Abstract
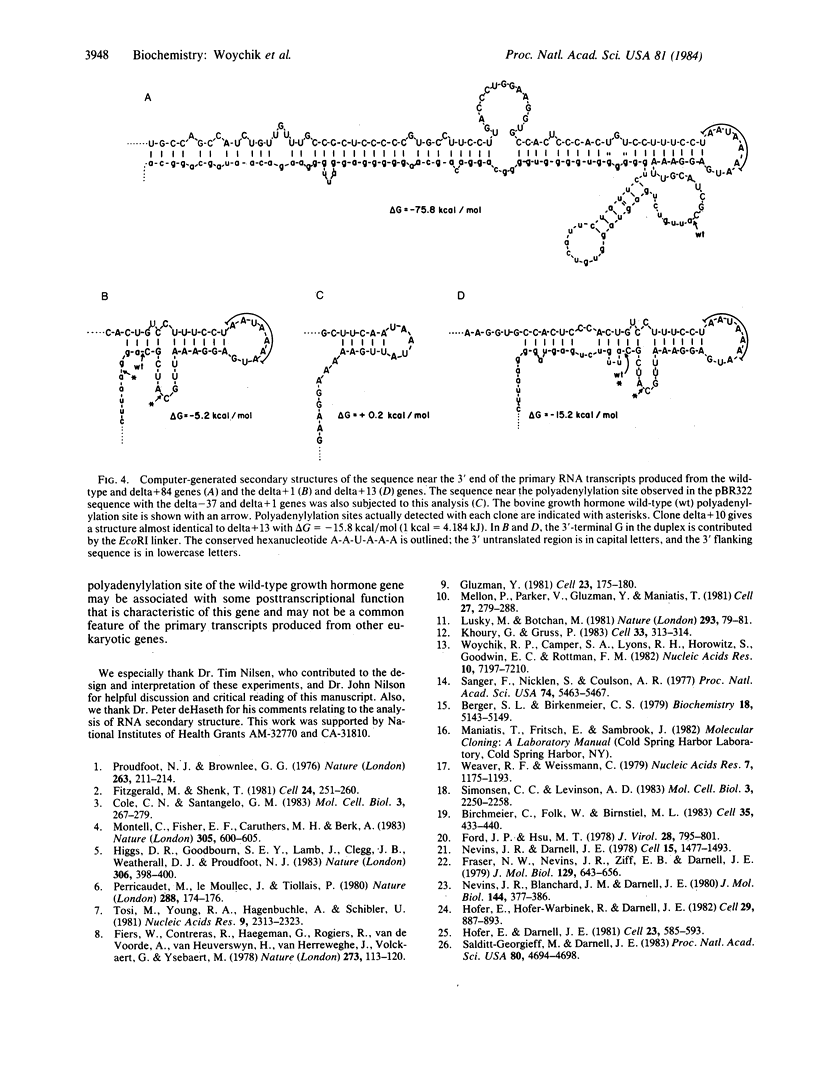
We examined whether the sequence extending 3' to the polyadenylylation site of the bovine growth hormone gene contains any signal that affects the polyadenylylation of the growth hormone mRNA. For this purpose, cloned copies of this gene, each containing a different length of growth hormone-specific sequence 3' to the wild-type polyadenylylation site, were used to transfect COS-1 cells. The polyadenylylation site on the mRNAs produced from the exogenously added growth hormone genes were analyzed with an S1 nuclease mapping procedure. We found that a gene containing 84 base pairs of its own 3' flanking sequence is capable of producing an accurately polyadenylylated mRNA. On the other hand, genes containing only 1, 10, or 13 base pairs of 3' flanking sequence were principally polyadenylylated at discrete sites either upstream or downstream from the wild-type position. Using a computer program, we examined whether secondary structures on the primary growth hormone transcript correlated with the site where the mRNA is polyadenylylated.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger S. L., Birkenmeier C. S. Inhibition of intractable nucleases with ribonucleoside--vanadyl complexes: isolation of messenger ribonucleic acid from resting lymphocytes. Biochemistry. 1979 Nov 13;18(23):5143–5149. doi: 10.1021/bi00590a018. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Folk W., Birnstiel M. L. The terminal RNA stem-loop structure and 80 bp of spacer DNA are required for the formation of 3' termini of sea urchin H2A mRNA. Cell. 1983 Dec;35(2 Pt 1):433–440. doi: 10.1016/0092-8674(83)90176-9. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Santangelo G. M. Analysis in Cos-1 cells of processing and polyadenylation signals by using derivatives of the herpes simplex virus type 1 thymidine kinase gene. Mol Cell Biol. 1983 Feb;3(2):267–279. doi: 10.1128/mcb.3.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Ford J. P., Hsu M. T. Transcription pattern of in vivo-labeled late simian virus 40 RNA: equimolar transcription beyond the mRNA 3' terminus. J Virol. 1978 Dec;28(3):795–801. doi: 10.1128/jvi.28.3.795-801.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser N. W., Nevins J. R., Ziff E., Darnell J. E., Jr The major late adenovirus type-2 transcription unit: termination is downstream from the last poly(A) site. J Mol Biol. 1979 Apr 25;129(4):643–656. doi: 10.1016/0022-2836(79)90474-1. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Higgs D. R., Goodbourn S. E., Lamb J., Clegg J. B., Weatherall D. J., Proudfoot N. J. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983 Nov 24;306(5941):398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Hofer E., Darnell J. E., Jr The primary transcription unit of the mouse beta-major globin gene. Cell. 1981 Feb;23(2):585–593. doi: 10.1016/0092-8674(81)90154-9. [DOI] [PubMed] [Google Scholar]
- Hofer E., Hofer-Warbinek R., Darnell J. E., Jr Globin RNA transcription: a possible termination site and demonstration of transcriptional control correlated with altered chromatin structure. Cell. 1982 Jul;29(3):887–893. doi: 10.1016/0092-8674(82)90450-0. [DOI] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Lusky M., Botchan M. Inhibition of SV40 replication in simian cells by specific pBR322 DNA sequences. Nature. 1981 Sep 3;293(5827):79–81. doi: 10.1038/293079a0. [DOI] [PubMed] [Google Scholar]
- Mellon P., Parker V., Gluzman Y., Maniatis T. Identification of DNA sequences required for transcription of the human alpha 1-globin gene in a new SV40 host-vector system. Cell. 1981 Dec;27(2 Pt 1):279–288. doi: 10.1016/0092-8674(81)90411-6. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Blanchard J. M., Darnell J. E., Jr Transcription units of adenovirus type 2. Termination of transcription beyond the poly(A) addition site in early regions 2 and 4. J Mol Biol. 1980 Dec 15;144(3):377–386. doi: 10.1016/0022-2836(80)90096-0. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Perricaudet M., le Moullec J. M., Tiollais P., Pettersson U. Structure of two adenovirus type 12 transforming polypeptides and their evolutionary implications. Nature. 1980 Nov 13;288(5787):174–176. doi: 10.1038/288174a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Salditt-Georgieff M., Darnell J. E., Jr A precise termination site in the mouse beta major-globin transcription unit. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4694–4698. doi: 10.1073/pnas.80.15.4694. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen C. C., Levinson A. D. Analysis of processing and polyadenylation signals of the hepatitis B virus surface antigen gene by using simian virus 40-hepatitis B virus chimeric plasmids. Mol Cell Biol. 1983 Dec;3(12):2250–2258. doi: 10.1128/mcb.3.12.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi M., Young R. A., Hagenbüchle O., Schibler U. Multiple polyadenylation sites in a mouse alpha-amylase gene. Nucleic Acids Res. 1981 May 25;9(10):2313–2323. doi: 10.1093/nar/9.10.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik R. P., Camper S. A., Lyons R. H., Horowitz S., Goodwin E. C., Rottman F. M. Cloning and nucleotide sequencing of the bovine growth hormone gene. Nucleic Acids Res. 1982 Nov 25;10(22):7197–7210. doi: 10.1093/nar/10.22.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]