Abstract
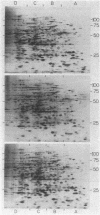
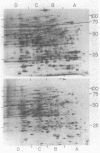
Two-dimensional gel electrophoresis resolved total cellular protein from Euglena gracilis Klebs var. bacillaris Cori into 640 polypeptides detectable by silver staining. The addition of 84 mM ethanol to dark-grown resting carbon-starved cells increased the relative amounts of 6 polypeptides and decreased the relative amounts of 3 polypeptides. The addition of 84 mM malate to resting cells increased the relative amounts of 3 of the ethanol-induced polypeptides, suggesting that the induction of these polypeptides represents a generalized response to the provision of a utilizable carbon source, a nutritional shift up, rather than a specific response to ethanol addition. Exposure of dark-grown resting Euglena to light increases the relative amounts of 79 polypeptides encoded by the nuclear as well as the chloroplast genome and decreases the relative amounts of 72 polypeptides. Ethanol but not malate specifically inhibited all of the light-dependent changes in polypeptide levels, indicating that chloroplast development in Euglena is a catabolite-sensitive process.
Keywords: two-dimensional, polyacrylamide gels, silver stain, light induced, ethanol
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beale S. I., Foley T., Dzelzkalns V. delta-Aminolevulinic acid synthase from Euglena gracilis. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1666–1669. doi: 10.1073/pnas.78.3.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovarnick J. G., Schiff J. A., Freedman Z., Egan J. M. Events surrounding the early development of Euglena chloroplasts: cellular origins of chloroplast enzymes in euglena. J Gen Microbiol. 1974 Jul;83(0):63–71. doi: 10.1099/00221287-83-1-63. [DOI] [PubMed] [Google Scholar]
- Chelm B. K., Hallick R. B. Changes in the expression of the chloroplast genome of Euglena gracilis during chloroplast development. Biochemistry. 1976 Feb 10;15(3):593–599. doi: 10.1021/bi00648a022. [DOI] [PubMed] [Google Scholar]
- Cohen D., Schiff J. A. Events surrounding the early development of Euglena chloroplasts. Photoregulation of the transcription of chloroplastic and cytoplasmic ribosomal RNAs. Arch Biochem Biophys. 1976 Nov;177(1):201–216. doi: 10.1016/0003-9861(76)90430-6. [DOI] [PubMed] [Google Scholar]
- Collins N., Merrett M. J. Microbody-marker Enzymes during Transition from Phototrophic to Organotrophic Growth in Euglena. Plant Physiol. 1975 Jun;55(6):1018–1022. doi: 10.1104/pp.55.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis S. E., Rawson J. R. Measurement of the transcription of nuclear single-copy deoxyribonucleic acid during chloroplast development in Euglena gracilis. Biochemistry. 1979 Nov 27;18(24):5299–5304. doi: 10.1021/bi00591a006. [DOI] [PubMed] [Google Scholar]
- Entian K. D. A carbon catabolite repression mutant of Saccharomyces cerevisiae with elevated hexokinase activity: evidence for regulatory control of hexokinase PII synthesis. Mol Gen Genet. 1981;184(2):278–282. doi: 10.1007/BF00272917. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Zimmermann F. K. New genes involved in carbon catabolite repression and derepression in the yeast Saccharomyces cerevisiae. J Bacteriol. 1982 Sep;151(3):1123–1128. doi: 10.1128/jb.151.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves L. B., Jr, Trelease R. N., Grill A., Becker W. M. Localization of glyoxylate cycle enzymes in glyoxysomes in Euglena. J Protozool. 1972 Aug;19(3):527–532. doi: 10.1111/j.1550-7408.1972.tb03521.x. [DOI] [PubMed] [Google Scholar]
- Handa A. K., Bressan R. A., Quader H., Filner P. Association of Formation and Release of Cyclic AMP with Glucose Depletion and Onset of Chlorophyll Synthesis in Poterioochromonas malhamensis. Plant Physiol. 1981 Aug;68(2):460–463. doi: 10.1104/pp.68.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. C., Kirk J. T. Control of chloroplast formation in Euglena gracilis. Antagonism between carbon and nitrogen sources. Biochem J. 1969 Jun;113(1):195–205. doi: 10.1042/bj1130195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrum M. A., Schwartzbach S. D. Nutritional Regulation of Organelle Biogenesis in Euglena: INDUCTION OF MICROBODIES. Plant Physiol. 1981 Aug;68(2):430–434. doi: 10.1104/pp.68.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrum M. A., Schwartzbach S. D. Nutritional Regulation of Organelle Biogenesis in Euglena: REPRESSION OF CHLOROPHYLL AND NADP-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE SYNTHESIS. Plant Physiol. 1980 Feb;65(2):382–386. doi: 10.1104/pp.65.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keirns J. J., Carritt B., Freeman J., Eisenstadt J. M., Bitensky M. W. Adenosine 3',5' cyclic monophosphate in Euglena gracilis. Life Sci. 1973 Aug 16;13(4):287–302. doi: 10.1016/0024-3205(73)90220-8. [DOI] [PubMed] [Google Scholar]
- Mahler H. R., Jaynes P. K., McDonough J. P., Hanson D. K. Catabolite repression in yeast: mediation by cAMP. Curr Top Cell Regul. 1981;18:455–474. doi: 10.1016/b978-0-12-152818-8.50033-5. [DOI] [PubMed] [Google Scholar]
- Nicolas P., Nigon V. Chloroplast and non-chloroplast adenosine-3':5'-cyclic-monophosphate-receptor-proteins in Euglena gracilis. FEBS Lett. 1974 Dec 15;49(2):254–259. doi: 10.1016/0014-5793(74)80524-7. [DOI] [PubMed] [Google Scholar]
- Pall M. L. Adenosine 3',5'-phosphate in fungi. Microbiol Rev. 1981 Sep;45(3):462–480. doi: 10.1128/mr.45.3.462-480.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastan I., Adhya S. Cyclic adenosine 5'-monophosphate in Escherichia coli. Bacteriol Rev. 1976 Sep;40(3):527–551. doi: 10.1128/br.40.3.527-551.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plattner H., Salpeter M. M., Saltzgaber J., Schatz G. Promitochondria of anaerobically grown yeast. IV. Conversion into respiring mitochondria. Proc Natl Acad Sci U S A. 1970 Aug;66(4):1252–1259. doi: 10.1073/pnas.66.4.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson J. R., Boerma C. L., Andrews W. H., Wilkerson C. G. Complexity and abundance of ribonucleic acid transcribed from restriction endonuclease fragments of Euglena chloroplast deoxyribonucleic acid during chloroplast development. Biochemistry. 1981 Apr 28;20(9):2639–2644. doi: 10.1021/bi00512a043. [DOI] [PubMed] [Google Scholar]
- Spector T. Refinement of the coomassie blue method of protein quantitation. A simple and linear spectrophotometric assay for less than or equal to 0.5 to 50 microgram of protein. Anal Biochem. 1978 May;86(1):142–146. doi: 10.1016/0003-2697(78)90327-5. [DOI] [PubMed] [Google Scholar]
- Thorner J. An essential role for cyclic AMP in growth control: the case for yeast. Cell. 1982 Aug;30(1):5–6. doi: 10.1016/0092-8674(82)90004-6. [DOI] [PubMed] [Google Scholar]
- Verdier G. Poly(adenylic acid)-containing RNA of Euglena gracilis during chloroplast development. I. Analysis of their complexity by hybridization to complementary DNA. Eur J Biochem. 1979 Feb 1;93(3):573–580. doi: 10.1111/j.1432-1033.1979.tb12857.x. [DOI] [PubMed] [Google Scholar]