Summary
Porphyromonas gingivalis a host-adapted opportunistic pathogen produces a serine phosphatase, SerB, known to affect virulence, invasion and persistence within the host cell. SerB induces actin filament rearrangement in epithelial cells, but the mechanistic basis of this is not fully understood. Here we investigated the effects of SerB on the actin depolymerizing host protein cofilin. P. gingivalis infection resulted in the dephosphorylation of cofilin in gingival epithelial cells. In contrast, a SerB deficient mutant of P. gingivalis was unable to cause cofilin dephosphorylation. The involvement of cofilin in P. gingivalis invasion was determined by quantitative image analysis of epithelial cells in which cofilin had been knocked-down or knocked-in with various cofilin constructs. siRNA-silencing of cofilin led to a significant decrease in numbers of intracellular P. gingivalis marked by an absence of actin colocalization. Transfection with wild-type cofilin or constitutively active cofilin both increased numbers of intracellular bacteria, while constitutively inactive cofilin abrogated invasion. Expression of LIM kinase resulted in reduced P. gingivalis invasion, an effect that was reversed by expression of constitutively active cofilin. These results show that P. gingivalis SerB activity induces dephosphorylation of cofilin, and that active cofilin is required for optimal invasion into gingival epithelial cells.
Introduction
The epithelial cells of the oral cavity are colonized by a diverse and complex microbiota that includes both commensals and pathogens (Rudney et al., 2001; Rudney et al., 2005; Kolenbrander et al., 2002; Avila et al., 2009). While health is the most common status in the oral cavity, in some cases the encounter between host and microbes becomes unbalanced and disease ensues (Jenkinson and Lamont, 2005; Paster et al., 2006). Indeed, periodontitis is one of the most prevalent human infections and it has been reported that more than 50% of adults over 30 years of age in the US have the some form of the disease (Albandar, 2011). Persistent bacterial infection of the periodontal tissues induces inflammation, and in severe cases culminates in the destruction of the periodontal tissues, resorption of the alveolar bone and, ultimately, tooth loss. The gram-negative anaerobe P. gingivalis is one of the most important members of the consortia of bacteria responsible for the initiation and progression of severe and chronic forms of periodontitis (Byrne et al., 2009; Feng and Weinberg, 2006; Holt and Ebersole, 2005; Lamont and Jenkinson, 1998). P. gingivalis expresses a multitude of virulence factors that enable colonization of the oral surfaces and that can initiate tissue destruction (Feng and Weinberg, 2006; Holt and Ebersole 2005; Lamont and Jenkinson, 1998). Paradoxically, however, P. gingivalis can also thrive in the subgingival area in the absence of overt disease (Columbo et al., 2006; Columbo et al., 2007; Rudney et al., 2005; Yilmaz, 2008). This commensal mode is accomplished through selective inhibition of components of the immune system and by localizing inside gingival epithelial cells (Darveau et al., 1998; Tribble and Lamont, 2010; Yilmaz, 2008; Hajishengallis, 2009).
P. gingivalis, which lacks type III secretion systems, utilizes more rudimentary invasion mechanisms involving a limited number of multifunctional effectors (Tribble and lamont, 2010). Intracellular invasion by P. gingivalis is initiated by the major fimbriae that engage integrin receptors; and the resulting signaling cascade remodels the host cytoskeleton to allow bacterial entry. Optimal P. gingivalis invasion also requires the activity of a bacterial serine phosphatase, SerB, a member of the haloacid dehalogenase (HAD) enzyme family (Tribble et al., 2006; Hasegawa et al., 2008). SerB can dephosphorylate host signaling proteins, and reprograms circuitry that converges on both actin microfilament and microtubule stability (Tribble et al., 2006; Hasegawa et al., 2008). Intracellular P. gingivalis accumulate in the perinuclear area where they remain viable and ultimately they can disseminate intercellularly without exposure to the intercellular space (Yilmaz et al., 2006; Lamont et al., 1995; Belton et al., 1999). In addition, P. gingivalis activates intrinsic JAK/Stat and Akt anti-apoptotic pathways in epithelial cells, and cell cycle progression is accelerated (Mao et al., 2007; Yilmaz et al., 2004; Kuboniwa et al., 2008).
The actin cytoskeleton is highly dynamic and tightly regulated, enabling rapid responses to extracellular stimuli while maintaining cell integrity. A number of regulatory proteins are involved in the temporal and spatial precision of actin filament assembly and disassembly (Bernstein and Bamburg, 2010) in many different cell types (Oser and Condeelis, 2009; Pritchard et al., 2004; Fazal et al., 2009; Nishita et al., 2002). One such group of actin filament rearranging proteins is the Actin depolymerizing factor (ADF) and cofilin family (Bamburg et al., 1999; Desmarais et al., 2005). ADF/cofilin is important for several key cellular processes, including mitosis, nuclear translocation, actin-mediated endocytosis and cell chemotaxis (Nishita et al., 2002; Fazal et al., 2009; Okreglak and Drubin, 2007; Amano et al., 2002). Cofilin binds both actin monomers and filaments, putatively utilizing two distinct binding sites on actin (Renoult et al., 1999). Actin severing follows and also the off rate of actin sub-units from the minus end of the filament is enhanced (Huang et al., 2006). Cofilin is inactivated by phosphorylation at Ser 3 by LIMK, the terminal kinase in a phospho-relay that includes upstream kinases ROCK and PAK (Pritchard et al., 2004). Conversely dephosphorylation by the slingshot (SSH) and chronophin phosphatases (HAD family members) activates cofilin (Arber et al., 1998; Gohla et al., 2005; Huang et al., 2006; DesMarais et al., 2005). Actin rearrangement involving cofilin can also directly contribute to the progression of disease and infection (Cameron et al., 2010; Amin et al., 2008; Bierne et al., 2001; Dai et al., 2004). We therefore investigated the possibility that SerB phosphatase activity can impact cofilin activity, and that activation of cofilin is a requirement for the efficient uptake of P. gingivalis into gingival epithelial cells. Consistent with this, P. gingivalis infection resulted in dephosphorylation of cofilin, a property lost with a SerB defective mutant. Numbers of internalizing bacteria were reduced following cofilin silencing, upregulation of LIM kinase or the expression of constitutively inactive cofilin. Conversely, internalization was promoted by the expression of exogenous cofilin or constitutively active cofilin. The results support the notion that activation of cofilin by the P. gingivalis SerB enzyme is required for the actin remodeling necessary for optimal invasion.
Results
SerB is required for P. gingivalis dephosphorylation of cofilin
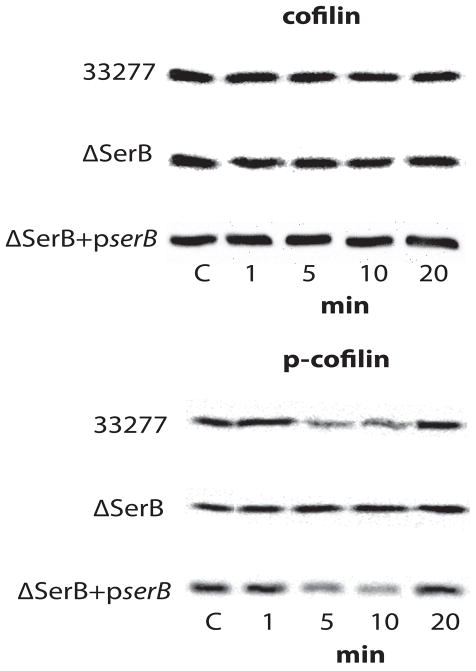
Actin remodeling is required for P. gingivalis invasion (Lamont et al., 1995) and activated cofilin can effect actin rearrangements in cells by depolymerizing actin filaments (Bamburg et al. 1999; Desmarais et al. 2005). Therefore, we first examined the possibility that SerB, through its phosphatase activity, can activate cofilin. Human immortalized gingival keratinocytes (HIGKs) were infected with P. gingivalis parental and SerB mutant strains, and the levels of phospho-cofilin and cofilin determined by western blotting. Wild type P. gingivalis caused transient dephosphorylation of cofilin after 5 min of bacterial challenge, with levels of phospho-cofilin returning to the levels in uninfected cells by 20 min (Figure 1). In contrast, the SerB deficient strain did not induce dephosphorylation of cofilin at any time period investigated. To ensure that the phenotype of the mutant was directly related to the loss of the serB gene, we tested a ΔserB+pserB mutant strain (Tribble et al., 2006) in which the mutation was complemented with the wild type allele in trans. Complementation restored the parental phenotype and the ΔserB+pserB strain was capable of inducing cofilin dephosphorylation (Figure 1).
Figure 1. Western blot analysis of cofilin and p-cofilin levels in P. gingivalis–infected HIGK cells.
Whole cell lysates of P. gingivalis 33277-infected, P. gingivalis ΔSerB-infected, or P. gingivalis ΔSerB+pserB-infected HIGK cells were examined by Western blotting with antibodies to cofilin (upper panel) or phospho(p)-cofilin (lower panel). Data are representative of three independent experiments.
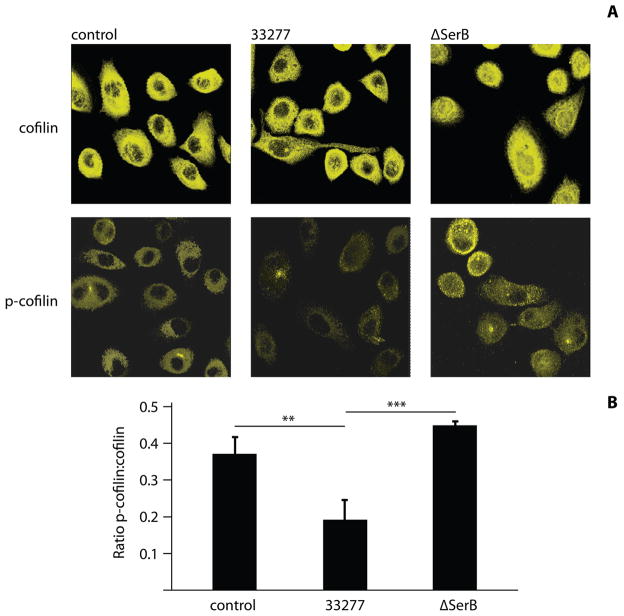
Cofilin phosphorylation levels were also examined by confocal laser scanning microscopy (CLSM). As shown in Figure 2A, phospho-cofilin levels were elevated in cells infected with the ΔserB mutant as compared to control or wild-type infected cells. Quantitative image analysis with Leica Confocal Software (LCS), quantifying relative fluorescence emission intensity (arbitrary units) to determine p-cofilin:cofilin fluorescence ratios confirmed the reduction in phospho-cofilin following infection with the P. gingivalis parental strain (Figure 2B). These experiments corroborate the role of SerB in the dephosphorylation, and hence activation, of host cell cofilin. The reduction in phospho-cofilin throughout the host cells may be due to release of SerB from P. gingivalis (Chen et al., 2001), and the ability of SerB to spread through the cell (Figure S1). Activation of cofilin by SerB may be direct or indirect. Evidence in favor of a direct interaction is provided by the localization of cofilin with secreted SerB (Figure S1) and invading bacteria (Figure S2), and the colocalization of ectopically expressed SerB with cofilin (Figure S3). However, SerB may act on a membrane receptor or a phosphoprotein in the signaling cascade that controls cofilin, and the matter requires further investigation.
Figure 2. Fluorescence analysis of cofilin and p-cofilin levels in P. gingivalis–infected HIGK cells.
A) P. gingivalis 33277-infected or ΔSerB-infected HIGK cells were labeled with antibodies to cofilin (upper panel) or p-cofilin (lower panel) and analyzed by CSLM. Controls were uninfected HIGKs. Magnification x63 of cells at 10 min time point. B) Fluorescence intensity analysis of p-cofilin:cofilin in infected or control HIGK cells. Intensity values were obtained using Leica Confocal software (arbitrary units) to calculate ratios. Results are representative of two independent experiments (n=3 coverslips/group). Data are means and error bars indicate standard deviation. **, P <0.01; ***, P <0.001 by Tukey-Kramer Multiple Comparison test.
Silencing cofilin reduces P. gingivalis invasion of HIGKs
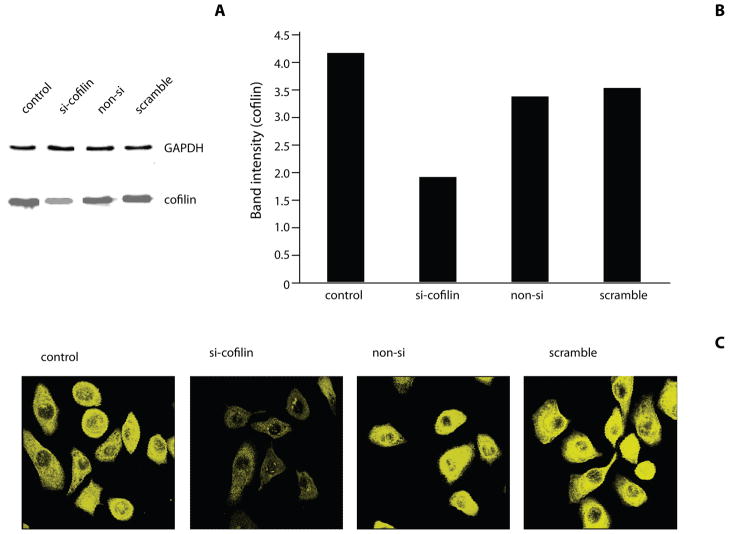
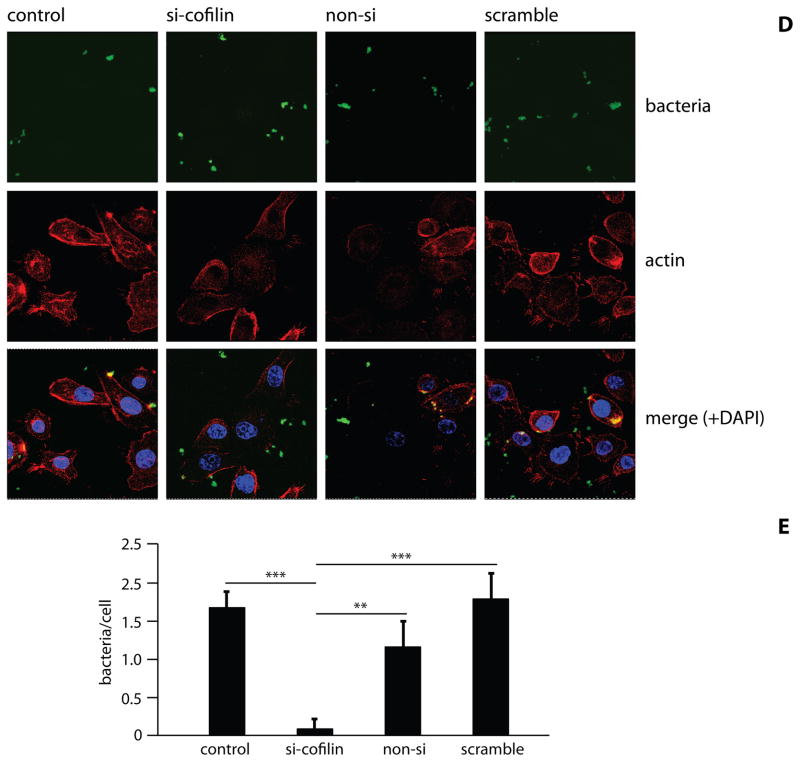
The ability of wild-type P. gingivalis to activate cofilin suggested the possibility that cofilin may be required for P. gingivalis entry into epithelial cells. To investigate the role of cofilin in P. gingivalis invasion, cofilin was knocked down using si-RNA. Reduction in the levels of cofilin following transfection was confirmed by Western blotting and densiometric analysis (Figure 3A & B). Controls of transfection agent only, non-silence and scrambled cofilin si-RNA did not affect levels cofilin. CSLM also showed a reduction in epithelial cell cofilin following specific si-RNA treatment (Figure 3C). Consistent with these results, knockdown of cofilin also caused an increase in F-actin (Figure S4). Transfected and control HIGKs were infected with P. gingivalis MOI 10 for 10 min and bacterial internalization was determined by CSLM (Figure 3D) with quantitative image analysis (Figure 3E). Cofilin knock-down resulted in a greater than 80% reduction in the level of P. gingivalis internalization compared to controls (p<0.01). Actin has been shown to co-localize with invading P. gingivalis, and SerB is involved in this process (Belton et al 1999; Hasegawa et al 2008). CSLM images of P. gingivalis invasion confirmed actin co-localization with bacteria in conditions where cofilin was at wild-type levels (Figure 3D). However, knockdown of cofilin resulted in failure of actin to co-localize with P. gingivalis. Taken together, these findings suggest that transient activation of cofilin by P. gingivalis is required for rapid (within 5 min) actin depolymerization that allows remodeling of actin and subsequently facilitates P. gingivalis entry. This concept is supported by the observation of a localized area of actin depolymerization around an invading P. gingivalis bacterium (Figure S5). As cofilin activation levels return to normal, actin is then polymerized and localizes with invading bacteria.
Figure 3. Internalization of P. gingivalis is inhibited by cofilin-silencing.
HIGK cells were transfected with silence(si)-cofilin, non-silence(non-si), or scramble cofilin. Control was transfection agent only. A) Western blot analysis to confirm cofilin-silencing in HIGK cells. Whole cell lysates were examined by Western blotting with antibodies to cofilin. GAPDH was used as a loading control. B) Blots were analysed by scanning densiometry to determine relative cofilin levels. Data are representative of two independent experiments. C) Cofilin-silenced HIGK cells (or non-si/scramble controls) were labeled with cofilin antibodies (yellow) and analyzed by CSLM. Magnification x63. D) Cofilin-silenced HIGK cells (or non-silence/scramble controls) were infected with P. gingivalis for 10 min and analyzed by CSLM. P. gingivalis (green) was detected with specific antibodies, actin (red) was stained with TRITC-phalloidin, and nuclei (blue) stained with DAPI. Magnification x63. Results are representative of three independent assays. Data shown are maximum projections of z-stacks (10 slices/z stack, 3 coverslips/group). E) Invasion levels of P. gingivalis in HIGK cells following cofilin-silencing. Numbers of intracellular bacteria were counted throughout z-stacks (10 slices/stack; 3 coverslips/group). Results are representative of three independent assays. Data are means and error bars indicate standard deviations. **, P <0.01; ***, P <0.001 by Tukey-Kramer Multiple Comparison test.
Activated cofilin is required for maximal invasion of P. gingivalis
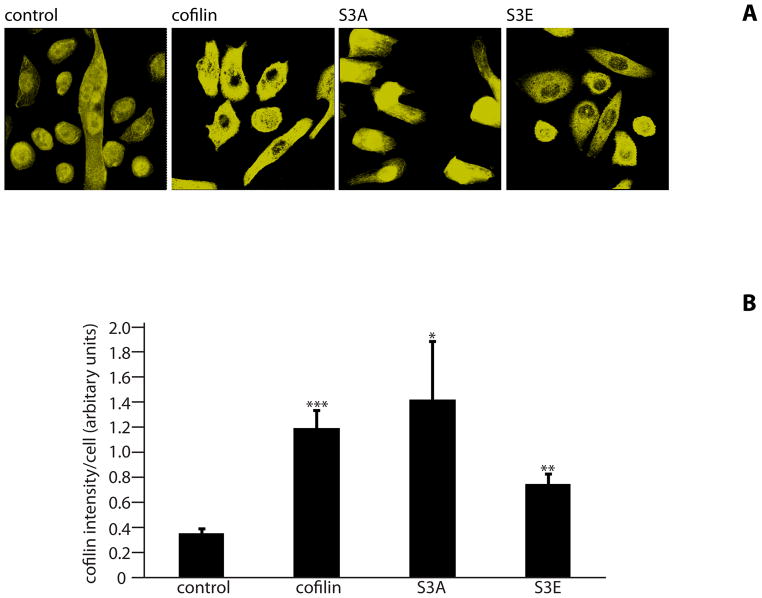
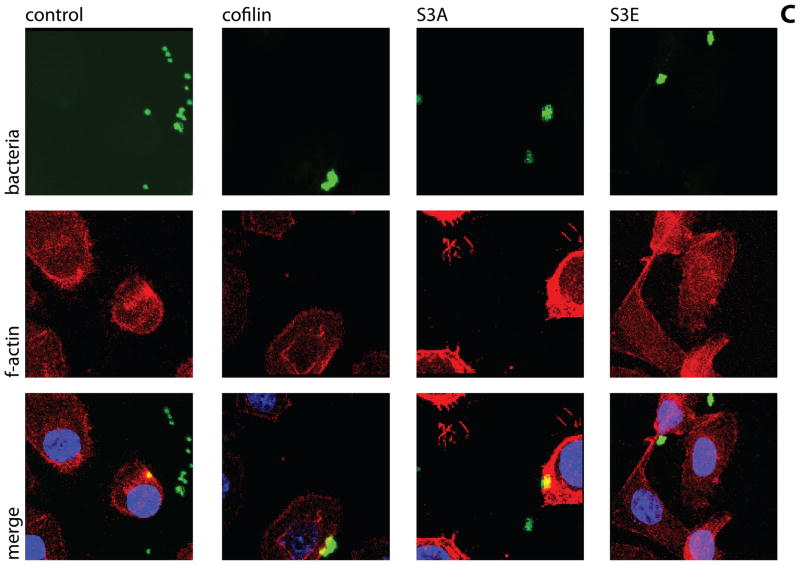
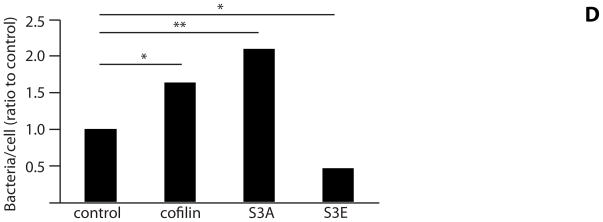
The phosphorylation state at the Ser-3 residue on cofilin controls activity (Moriyama et al., 1996). We sought to corroborate the significance of cofilin phosphorylation for P. gingivalis internalization by transfecting HIGKs with cofilin constructs displaying different activation states. Exogenous wild type cofilin, its constitutively active S3A mutant, and its minimally active S3E mutant, were expressed from the mammalian vector pCMV-Myc. In the S3A mutant the Ser-3 is replaced by an Ala that cannot be phosphorylated, whereas in the S3E mutant Ser-3 is replace by Glu that mimics the phosphorylated state (Kuhn et al., 2000). Overexpression of cofilin was confirmed by confocal microscopy with quantitative image analysis (Figure 4A & B). Transfected cells were infected with wild type P. gingivalis at MOI 10 for 10 min and the level of internalization determined by CSLM. Expression of exogenous wild type cofilin increased numbers of intracellular P. gingivalis by over 50% compared with controls (Figure 4C & D). Moreover, expression of constitutively active (S3A) cofilin resulted in a greater than twofold increase in P. gingivalis invasion. In contrast, expression of the inactive S3E construct decreased P. gingivalis invasion, with fewer than half the number of intracellular bacteria compared to control cells (p<0.001). Examination of the CSLM images showed association of P. gingivalis with actin microfilaments in HIGKs that contain native cofilin, exogenously expressed cofilin or constitutive active cofilin. A greater degree of actin rearrangement compared to control was also visible in the cofilin and S3A transfected cells, but not in the S3E transfected cells. While there are other mechanisms that regulate cofilin activity, including pH and membrane sequestering PIP2, the results with these phospho/dephospho mimics support the concept that activation of cofilin by dephosphorylation of Ser-3 is required for maximal P. gingivalis invasion, and that the remodeling initiated by cofilin causes greater localization of filamentous actin with P. gingivalis.
Figure 4. Expression of exogenous or constitutively active cofilin increases internalization of P. gingivalis.
HIGK cells were transfected with exogenous cofilin, constitutively active cofilin (S3A), or constitutively inactive cofilin (S3E). Control was transfection agent only. A) Confirmation of an increase in cofilin expression in transfected cells. Cofilin-transfected HIGK cells (or control) were labeled with cofilin antibodies (yellow) and analyzed by CSLM. Magnification x63. B) Fluorescence intensity analysis of cofilin HIGK cells. Intensity values were obtained using Leica Confocal software (arbitrary units) to calculate cofilin fluorescence intensity. Results are representative of two independent experiments (n=3 coverslips/group). Data are means and error bars indicate standard deviations. *, P <0.05; **, P <0.01; ***, P <0.001 by unpaired t-test to control. C) Cofilin-transfected cells were infected with P. gingivalis for 10 min and analyzed by CSLM. P. gingivalis (green) was detected with specific antibodies, actin (red) was stained with TRITC-phalloidin, and nuclei (blue) stained with DAPI. Magnification x63. Results are representative of three independent assays. Data shown are maximum projections of z-stacks (10 slices/z stack, 3 coverslips/group). D) Invasion levels of P. gingivalis following cofilin-transfection. Numbers of intracellular bacteria were counted throughout z-stacks (10 slices/stack; 3 coverslips/group). Results are representative of three independent assays. Data are ratios of internalization in transfected-infected groups compared with non-transfected control-infected cells. *, P <0.05; **, P <0.01 by unpaired t-test to control.
LIMK-1 inhibits P. gingivalis invasion
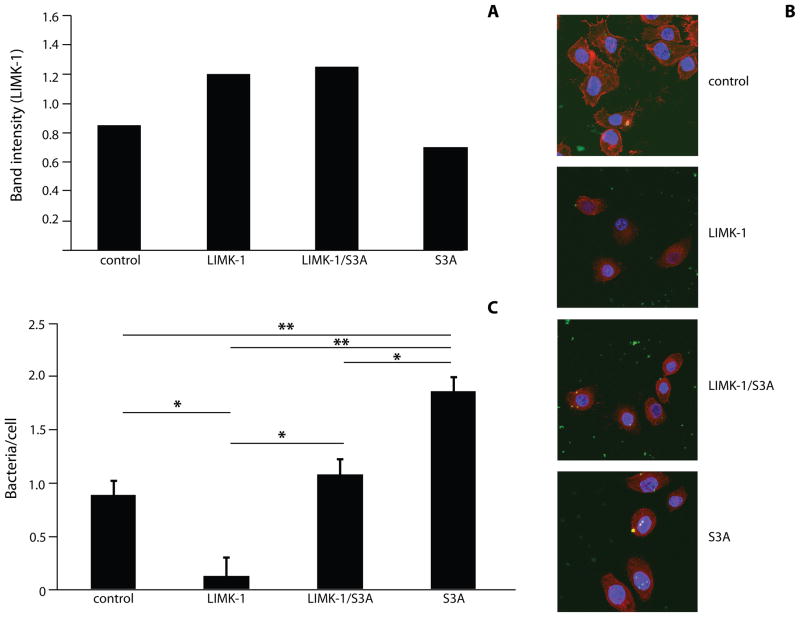
In host cells cofilin is phosphorylated primarily by LIMK (Pritchard et al., 2004) and overexpression of LIMK increases phosphorylation, and thereby inactivation, of cofilin (Bernard, 2007). To provide additional insight into the role of cofilin in P. gingivalis invasion, HIGKs were transfected with a plasmid expressing LIMK-1 as a means of modulating the activity of endogenous cofilin in vivo. Transfected cells were infected with P. gingivalis and the level of invasion assessed by confocal microscopy. Increased LIMK-1 expression in transfected cells was confirmed by Western blotting followed by densiometric analysis (Figure 5A). Western blotting also demonstrated that ectopic LIMK elevated the amount of phospho-cofilin by two fold. Increased LIMK levels significantly reduced the numbers of intracellular bacteria (p<0.05) (Figure 5B & C). Co-transfection with both LIMK and S3A (constitutively active) cofilin, reversed this effect and intracellular P. gingivalis numbers returned to control levels. Confocal images revealed visible reduction of intracellular P. gingivalis in LIMK-1 transfected cells. Expression of constitutively active cofilin completely compensated for the effect of LIMK-1, with more bacteria internalized in HIGKs. In S3A-only transfected cells, there were higher numbers of intracellular P. gingivalis compared with all groups. These investigations demonstrated that LIMK-1 deactivation of cofilin inhibits P. gingivalis invasion.
Figure 5. Expression of LIMK-1 in HIGK cells reduces invasion levels of P. gingivalis.
HIGK cells were transfected with LIMK-1 and/or S3A. Control was transfection agent only. A) Scanning densiometry of Western blots of LIMK-1 levels in transfected cells expressed as ratio to GAPDH. Data are representative of two independent experiments. B) LIMK-1/S3A-transfected HIGK cells were infected with P. gingivalis for 10 min and analyzed by CSLM. Control was transfection agent. P. gingivalis (green) was detected with specific antibodies, actin (red) was stained with TRITC-phalloidin, and nuclei (blue) stained with DAPI. Magnification x63. Results are representative of three independent assays. Data shown are maximum projections of z-stacks (10 slices/z stack, 3 coverslips/group). C) Invasion levels of P. gingivalis following LIMK-1/S3A transfection. Numbers of intracellular bacteria were counted throughout z-stacks (10 slices/stack; 3 coverslips/group). Results are representative of three independent assays. Data are means and error bars indicate standard deviations. *, P <0.05; **, P <0.01 by Tukey-Kramer Multiple Comparison test.
Discussion
The necessity for remodeling host cell actin filaments in order to effectuate bacteria-directed intracellular invasion is well established (Bourdet-Sicard et al., 1999; McGhie et al., 2004; Lamont et al., 1995; Cossart and Bierne, 2001). Actin polymerization and depolymerization is tightly regulated thus allowing cells to respond rapidly to external stimuli. ADF/cofilin family proteins are effectors that enhance the level of depolymerization of actin, a first step in cytoskeletal rearrangements (Moon and Drubin, 1995; Bamburg et al., 1999). Morover, cofilin responds to an array of inputs and not only feeds back to the dynamics of actin, but also to apoptosis cascades, phospholipid metabolism, and gene expression. Consequently, it has been proposed that the ADF/cofilin protein family constitutes a homeostatic regulator or ‘functional node’ in cell biology (Berstein and Bamburg, 2010). Cofilin has been shown to play a role in the internalization of Listeria and Salmonella, through modification of host cell actin filaments (Bierne et al., 2001; Dai et al., 2004), although the bacterial effector molecules have yet to be defined. The entry of P. gingivalis into epithelial cells requires modulation of the actin cytoskeleton which ultimately contracts and condenses in the cortical area (Lamont et al., 1995, Belton et al., 1999), a process that involves the serine phosphatase, SerB (Hasegawa et al., 2008). Moreover, SerB activity is necessary for maximal invasion of P. gingivalis and SerB can directly impact the progression of disease in vivo. In a rat model of periodontitis a SerB deletion mutant displays abrogated host bone-resorption as compared to wild type P. gingivalis (Bainbridge et al., 2010). Internalization of P. gingivalis into epithelial cells is a rapid process that is complete in under 20 min (Belton et al., 1999). Hence we predicted that the initial stages of actin restructuring, which involve cofilin-mediated transient depolymerization, would occur rapidly in epithelial cells upon challenge with P. gingivalis.
In this work we show by both western blotting and confocal microscopy that P. gingivalis dephosphorylates cofilin in epithelial cells, and that deletion of serB ablates this response. Cofilin was ubiquitously expressed throughout the cytoplasm, with an absence of cofilin in the nuclear region of the cells. These expression patterns resembled those previously shown in other cell types (Nebl et al., 1996; Sarmiere & Bamburg, 2004). SerB-mediated dephosphorylation of cofilin occurred rapidly, as early as a few minutes post-infection. One function of SerB, therefore, relates to transiently increasing the level of cofilin-mediated actin depolymerization. The pool of monomeric G-actin (Kiuchi et al., 2007) and free barbed ends thus produced will allow the regulated re-polymerization of actin and colocalization with P. gingivalis cells, features that characterize P. gingivalis entry into epithelial cells. Levels of phospho-cofilin returned to control amounts by 20 min after infection, possibly the result of the cell compensating for the additional phosphatase activity by the upregulation of a kinase. Alternatively, SerB may be inactivated within the host cell over time, or P. gingivalis may secrete another effector that reverses the action of SerB.
Having established that SerB is required for dephosphorylation of cofilin, we next sought to confirm that active cofilin is participates in the invasion process. Cofilin levels were knocked down by siRNA and, consistent with our hypothesis, silencing cofilin severely curtailed P. gingivalis invasion into cells. Confocal images also revealed the absence of actin localization around cell-associated bacteria in cofilin-silenced cells. In addition, bacteria associated with cells in control groups aggregated more than bacteria in the cofilin-silenced group. Actin has been shown to play a role in signal transduction (Maniotis et al., 1997), including the spatial reorganization of signaling enzymes such as ERK (Rosengart et al., 2002). This could infer that cofilin can also induce inside-out signaling that may influence receptor availability for P. gingivalis.
Following confirmation that cofilin is indeed involved in the modification of actin filaments required for P. gingivalis invasion, we next investigated the importance of cofilin activation state on invasion levels. Our results showed that expression of either exogenous cofilin or constitutively active cofilin resulted in an increase in the numbers of intracellular P. gingivalis. In contrast, expression of constitutively phosphorylated mimic cofilin (S3E) reduced the numbers of intracellular P. gingivalis. As endogenous cofilin was still present in these cells, this finding indicates that S3E cofilin can act as a dominant negative mutant. Similar results have been reported in smooth muscle cells where S3E cofilin inhibited the dephosphorylation of endogenous cofilin, thereby preventing activation (Zhao et al., 2008).
The effect of LIMK-1 on P. gingivalis internalization provided additional confirmation of the importance of cofilin in the entry process. LIMK-1 inhibits the activity of cofilin by phosphorylating the Ser3 residue (Agnew et al., 1995; Arber et al., 1998; Yang et al., 1998). The expression of exogenous LIMK-1 in epithelial cells reduced the levels of intracellular bacteria. Moreover, there were few P. gingivalis attached to the epithelial cells suggesting that in the absence of invasion the bacteria cycle off the cell surfaces. The effect of LIMK-1 could be reversed by cotransfection with constitutively active S3A cofilin, indicating that the inhibition of P. gingivalis invasion was a direct result of the action of LIMK-1 on cofilin.
This study shows that SerB dephosphorylation of cofilin is required for efficient internalization of P. gingivalis into HIGK cells. SerB may act on cofilin directly, on another phosphoprotein in the regulatory pathway, on a membrane receptor, or any combination of these actions. Work is currently underway in our laboratory to distinguish between these possibilities. In addition, at later time points SerB activity can result in modulation of expression of genes that are involved in the regulation of actin (Hasegawa et al., 2008). Indeed, numerous interactions occur within host cells, involving both regulatory proteins and bacterial components, and there is redundancy and cross-talk across pathways in cytoskeletal regulation. It is unlikely that cofilin dephosphorylation is the only pathway that can affect P. gingivalis invasion. For example, interaction of Chronophin with the chaperone Hsp90 has been shown to form a biosensor that mediates cofilin/actin rod formation (Huang et al., 2008). SerB also interacts with cytosolic glyceraldehydes-3-phosphate dehydrogenase and Hsp90, impacting the integrity of the microtubule cytoskeleton (Tribble et al., 2006). Activated cofilin sequesters microtubule-associated protein by the formation of cofilin/actin rods (Whiteman et al., 2009). Thus, the microtubule/actin network in cells is likely regulated in concert, and modulating one aspect could reverberate through several pathways. Additionally, cofilin itself is known to have more than one role in cells. While newly generated actin barbed ends provide sites for actin filament assembly, cofilin has also been shown to act as a pH sensor modifying pH-dependent actin filament dynamics through a histidine residue on the C-terminal of cofilin (Franz et al., 2008). Cofilin is thus a multi-functional molecule, with more than one residue/active site capable of inducing a biological response.
In conclusion, we established a role for cofilin in the internalization of P. gingivalis in human gingival epithelial cells. While cofilin has been shown to be involved in the invasion of other bacteria, our studies are the first to establish the function of a specific bacterial invasin, the serine phosphatase SerB, in the activation of cofilin.
Experimental Procedures
Bacterial and Eukaryotic Cell Culture
Wild type P. gingivalis ATCC 33277, isogenic ΔserB, ΔserB+pserB (Tribble et al., 2006) and serB::FLAG were used in this study. In the complemented strain, the SerB protein is expressed from the pT-COW low copy number plasmid under the control of the serB promoter. In the serB::FLAG strain, a three tandem FLAG-tag sequence was inserted at the C-terminal end of SerB protein. Bacteria were cultured in trypticase soy broth supplemented with yeast extract (1mg ml−1), hemin (5 μg ml−1), and menadione(1 μg ml−1), anaerobically, at 37°C. Overnight cultures were diluted 1:100 and grown to mid-exponential phase (OD600 nm 0.7). Human immortalized gingival keratinocytes (HIGKs) (Oda et al., 1996), derived from the gingival epithelium, were cultured at 37°C, 5% CO2 in keratinocyte serum-free medium supplemented with 0.05 mM calcium chloride and 200 mM L-glutamine (Invitrogen).
Confocal Laser Scanning Microscopy
HIGKs were seeded at 1.0 × 105 cells on glass coverslips in 12-well plates and grown until ≈40% confluent. Cells were infected with P. gingivalis at MOI 10 for 10 min unless otherwise stated. Coverslips were washed 4 times in phosphate-buffered saline (PBS) and fixed for 10 min in 4% paraformaldehyde. Permeabilization was with 0.2% Triton X-100 for 10 min at room temperature, prior to blocking in 10% goat serum for 20 min. Cofilin and phospho-cofilin were detected by reacting with primary antibodies (Sigma) at 1:100 dilution for 1 h, followed by Alexa-647-conjugated anti-rabbit secondary antibody (1:200) for 1 h in the dark. Bacteria were detected using 1:200 P. gingivalis antibodies and 1:200 Alexa-488-conjugated secondary antibodies. FLAG was detected with monoclonal antibody (1:1000; Sigma) and Alexa-647 conjugate anti-mouse secondary antibody. Myc was detected with FITC-conjugated antibody (1:1000; Sigma). Actin was labeled using 1:100 TRITC-phalloidin (Sigma) for 40 min at room temperature. After 4 washes in PBS, coverslips were mounted using ProLong Gold with DAPI mounting medium (Invitrogen).
Images were acquired on a Leica DM IRE2 inverted fluorescent microscope, with a Leica TCS SP2 AOBS spectral confocal scanner and Leica LCS software, using a 63x water immersion HCX PL APO WCORR objective. Briefly, z-stacks were obtained (10 layers/stack, 2 μm between layers) through the z-axis of cells (3 z-stacks/coverslip). Internal P. gingivalis were enumerated by counting intracellular bacteria for each z-stack, thus allowing bacteria to be visualized throughout cytoplasmic layers and ensuring that the bacteria counted were within the host cell cytoplasm. To calculate cofilin:p-cofilin ratios, and levels of F-actin, fluorescence intensity was measured using Leica Lite Confocal software, and values (arbitrary fluorescence units) obtained.
Plasmid Construction
Wild type, constitutively active (S3A) or phosphorylated mimic (S3E, inactive) cofilin constructs were generated by PCR using primers previously described (Dai et al., 2004), and cloned into the pCMV-Myc vector (Clontech). Plasmid pWZLNeoMyrFlagLIMK-1 plasmid containing wild type LIMK-1 was obtained from Addgene
HIGK transfections
Cells were transfected using siPORT NeoFX transfection agent (Applied Biosytems). Cofilin and LIMK constructs were used at 2 μg/ml and 10 μg/ml respectively. For cofilin silencing assays, 100 nM of si-cofilin, non-silence control (a non-targeting RNA negative control, no significant homology to any known human gene sequence), or scramble cofilin (Qiagen) were utilized. For ectopic expression of SerB, pCMV-Myc containing serB was transfected at 1 μg/ml. Following 24 h in transfection media, cells were returned to regular culture medium for a further 24 h. Transfected cells were immuno-labeled and examined by CSLM, or analyzed by Western blot analyses.
Western Immunoblotting
Infected and control HIGKs were lysed in SDS-PAGE sample buffer, separated by SDS-PAGE, and transferred onto nitrocellulose membranes by electroblotting. Blots were blocked in 10% skimmed dry milk in Tris-buffered saline (TBS) overnight at 4°C. Primary antibody was rabbit anti-cofilin, rabbit anti-phospho-cofilin, or rabbit anti-LIMK-1 (Sigma), diluted 1:1000 and incubated for 2 h at room temperature. Antigen-antibody binding was detected using horseradish peroxidase-conjugated species-specific secondary antibodies followed by ECL Western Blotting detection reagents (Thermo). Blots were stripped and probed with GAPDH antibodies as a loading control.
Supplementary Material
Acknowledgments
The support of the NIH/NIDCR through DE11111 is gratefully acknowledged
References
- Agnew BJ, Minamide LS, Bamburg JR. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J Biol Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- Albandar JM. Underestimation of Periodontitis in NHANES Surveys. J Periodontol. 2011;82:337–341. doi: 10.1902/jop.2011.100638. [DOI] [PubMed] [Google Scholar]
- Amano T, Kaji N, Ohashi K, Mizuno K. Mitosis-specific activation of LIM motif-containing protein kinase and roles of cofilin phosphorylation and dephosphorylation in mitosis. J Cell Biol. 2002;277:22093–22102. doi: 10.1074/jbc.M201444200. [DOI] [PubMed] [Google Scholar]
- Amin M, Magnusson K, Kapus A, Glogauer M, Ellen RP. Treponema denticola Msp-deduced peptide conjugate, P34BSA, promotes RhoA-dependent actin stress fiber formation independent of its internalization by fibroblasts. Cell Motil Cytoskeleton. 2008;65:406–421. doi: 10.1002/cm.20270. [DOI] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- Avila M, Ojcius DM, Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 2009;28:405–411. doi: 10.1089/dna.2009.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge B, Verma RK, Eastman C, Yehia B, Rivera M, Moffatt CE, et al. Role of Porphyromonas gingivalis phosphoserine phosphatase enzyme SerB in inflammation, immune response, and induction of alveolar bone resorption in rats. Infect Immun. 2010;78:4560–4569. doi: 10.1128/IAI.00703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg JR, McGough A, Ono S. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 1999;9:364–370. doi: 10.1016/s0962-8924(99)01619-0. [DOI] [PubMed] [Google Scholar]
- Belton CM, Izutsu KT, Goodwin PC, Park Y, Lamont RJ. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell Microbiol. 1999;1:215–223. doi: 10.1046/j.1462-5822.1999.00022.x. [DOI] [PubMed] [Google Scholar]
- Bernard O. Lim kinases, regulators of actin dynamics. Int J Biochem Cell Biol. 2007;39:1071–1076. doi: 10.1016/j.biocel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Gouin E, Roux P, Caroni P, Yin HL, Cossart P. A role for cofilin and LIM kinase in Listeria-induced phagocytosis. J Cell Biol. 2001;155:101–112. doi: 10.1083/jcb.200104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdet-Sicard R, Rudiger M, Jockusch BM, Gounon P, Sansonetti PJ, Nhieu GT. Binding of the Shigella protein IpaA to vinculin induces F-actin depolymerization. EMBO J. 1999;18:5853–5862. doi: 10.1093/emboj/18.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SJ, Dashper SG, Darby IB, Adams GG, Hoffmann B, Reynolds EC. Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol Immunol. 2009;24:469–477. doi: 10.1111/j.1399-302X.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- Cameron PU, Saleh S, Sallmann G, Solomon A, Wightman F, Evans VA, et al. Establishment of HIV-1 latency in resting CD4+ T cells depends on chemokine-induced changes in the actin cytoskeleton. Proc Natl Acad Sci USA. 2010;107:16934–16939. doi: 10.1073/pnas.1002894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Laidig KE, Park Y, Park K, Yates JR, Lamont RJ, Hackett M. Searching the Porphyromonas gingivalis genome with peptide fragmentation mass spectra. The Analyst. 2001;126:52–57. doi: 10.1039/b008012h. [DOI] [PubMed] [Google Scholar]
- Colombo AV, Silva CM, Haffajee A, Colombo APV. Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J Med Microbiol. 2006;55:609–615. doi: 10.1099/jmm.0.46417-0. [DOI] [PubMed] [Google Scholar]
- Colombo AV, Silva CM, Haffajee A, Colombo APV. Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J Periodont Res. 2007;42:236–243. doi: 10.1111/j.1600-0765.2006.00938.x. [DOI] [PubMed] [Google Scholar]
- Cossart P, Bierne H. The use of host cell machinery in the pathogenesis of Listeria monocytogenes. Curr Opin Immunol. 2001;13:96–103. doi: 10.1016/s0952-7915(00)00188-6. [DOI] [PubMed] [Google Scholar]
- Dai S, Sarmiere PD, Wiggan O, Bamburg JR, Zhou D. Efficient Salmonella entry requires activity cycles of host ADF and cofilin. Cell Microbiol. 2004;6:459–471. doi: 10.1111/j.1462-5822.2004.00375.x. [DOI] [PubMed] [Google Scholar]
- Darveau RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais V, Ghosh M, Eddy R, Condeelis J. Cofilin takes the lead. J Cell Sci. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- Fazal F, Bijli KM, Minhajuddin M, Rein T, Finkelstein JN, Ranman A. Essential role of cofilin-1 in regulating thrombin-induced RelA/p65 nuclear translocation and intercellular adhesion molecule 1 (ICAM-1) expression in endothelial cells. J Biol Chem. 2009;284:21047–21056. doi: 10.1074/jbc.M109.016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Weinberg A. Role of bacteria in health and disease of periodontal tissues. Periodontol 2000. 2006;40:50–76. doi: 10.1111/j.1600-0757.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- Frantz C, Barreiro G, Dominguez L, Chen X, Eddy R, Condeelis J, et al. Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J Cell Biol. 2008;183:865–879. doi: 10.1083/jcb.200804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohla A, Birkenfeld J, Bokoch GM. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat Cell Biol. 2005;7:21–29. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 2009;11:637–645. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Tribble GD, Baker HV, Mans JJ, Handfield M, Lamont RJ. Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect Immun. 2008;76:2420–2427. doi: 10.1128/IAI.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Huang TY, DerMardirossian C, Bokoch GM. Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Huang TY, Minamide LS, Bamburg JR, Bokoch GM. Chronophin mediates an ATP-sensing mechanism for cofilin dephosphorylation and neuronal cofilin-actin rod formation. Dev Cell. 2008;15:691–703. doi: 10.1016/j.devcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Kiuchi T, Ohashi K, Kurita S, Mizuno K. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J Cell Biol. 2007;177:465–476. doi: 10.1083/jcb.200610005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P, Andersen R, Blehert D, Egland P, Foster J, Palmer R., Jr Communication among oral bacteria. Micro Mol Biol Rev. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M, Hasegawa Y, Mao S, Shizukuishi S, Amano A, Lamont RJ, Yilmaz O. P. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 2008;10:122–128. doi: 10.1016/j.micinf.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn TB, Meberg PJ, Brown MD, Bernstein BW, Minamide LS, Jensen JR, et al. Regulating actin dynamics in neuronal growth cones by ADF/cofilin and rho family GTPases. J Neurobiol. 2000;44:126–144. [PubMed] [Google Scholar]
- Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Micro Mol Bio Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhie EJ, Hayward RD, Koronakis V. Control of actin turnover by a Salmonella invasion protein. Mol Cell. 2004;13:497–510. doi: 10.1016/s1097-2765(04)00053-x. [DOI] [PubMed] [Google Scholar]
- Maniotis AJ, Chen CS, Ingbar DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, et al. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9:1997–2007. doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon A, Drubin DG. The ADF/cofilin proteins: stimulus-responsive modulators of actin dynamics. Mol Biol Cell. 1995;6:1423–1431. doi: 10.1091/mbc.6.11.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K, Iida K, Yahara I. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells. 1996;1:73–86. doi: 10.1046/j.1365-2443.1996.05005.x. [DOI] [PubMed] [Google Scholar]
- Nebl G, Meuer SC, Samstag Y. Dephosphorylation of serine 3 regulates nuclear translocation of cofilin. J Biol Chem. 1996;271:26276–26280. doi: 10.1074/jbc.271.42.26276. [DOI] [PubMed] [Google Scholar]
- Nishita M, Aizawa H, Mizuno K. Stromal cell-derived factor 1α activates LIM kinase 1 and induces cofilin phosphorylation for T-cell chemotaxis. Mol Cell Biol. 2002;22:774–783. doi: 10.1128/MCB.22.3.774-783.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda D, Bigler L, Lee P, Blanton R. HPV immortalization of human oral epithelial cells: a model for carcinogenesis. Exp Cell Res. 1996;226:164–169. doi: 10.1006/excr.1996.0215. [DOI] [PubMed] [Google Scholar]
- Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M, Condeelis J. The cofilin activity cycle in lamellipodia and invadopodia. J Cell Biol. 2009;108:1252–1262. doi: 10.1002/jcb.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Olasen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Pritchard CA, Hayes L, Wojnowski L, Zimmer A, Marais RM, Norman JC. B-Raf acts via the ROCKII/LIMK/cofilin pathway to maintain actin stress fibers in fibroblasts. Mol Cell Biol. 2004;24:5937–5952. doi: 10.1128/MCB.24.13.5937-5952.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renoult C, Ternent D, Maciver SK, Fattoum A, Astier C, Benyamin Y, Roustan C. The identification of a second cofilin binding site on actin suggests a novel, intercalated arrangement of F-actin binding. J Biol Chem. 1999;274:28893–28899. doi: 10.1074/jbc.274.41.28893. [DOI] [PubMed] [Google Scholar]
- Rosengart MR, Arbabi S, Bauer GJ, Garcia I, Jelacic S, Maier RV. The actin cytoskeleton: an essential component for enhanced TNF-α production by adherent monocytes. Shock. 2002;17:109–113. doi: 10.1097/00024382-200202000-00005. [DOI] [PubMed] [Google Scholar]
- Rudney J, Chen R, Sedgewick G. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect Immun. 2001;69:2700–2707. doi: 10.1128/IAI.69.4.2700-2707.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudney J, Chen R, Zhang G. Streptococci dominate the diverse flora within buccal cells. J Dent Res. 2005;84:1165–1171. doi: 10.1177/154405910508401214. [DOI] [PubMed] [Google Scholar]
- Sarmiere PD, Bamburg JR. Regulation of the neuronal actin cytoskeleton by ADF/cofilin. J Neurobiol. 2004;58:103–117. doi: 10.1002/neu.10267. [DOI] [PubMed] [Google Scholar]
- Tribble GD, Lamont RJ. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol 2000. 2010;52:68–83. doi: 10.1111/j.1600-0757.2009.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribble GD, Mao S, James CE, Lamont RJ. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc Natl Acad Sci USA. 2006;103:11027–11032. doi: 10.1073/pnas.0509813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman I, Gervasio OL, Cullen KM, Guillemin GJ, Jeong EV, Witting PK, et al. Activated ADF/cofilin sequesters phosphorylated microtubuleassociated-protein during the assembly of Alzheimer-like neuritic cytoskeletal striations. J Neurosci. 2009;29:12994–13005. doi: 10.1523/JNEUROSCI.3531-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, et al. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- Yilmaz O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiol. 2008;154:2897–2903. doi: 10.1099/mic.0.2008/021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O, Jungas T, Verbeke P, Ojcius DM. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 2004;72:3743–3751. doi: 10.1128/IAI.72.7.3743-3751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O, Verbeke P, Lamont RJ, Ojcius DM. Intercellular spreading of Porphyromonas gingivalis infection in primary gingival epithelial cells. Infect Immun. 2006;74:703–710. doi: 10.1128/IAI.74.1.703-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Du L, Huang Y, Wu Y, Gunst SJ. Actin depolymerization factor/cofilin activation regulates actin polymerization and tension development in canine tracheal smooth muscle. J Biol Chem. 2008;283:36522–36531. doi: 10.1074/jbc.M805294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.