Abstract
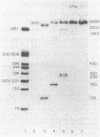
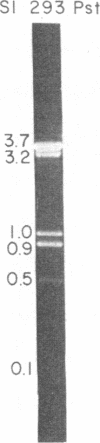
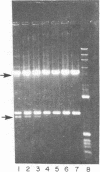
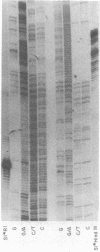
A site that is preferentially cleaved by the single-strand-specific endonuclease from Aspergillus oryzae was located in vitro 180 base pairs upstream from the 5' end of the chicken pro-alpha 2(I) collagen gene. It is found in supercoiled plasmids with a negative superhelical density of -0.024 or more but not in linear DNA molecules. The nuclease S1 sensitivity is retained in plasmids containing genomic fragments extending from position +8 to -285 (where +1 is the first transcribed base) and from -147 to -351 and also in a 5.7-kilobase EcoRI fragment that extends 1.6 kilobases 5' and 4.1 kilobases 3' to the 5' end of the gene. Analysis at the nucleotide level on a DNA sequence gel places the site at -181 to -182 on the sense strand and at -182 to -184 and -192 to -195 on the nonsense strand. These sites lie within a stretch of 42 pyrimidines interrupted by a single guanine and within the sequence T-C-C-C-T-C-C-C-T-T-C-C-T-C-C-C-T-C-C-C-T.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goding C. R., Russell W. C. S1 sensitive sites in adenovirus DNA. Nucleic Acids Res. 1983 Jan 11;11(1):21–36. doi: 10.1093/nar/11.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Weintraub H. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell. 1982 Aug;30(1):131–139. doi: 10.1016/0092-8674(82)90019-8. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Keene M. A., Corces V., Lowenhaupt K., Elgin S. C. DNase I hypersensitive sites in Drosophila chromatin occur at the 5' ends of regions of transcription. Proc Natl Acad Sci U S A. 1981 Jan;78(1):143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace H. A., Pelham H. R., Travers A. A. Association of an S1 nuclease-sensitive structure with short direct repeats 5' of Drosophila heat shock genes. Nature. 1983 Aug 11;304(5926):555–557. doi: 10.1038/304555a0. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Merlino G. T., McKeon C., de Crombrugghe B., Pastan I. Regulation of the expression of genes encoding types I, II, and III collagen during chick embryonic development. J Biol Chem. 1983 Aug 25;258(16):10041–10048. [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Nordheim A., Rich A., Wang J. C. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Künzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 1979 Apr;6(4):1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Tate V. E., Finer M. H., Boedtker H., Doty P. Chick pro alpha 2 (I) collagen gene: exon location and coding potential for the prepropeptide. Nucleic Acids Res. 1983 Jan 11;11(1):91–104. doi: 10.1093/nar/11.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate V., Finer M., Boedtker H., Doty P. Procollagen genes: further sequence studies and interspecies comparisons. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1039–1049. doi: 10.1101/sqb.1983.047.01.117. [DOI] [PubMed] [Google Scholar]
- Vogeli G., Ohkubo H., Sobel M. E., Yamada Y., Pastan I., de Crombrugghe B. Structure of the promoter for chicken alpha 2 type I collagen gene. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5334–5338. doi: 10.1073/pnas.78.9.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Interactions between twisted DNAs and enzymes: the effects of superhelical turns. J Mol Biol. 1974 Aug 25;87(4):797–816. doi: 10.1016/0022-2836(74)90085-0. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]