3 N/m2 (DVS+). However, the modified paper demonstrated no significant difference under routine handling, printing, and assay development.
Detection of Carbohydrate-Protein Interactions
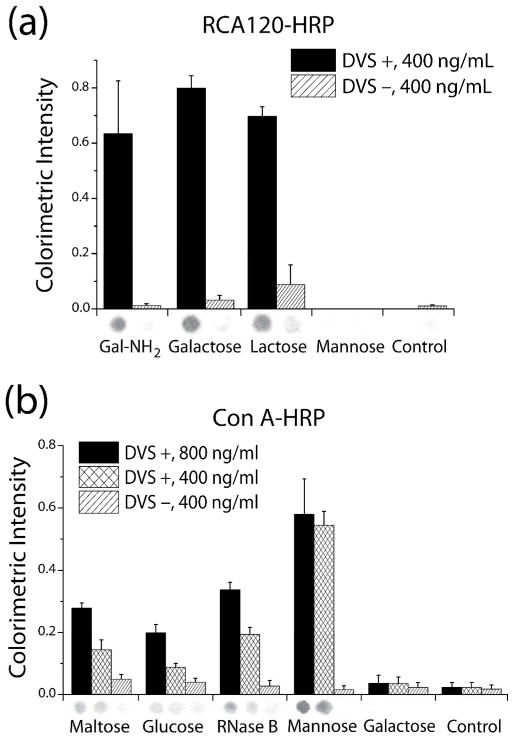
To examine the capabilities and versatility of DVS-activated cellulose in conjugating bioactive molecules, we immobilized unmodified, aminated oligosaccharides and glycoprotein on cellulose membranes and determined the bioactivity via colorimietric and fluorometric lectin-binding assays. The results of the colorimetric assays in Figure 2 show the binding response of two lectins to DVS-immobilized glycans vs. glycan non-specifically adsorbed on DVS− (control) paper. On the DVS+ papers, ricin (RCA120) exhibited specific binding to the carbohydrates containing a β-galactose moiety (including aminopropyl galactose, galactose, and lactose), but no response to glycans with mannose residues (including mannose and RNaseB, Figure 2a). By contrast, Con A only shows binding to Con A-specific glycans (including mannose, glucose, maltose and RNase B, Figure 2b). The control rows on DVS+ membranes show little to no binding of either lectin, which demonstrate that non-specific binding is negligible. On the control membrane (DVS−) both lectins show minimal non-specific binding to physisorbed glycans. These results suggest that the carbohydrate-protein interactions documented on the DVS+ membranes are specific.
Figure 2.
(a) Galactose, lactose, and aminopropyl galactose (gal-NH2) were probed with 400 ng/mL RCA120-HRP; glycan bioactivity was observed on the DVS+ membrane but not on the DVS− membrane. There was no signal from the mannose and control (no glycan functionalized) spots, indicating that non-specific binding of RCA120 was negligible. (b) Maltose, glucose, RNase B, and mannose were bound by Con A-HRP on the DVS+ membrane but not the DVS− membrane. There was little signal from the galactose and control spots, indicating that non-specific binding of Con A was negligible. Representative colorimetric spots from the membranes appear below each corresponding bar (data were averaged over 6 spots for each carbohydrate).
Dose-dependent responses from the galactose-RCA120 and mannose-Con A interactions were observed when varying the concentration of both lectins. For RCA120-HRP, response saturated at approximately 500 ng/mL (4.8 nM), with the response of the lowest concentration tested (100 ng/mL, or 0.96 nM) well above background (Figure S8a). For Con A-HRP, response saturated at approximately 1000 ng/mL (9.6 nM), while the limit of detection (LOD) was determined to be 10 ng/mL (96 pM) (Figure S8b).
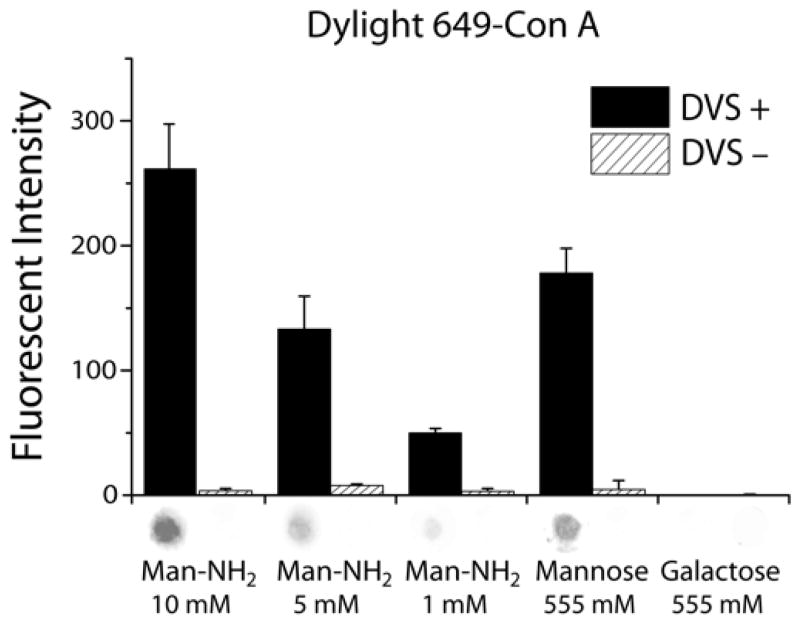
The fluorometric assays for immobilized carbohydrates and glycoprotein demonstrated the same specific binding observed in the colorimetric analysis. Figure 3 depicts specific binding of aminopropyl mannoside, mannose and galactose (negative control) by Dylight 649-labeled Con A. As previously observed, amino-modified glycans show significantly higher conjugation efficiency, as indicated by their increased bioactivity at a given immobilization concentration.29
Figure 3.
Aminopropyl mannoside (man-NH2) and mannose were probed with 50 nM Dylight 649-labeled Con A; bioactivity was observed on the DVS+ membrane but not on the DVS− membrane. There was no signal from the galactose spots, as expected. Representative fluorometric spots from the membranes appear below each corresponding bar (data were averaged over 6 spots for each carbohydrate).
Both the colorimetric and fluorometric assays demonstrate that DVS-modified cellulose paper is suitable for conjugation of carbohydrates and analysis of biological interactions using various means of detection. Aminated and hydroxylated biomolecules were successfully conjugated onto the paper with apparent efficiencies reflecting their nucleophilicity. These interactions have been demonstrated to be semi-quantitative, as variations in the concentration of immobilized biomolecules and the probing lectins yield statistically and visually significant differences in response.
It is worth noting that the DVS chemistry enables direct conjugation of free oligosaccharides onto cellulose through a vinylsulfone-hydroxyl reaction, dramatically reducing the burden of chemical modification of carbohydrates. While the selectivity of this reaction could be complicated by the multiple hydroxyls presented by a carbohydrate molecule, we previously conducted work demonstrating that the majority of free sugars are linked via the anomeric hydroxyl.29 To investigate the vinyl sulfone reaction preference for hydroxyl groups in saccharides we studied a model reaction of ethyl vinyl sulfone (EVS) and free mannose in solution. Using thin-layer chromatography and Nuclear Magnetic Resonance spectroscopy we found that ~90% of EVS-mannose products react at anomeric position, and the ratio of α- and β- isomers is around 3:1, which is close to the ratio of free mannose in solution.29
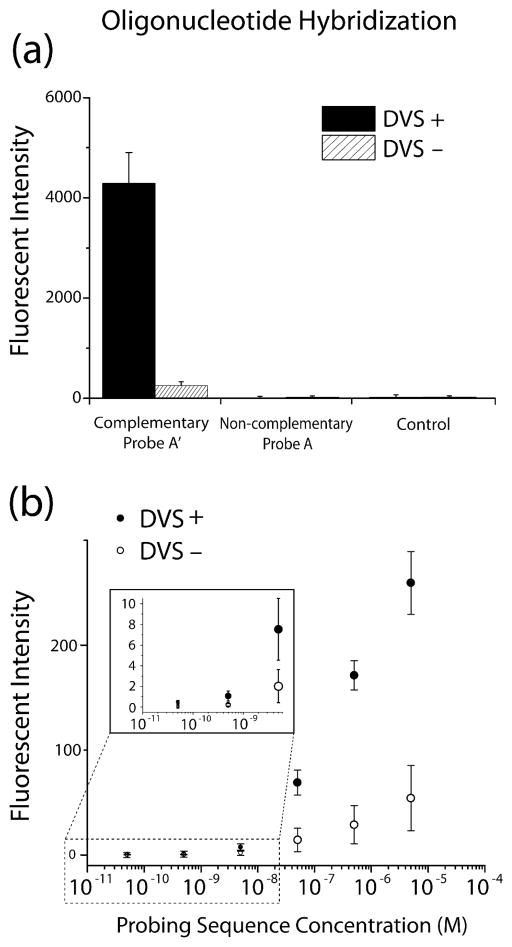
Detection of DNA Oligonucleotide Hybridization
Nucleic acid tests (NATs) are valued in clinical diagnostics for their ability to sense pathogens, by detecting the genetic material of viruses, bacteria and protozoa. The development of NATs in a lab-on-a-chip or paper-based format for low-resource settings has received considerable attention.2,6,22,34 To demonstrate the suitability of DVS-modified cellulose for DNA detection, we performed a simple hybridization assay (Scheme S2). We observed binding of a labeled sequence (A′) to spotted rows of the immobilized sequence (A) but not to the control row on DVS+ membranes (Figure 4a), indicating that DNA hybridization can be detected on the DVS-activated cellulose and non-specific binding of sequence A′ to the surface was negligible. A labeled non-complementary sequence (A) was incubated as a negative control on both the DVS+ and DVS− membranes, with no observed binding to the immobilized sequence (A) on either membrane. These results suggest that the incubated sequence A′ was specifically hybridized to immobilized sequence A on the DVS+ membrane. In a separate experiment, a dose-dependent response of the DNA-functionalized membrane was observed by varying the concentration of the probing sequence (A′) (Figure 4b). We observed a significant signal difference between the DVS+ and DVS− membranes, in accordance with the role played by DVS in covalent immobilization of the oligonucleotide target. The hybridization observed on the DVS− paper can be attributed to a low-level of physisorbed target sequence (non-covalent) to the unmodified cellulose. However, DVS yielded an order of magnitude higher response for the hybridization assay. The limit of detection (LOD) of this non-optimized system was found to be 500 pM. This result is on-par with those reported for short oligonucleotide hybridization on zirconia-modified filter paper (using FITC-labeled probe),35 and on gold using surface plasmon resonance imaging (SPRi) (unlabeled probe).36
Figure 4.
(a) Immobilized sequence (A) was probed by 5 μM of the complementary probe sequence (A′); bioactivity was observed on the DVS+ membrane but not the DVS− membrane. Labeled sequence (A) was also incubated as a negative control, and was not detected on either membrane. The control row (no oligonucleotides functionalized) also showed no response to incubated A′, indicating that non-specific binding of the sequence was negligible. (b) Dose-dependent detection of immobilized sequence (A) was observed for concentrations of probe (A′) from 0.5 nM to 5 μM.
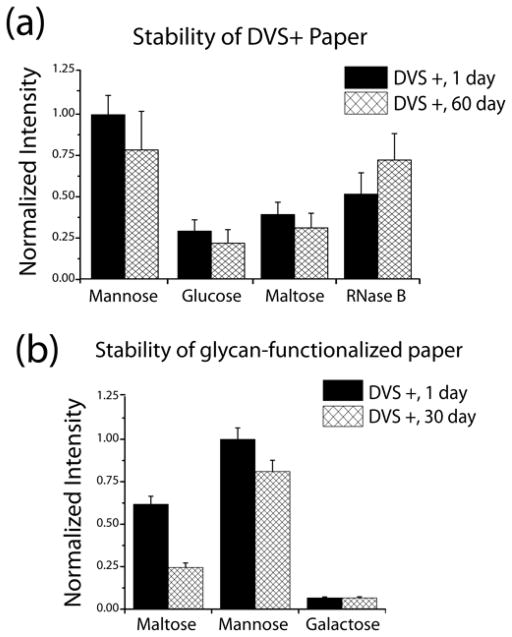
Stability of DVS-activated Cellulose
To develop a reliable paper-based platform suitable for future point-of-care applications, the stability of DVS-activated and biofunctional cellulose is critical. A cellulose-based LF platform for low-resource settings needs to maintain its stability for extended periods of time. To gain a better understanding of the stability of our DVS-cellulose platform, we tested the stability of DVS-activated membranes after 60 days of storage in a dry, non-sterile, nitrogen-filled environment. Carbohydrates and the glycoprotein RNase B were spotted on the DVS-modified cellulose membrane after the storage period and tested for specific binding to labeled Con A. The results in Figure 5a indicate that the activity of carbohydrates and glycoprotein spotted on the aged DVS-cellulose was very similar to that of freshly activated DVS-paper. Only minor variability was observed for the immobilized mannose. We ascribe these minor differences to physical irregularities within the manufactured chromatography paper used for this study; the inconsistencies within the Chr 1 paper are apparent on visual inspection and may affect the degree of DVS activation, biomolecule immobilization, and therefore probe response. However, this heterogeneity results in only modest standard deviations for the bioactivity of the immobilized carbohydrates, glycoprotein and oligonucleotide. An experiment involving a membrane stored for 30 days in ambient conditions (on the lab bench) yielded similar results (Figure S9). These observations compare favorably with previous investigations by our lab regarding the stability of DVS-modified surfaces (80% activity retained after 1 month of storage), 29 and demonstrate that DVS-activated cellulose is stable to prolonged storage while retaining reactivity for subsequent biomolecule immobilization.
Figure 5.
(a) DVS-activated membranes were stored for 60 days and subsequently functionalized with mannose, glucose, maltose, and RNase B. The bioactivity, as observed by fluorescent ConA detection, was minimally affected by storage of the DVS-activated paper. (b) Biofunctional DVS+ membranes stored for 30 days retained significant bioactivity. The aged membranes experienced slight bioactivity loss for mannose and a more significant loss for maltose, when compared to a membrane freshly functionalized and assayed. Loss in bioactivity for the maltose-functionalized membrane may be ascribed to its susceptibility to degradation by microbiota in the non-sterile storage environment.
Having established that DVS-activated membranes retain their reactivity after prolonged storage, we investigated the ability of biofunctional DVS+ membranes to retain their bioactivity following storage at room temperature. We tested the stability of a DVS-activated membrane functionalized with carbohydrates and glycoprotein and stored in a dry, non-sterile, nitrogen-filled environment for 30 days. Following storage, Con A binding showed preservation of the bioactivity with differing amounts of loss, varying by specific glycans (Figure 5b). These results demonstrate that biofunctional DVS-cellulose has potential as a robust platform with an extended shelf-life for point-of-care diagnostics and other applications.
Based on the data presented in this study, we summarize several important advantages of the DVS activation strategy over previously published methods for cellulose functionalization. First, DVS-activated cellulose can immobilize a variety of biomolecules presenting nucleophilic groups, while most of the existing immobilization methods target a specific nucleophile (e.g. amines) or class of biomolecules.17–21 Second, the facile DVS method requires only two steps for functionalization. Unmodified carbohydrates and other biomolecules bearing hydroxyl, amine or thiol groups can be directly immobilized onto cellulose, which dramatically reduces the burden of chemical modification of biomolecules. Third, the DVS chemistry on cellulose is stable over time. Many reported amine-reactive linkers (e.g., carbonyldiimidazole16 and isothiocyanate23) are sensitive to hydrolysis and require specialized handling and storage, whereas the DVS-activated cellulose retains its reactivity after 1-month storage under ambient conditions. Overall, the advantages presented by DVS chemistry could significantly facilitate the fabrication and storage of bioactive paper.
Printed Biomolecular Patterning
Fabrication of biomolecular patterns on LF-based cellulosic diagnostic devices may benefit from the development of straightforward bioprinting techniques.37, 38 To demonstrate the adaptability of the DVS functionalization technique to bioprinting we have visualized patterns of immobilized galactose on DVS-activated cellulose membranes using fluorescently labeled RCA120. An Epson R280 inkjet printer was modified for printing biological solutions. The printing solution we formulated (10% w/v galactose, 40% w/v sucrose and 5% w/v TWEEN® 20 in 0.1 M pH 10 sodium carbonate buffer) was previously immobilized and assayed on a DVS-activated membrane without the galactose component to ensure that the non-reducing sugar sucrose did not interfere with binding by RCA120 (not shown). Examples of the printed patterns are depicted in Scheme 1c. These images show that DVS-activated cellulose is amenable to accurate, sub-millimeter patterning of biomolecules through bioprinting, and that the printed biomolecules retain their bioactivity following the printing process.
Integrating the DVS strategy with bioprinting could benefit the design of complex patterns on lateral flow-based devices, providing reliable and stable biofunctionality and multiplexed detection capability. For instance, multiple biomolecules could be covalently patterned separately onto a test line in a fashion similar to the immobilization of proteins on test lines for nitrocellulose-based assays. Bioprinting is also highly scalable for mass production of lateral flow devices. Separation of the physical printing and digitized design processes also permits the rapid implementation of design changes during fabrication. In particular, production of muiltiplexed paper devices could be greatly expedited by the accuracy and automation presented by bioprinting.
CONCLUSIONS
We have presented a simple covalent immobilization method for cellulose using DVS chemistry. DVS-activated cellulose membranes exhibit a versatile capacity for conjugation of biomolecules presenting nucleophilic functional groups. Immobilized carbohydrates and oligonucleotides on DVS-activated cellulose membranes exhibit specific response to lectins and complementary nucleotides, respectively. This membrane exhibits trivial amounts of non-specific binding of the aforementioned biomolecular analytes. Probing of the functionalized surface with either lectins or oligonucleotides is dose-dependent, and the LOD of nucleic acid hybridization is comparable to results observed previously using SPR and zirconia-modified paper. In addition, DVS-activated and biofunctional cellulose membranes maintain their activity for prolonged storage under ambient conditions, which supports the suitability of this technique for the fabrication of paper-based bioassays for low-cost and portable point-of-care diagnostics. Finally, the DVS strategy can be combined with inkjet bioprinting to accurately produce biomolecular patterns, highlighting the potential of cellulose for sophisticated bioassay design and multiplexed detection. Overall, the findings in this study demonstrate the facile generation of biofunctional paper with potential applications in lateral flow-based diagnostic devices for use in molecular detection in research, diagnostics, environmental monitoring, and limited-resource molecular sensing.
Supplementary Material
Acknowledgments
We thank the Washington Research Foundation, the University of Washington Royalty Research Fund, and the Department of Bioengineering for their support. Surface analysis was performed by NESAC/BIO (NIH Grant P41 EB002027). We thank Mr. James Kirk for data plot creation, Lauren Cummings for the scanning electron microscopy, and Mark Lewis for assistance with the tensile strength experiments. Additionally, we gratefully acknowledge the Life Sciences Discovery Fund Center for Intracellular Delivery of Biologics (Life Sciences Discovery Fund Grant 2496490) for access to the Storm scanner.
Footnotes
Supporting Information Available: Synthesis of hexafluorobenzamide, Biomolecules used in this work, Experimental Scheme for Oligonucleotide Hybridization, Assembly of the Bio-Dot apparatus, dose-dependent response of RCA120-galactose and Con A-mannose interactions (varying lectin concentration), activity of DVS-activated membrane after 30 days of storage in aluminum foil,. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Arthur Yu, Email: arthuryu@u.washington.edu.
Jing Shang, Email: shangj@u.washington.edu.
Fang Cheng, Email: ffcheng@dlut.edu.cn.
Bradford A. Paik, Email: bradford.paik@gmail.com.
Justin M. Kaplan, Email: jkaplan1@temple.edu.
Rodrigo B. Andrade, Email: randrade@temple.edu.
Daniel M. Ratner, Email: dratner@u.washington.edu.
References
- 1.von Lode P. Point-of-Care Immunotesting: Approaching the Analytical Performance of Central Laboratory Methods. Clin Biochem. 2005;38:591–606. doi: 10.1016/j.clinbiochem.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Microfluidic diagnostic technologies for global public health. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 3.Yager P, Domingo GJ, Gerdes J. Annu Rev Biomed Eng Point-of-Care diagnostics for Global Health. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 4.Tonkinson JL, Stillman BA. Nitrocellulose: A Tried and True Polymer Finds Utility as a Post-Genomic Substrate. Front Biosci. 2002;7:C1–C12. doi: 10.2741/A755. [DOI] [PubMed] [Google Scholar]
- 5.Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Patterned Paper as a Platform for Inexpensive, Low-Volume, Portable Bioassays. Angew Chem Int Edit. 2007;46:1318–1320. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pelton R. Bioactive Paper Provides a Low-Cost Platform for Diagnostics. Trac-Trend Anal Chem. 2009;28:925–942. doi: 10.1016/j.trac.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez AW, Phillips ST, Whitesides GM. Three-Dimensional Microfluidic Devices Fabricated in Layered Paper and Tape. Proc Natl Acad Sci U S A. 2008;105:19606–19611. doi: 10.1073/pnas.0810903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Diagnostics for the Developing World: Microfluidic Paper-Based Analytical Devices. Anal Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 9.Fenton EM, Mascarenas MR, Lopez GP, Sibbett SS. Multiplex Lateral-Flow Test Strips Fabricated by Two-Dimensional Shaping. Acs Appl Mater Inter. 2009;1:124–129. doi: 10.1021/am800043z. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Tian JF, Shen W. Thread As a Versatile Material for Low-Cost Microfluidic Diagnostics. Acs Appl Mater Inter. 2010;2:1–6. doi: 10.1021/am9006148. [DOI] [PubMed] [Google Scholar]
- 11.Reches M, Mirica KA, Dasgupta R, Dickey MD, Butte MJ, Whitesides GM. Thread as a Matrix for Biomedical Assays. Acs Appl Mater Inter. 2010;2:1722–1728. doi: 10.1021/am1002266. [DOI] [PubMed] [Google Scholar]
- 12.Towbin H, Staehelin T, Gordon J. Electrophoretic Transfer of Proteins from Polyacrylamide Gels to Nitrocellulose Sheets - Procedure and Some Applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bode L, Beutin L, Kohler H. Nitrocellulose-Enzyme-Linked Immunosorbent-Assay (NC-ELISA) - A Sensitive Technique for the Rapid Visual Detection of Both Viral-Antigens and Antibodies. J Virol Methods. 1984;8:111–121. doi: 10.1016/0166-0934(84)90045-4. [DOI] [PubMed] [Google Scholar]
- 14.Tovey ER, Baldo BA. Protein-Binding to Nitrocellulose, Nylon and PVDF Membranes in Immunoassays and Electroblotting. J Biochem Bioph Methods. 1989;19:169–183. doi: 10.1016/0165-022x(89)90024-9. [DOI] [PubMed] [Google Scholar]
- 15.Jones KL, O’Melia CR. Protein and Humic Acid Adsorption onto Hydrophilic Membrane Surfaces: Effects of pH and Ionic Strength. J Membrane Sci. 2000;165:31–46. [Google Scholar]
- 16.Stollner D, Scheller FW, Warsinke A. Activation of Cellulose Membranes with 1,1′-carbonyldiimidazole or 1-cyano-4-dimethylaminopyridinium tetrafluoroborate as a Basis for the Development of Immunosensors. Anal Biochem. 2002;304:157–165. doi: 10.1006/abio.2001.5603. [DOI] [PubMed] [Google Scholar]
- 17.Bora U, Sharma P, Kannan K, Nahar P. Photoreactive Cellulose Membrane - A Novel Matrix for Covalent Immobilization of Biomolecules. J Biotechnol. 2006;126:220–229. doi: 10.1016/j.jbiotec.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Sharma P, Basir SF, Nahar P. Photoimmobilization of unmodified carbohydrates on activated surface. J Colloid Interface Sci. 2010;342:202–204. doi: 10.1016/j.jcis.2009.10.038. [DOI] [PubMed] [Google Scholar]
- 19.Brumer H, Zhou Q, Baumann MJ, Carlsson K, Teeri TT. Activation of Crystalline Cellulose Surfaces through the Chemoenzymatic Modification of Xyloglucan. J Am, Chem Soc. 2004;126:5715–5721. doi: 10.1021/ja0316770. [DOI] [PubMed] [Google Scholar]
- 20.Bryjak J, Liesiene J, Stefuca V. Man-Tailored Cellulose-Based Carriers for Invertase Immobilization. Cellulose. 2008;15:631–640. [Google Scholar]
- 21.Kong DL, Schuett W, Dai J, Kunkel S, Holtz M, Yamada R, Yu YT, Klinkmann H. Development of Cellulose-DNA Immunoadsorbent. Artif Organs. 2002;26:200–208. doi: 10.1046/j.1525-1594.2002.06721.x. [DOI] [PubMed] [Google Scholar]
- 22.Su SX, Nutiu R, Filipe CDM, Li YF, Pelton R. Adsorption and Covalent Coupling of ATP-Binding DNA Aptamers onto Cellulose. Langmuir. 2007;23:1300–1302. doi: 10.1021/la060961c. [DOI] [PubMed] [Google Scholar]
- 23.Araujo AC, Song Y, Lundeberg J, Stahl PL, Brumer H. Activated Paper Surfaces for the Rapid Hybridization of DNA through Capillary Transport. Anal Chem. 2012 doi: 10.1021/ac300025v. in press. [DOI] [PubMed] [Google Scholar]
- 24.Xu C, Spadiut O, Araujo AC, Nakhai A, Brumer H. Chemo-enzymatic Assembly of Clickable Cellulose Surfaces via Multivalent Polysaccharides. ChemSusChem. 2012;5:661–665. doi: 10.1002/cssc.201100522. [DOI] [PubMed] [Google Scholar]
- 25.Filpponen I, Kontturi E, Nummelin S, Rosilo H, Kolehmainen E, Ikkala O, Laine J. Generic Method for Modular Surface Modification of Cellulosic Materials in Aqueous Medium by Sequential “Click” Reaction and Adsorption. Biomacromolecules. 2012;13:736–742. doi: 10.1021/bm201661k. [DOI] [PubMed] [Google Scholar]
- 26.Sereikaite J, Bassus D, Bobnis R, Dienys G, Bumeliene Z, Bumelis VA. Divinyl Sulfone as a Crosslinking Reagent for Oligomeric Proteins. uss. J Bioorg Chem. 2003;29:227–230. [PubMed] [Google Scholar]
- 27.Porath J, Laas T, Janson JC. Agar Derivatives for Chromatography, Electrophoresis, and Gel-Bound Enzymes. III. Rigid Agarose Gels Cross-Linked with Divinyl Sulfone (DVS) J Chromatogr. 1975;103:49–62. doi: 10.1016/s0021-9673(00)83800-5. [DOI] [PubMed] [Google Scholar]
- 28.Fornstedt N, Porath J. Characterization Studies on a New Lectin Found in Seeds of Vicia-Ervilia. FEBS Lett. 1975;57:187–191. doi: 10.1016/0014-5793(75)80713-7. [DOI] [PubMed] [Google Scholar]
- 29.Cheng F, Shang J, Ratner DM. A Versatile Method for Functionalizing Surfaces with Bioactive Glycans. Bioconjugate Chem. 2011;22:50–57. doi: 10.1021/bc1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang J, Cheng F, Dubey M, Kaplan JM, Rawal M, Jiang X, Newburg DS, Sullivan PA, Andrade RB, Ratner DM. An organophosphonate strategy for functionalizing silicon photonic biosensors. Langmuir. 2012;28:3338–3344. doi: 10.1021/la2043153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayes W, Osborn HMI, Osborne SD, Rastall RA, Romagnoli B. One-Pot Synthesis of Multivalent Arrays of Mannose Mono-and Disaccharides. Tetrahedron. 2003;59:7983–7996. [Google Scholar]
- 32.Goldstein IJ, Hollerman CE, Smith EE. Protein-Carbohydrate Interaction. 2. Inhibition Studies on Interaction of Concanavalin A with Polysaccharides. Biochemistry. 1965;4:876. doi: 10.1021/bi00881a013. [DOI] [PubMed] [Google Scholar]
- 33.Olsnes S, Pihl A. Different Biological Properties of 2 Constituent Peptide Chains of Ricin, a Toxic Protein Inhibiting Protein-Synthesis. Biochemistry (Mosc) 1973;12:3121–3126. doi: 10.1021/bi00740a028. [DOI] [PubMed] [Google Scholar]
- 34.Lutz S, Weber P, Focke M, Faltin B, Hoffmann J, Muller C, Mark D, Roth G, Munday P, Armes N, Piepenburg O, Zengerle R, von Stetten F. Microfluidic Lab-On-a-Foil for Nucleic Acid Analysis Based on Isothermal Recombinase Polymerase Amplification (RPA) Lab Chip. 2010;10:887–893. doi: 10.1039/b921140c. [DOI] [PubMed] [Google Scholar]
- 35.Xiao W, Huang JG. Immobilization of Oligonucleotides onto Zirconia-Modified Filter Paper and Specific Molecular Recognition. Langmuir. 2011;27:12284–12288. doi: 10.1021/la203150f. [DOI] [PubMed] [Google Scholar]
- 36.Nelson BP, Grimsrud TE, Liles MR, Goodman RM, Corn RM. Surface Plasmon Resonance Imaging Measurements of DNA and RNA Hybridization Adsorption onto DNA Microarrays. Anal Chem. 2001;73:1–7. doi: 10.1021/ac0010431. [DOI] [PubMed] [Google Scholar]
- 37.Derby B. Bioprinting: Inkjet Printing Proteins and Hybrid Cell-Containing Materials and Structures. J Mater Chem. 2008;18:5717–5721. [Google Scholar]
- 38.Khan MS, Fon D, Li X, Tian JF, Forsythe J, Garnier G, Shen W. Biosurface Engineering Through Inkjet Printing. Colloid Surface B. 2010;75:441–447. doi: 10.1016/j.colsurfb.2009.09.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.