Abstract
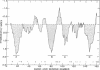
The nucleotide sequence of the prolipoprotein signal peptidase (lsp) gene has been determined. The lsp gene was found to be adjacent to the isoleucyl-tRNA synthetase ( ileS ) gene, such that the termination codon of the ileS gene overlaps with the initiation codon of lsp. These two genes are transcribed in the same direction and the major promotor for the lsp gene appears to be upstream of ileS . Identification of the lsp gene was established by amplification of prolipoprotein signal peptidase activity in strains carrying a subcloned 1.1-kilobase Stu I-Acc I fragment and was further confirmed by introducing mutational alterations in the COOH terminus of the protein that caused a decrease in prolipoprotein signal peptidase activity. The deduced amino acid sequence indicates that prolipoprotein signal peptidase contains 164 residues. Unlike most exported proteins, there is no apparent signal peptide sequence for the lsp protein. Computer-assisted secondary structure analysis of the deduced amino acid sequence identified four hydrophobic regions that share features common to transmembrane segments in integral membrane proteins.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Date T., Wickner W. Isolation of the Escherichia coli leader peptidase gene and effects of leader peptidase overproduction in vivo. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6106–6110. doi: 10.1073/pnas.78.10.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisolia V., Riccio A., Bruni C. B. Structure and function of the internal promoter (hisBp) of the Escherichia coli K-12 histidine operon. J Bacteriol. 1983 Sep;155(3):1288–1296. doi: 10.1128/jb.155.3.1288-1296.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hussain M., Ichihara S., Mizushima S. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J Biol Chem. 1980 Apr 25;255(8):3707–3712. [PubMed] [Google Scholar]
- Hussain M., Ichihara S., Mizushima S. Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J Biol Chem. 1982 May 10;257(9):5177–5182. [PubMed] [Google Scholar]
- Inokuchi K., Mutoh N., Matsuyama S., Mizushima S. Primary structure of the ompF gene that codes for a major outer membrane protein of Escherichia coli K-12. Nucleic Acids Res. 1982 Nov 11;10(21):6957–6968. doi: 10.1093/nar/10.21.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai M., Takeuchi M., Shimizu K., Arai M. Mechanism of action of globomycin. J Antibiot (Tokyo) 1978 Nov;31(11):1203–1205. doi: 10.7164/antibiotics.31.1203. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Mackie G. A., Parsons G. D. Identification of gene products from cloned fragments of the left arm of lambda dapB2. Can J Biochem. 1982 Mar;60(3):338–346. doi: 10.1139/o82-040. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Chang S. Y., Chang S. Transcriptional analyses of the Bacillus licheniformis penP gene. Nucleic Acids Res. 1982 Jul 10;10(13):3905–3919. doi: 10.1093/nar/10.13.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Michaelis S., Beckwith J. Mechanism of incorporation of cell envelope proteins in Escherichia coli. Annu Rev Microbiol. 1982;36:435–465. doi: 10.1146/annurev.mi.36.100182.002251. [DOI] [PubMed] [Google Scholar]
- Platt T., Yanofsky C. An intercistronic region and ribosome-binding site in bacterial messenger RNA. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2399–2403. doi: 10.1073/pnas.72.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regue M., Remenick J., Tokunaga M., Mackie G. A., Wu H. C. Mapping of the lipoprotein signal peptidase gene (lsp). J Bacteriol. 1984 May;158(2):632–635. doi: 10.1128/jb.158.2.632-635.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M., Loranger J. M., Wolfe P. B., Wu H. C. Prolipoprotein signal peptidase in Escherichia coli is distinct from the M13 procoat protein signal peptidase. J Biol Chem. 1982 Sep 10;257(17):9922–9925. [PubMed] [Google Scholar]
- Tokunaga M., Loranger J. M., Wu H. C. A distinct signal peptidase for prolipoprotein in Escherichia coli. J Cell Biochem. 1984;24(2):113–120. doi: 10.1002/jcb.240240203. [DOI] [PubMed] [Google Scholar]
- Tokunaga M., Loranger J. M., Wu H. C. Isolation and characterization of an Escherichia coli clone overproducing prolipoprotein signal peptidase. J Biol Chem. 1983 Oct 25;258(20):12102–12105. [PubMed] [Google Scholar]
- Wolfe P. B., Wickner W., Goodman J. M. Sequence of the leader peptidase gene of Escherichia coli and the orientation of leader peptidase in the bacterial envelope. J Biol Chem. 1983 Oct 10;258(19):12073–12080. [PubMed] [Google Scholar]
- Yamagata H., Taguchi N., Daishima K., Mizushima S. Genetic characterization of a gene for prolipoprotein signal peptidase in Escherichia coli. Mol Gen Genet. 1983;192(1-2):10–14. doi: 10.1007/BF00327640. [DOI] [PubMed] [Google Scholar]
- Zwizinski C., Wickner W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem. 1980 Aug 25;255(16):7973–7977. [PubMed] [Google Scholar]
- von Heijne G. Membrane proteins: the amino acid composition of membrane-penetrating segments. Eur J Biochem. 1981 Nov;120(2):275–278. doi: 10.1111/j.1432-1033.1981.tb05700.x. [DOI] [PubMed] [Google Scholar]