Abstract
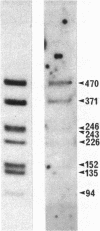
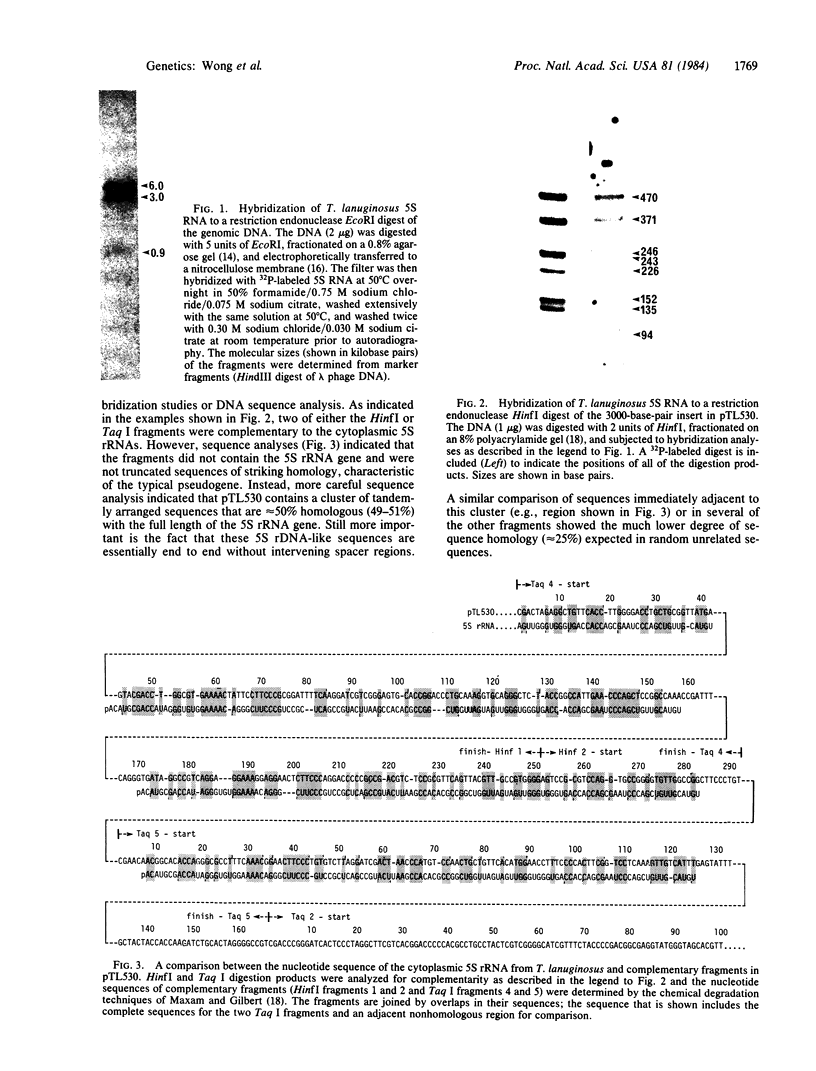
The genome of a thermophilic fungus, Thermomyces lanuginosus, contains a tandemly arranged cluster of sequences that are approximately equal to 50% homologous with the cytoplasmic 5S RNA and that are selectable by hybridization techniques. Unlike typical pseudogenes, these sequences are not truncated; rather, they bear a limited sequence homology with the entire length of the 5S RNA and are oriented end to end without significant intervening sequences. We suggest that these are gene relics that were duplicated by a rolling circle-like mechanism and that have evolutionarily drifted to become gene spacers. Accordingly, we raise the possibility that this offers a fortuitous glimpse at the origins for many of the gene spacers in the eukaryotic genome.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J. RNA processing and the intervening sequence problem. Annu Rev Biochem. 1979;48:1035–1069. doi: 10.1146/annurev.bi.48.070179.005131. [DOI] [PubMed] [Google Scholar]
- Bernstein L. B., Mount S. M., Weiner A. M. Pseudogenes for human small nuclear RNA U3 appear to arise by integration of self-primed reverse transcripts of the RNA into new chromosomal sites. Cell. 1983 Feb;32(2):461–472. doi: 10.1016/0092-8674(83)90466-x. [DOI] [PubMed] [Google Scholar]
- Bittner M., Kupferer P., Morris C. F. Electrophoretic transfer of proteins and nucleic acids from slab gels to diazobenzyloxymethyl cellulose or nitrocellulose sheets. Anal Biochem. 1980 Mar 1;102(2):459–471. doi: 10.1016/0003-2697(80)90182-7. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Cryer D. R., Eccleshall R., Marmur J. Isolation of yeast DNA. Methods Cell Biol. 1975;12:39–44. doi: 10.1016/s0091-679x(08)60950-4. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. V. On spacers. Cell. 1979 Apr;16(4):697–710. doi: 10.1016/0092-8674(79)90086-2. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran P., Forget B. G., Weissman S. M. Short interspersed repetitive DNA elements in eucaryotes: transposable DNA elements generated by reverse transcription of RNA pol III transcripts? Cell. 1981 Oct;26(2 Pt 2):141–142. doi: 10.1016/0092-8674(81)90296-8. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Tizard R., Skryabin K. G., Gilbert W. Promotor region for yeast 5S ribosomal RNA. Nature. 1977 Jun 16;267(5612):643–645. doi: 10.1038/267643a0. [DOI] [PubMed] [Google Scholar]
- Miller J. R., Cartwright E. M., Brownlee G. G., Fedoroff N. V., Brown D. D. The nucleotide sequence of oocyte 5S DNA in Xenopus laevis. II. The GC-rich region. Cell. 1978 Apr;13(4):717–725. doi: 10.1016/0092-8674(78)90221-0. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Free S. J., Metzenberg R. L., Yanofsky C. An isolated pseudogene related to the 5S RNA genes in Neurospora crassa. Nature. 1981 Dec 10;294(5841):576–578. doi: 10.1038/294576a0. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Yanofsky C., Driftmier K., Metzenberg R. L., Alzner-DeWeerd B., RajBhandary U. L. Dispersed 5S RNA genes in N. crassa: structure, expression and evolution. Cell. 1981 Jun;24(3):819–828. doi: 10.1016/0092-8674(81)90107-0. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Wildeman A. G., Nazar R. N. Structural studies of 5 S ribosomal RNAs from a thermophilic fungus, Thermomyces lanuginosus. A comparison of generalized models for eukaryotic 5 S RNAs. J Biol Chem. 1982 Oct 10;257(19):11395–11404. [PubMed] [Google Scholar]