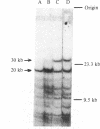
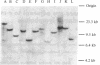
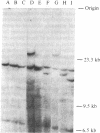
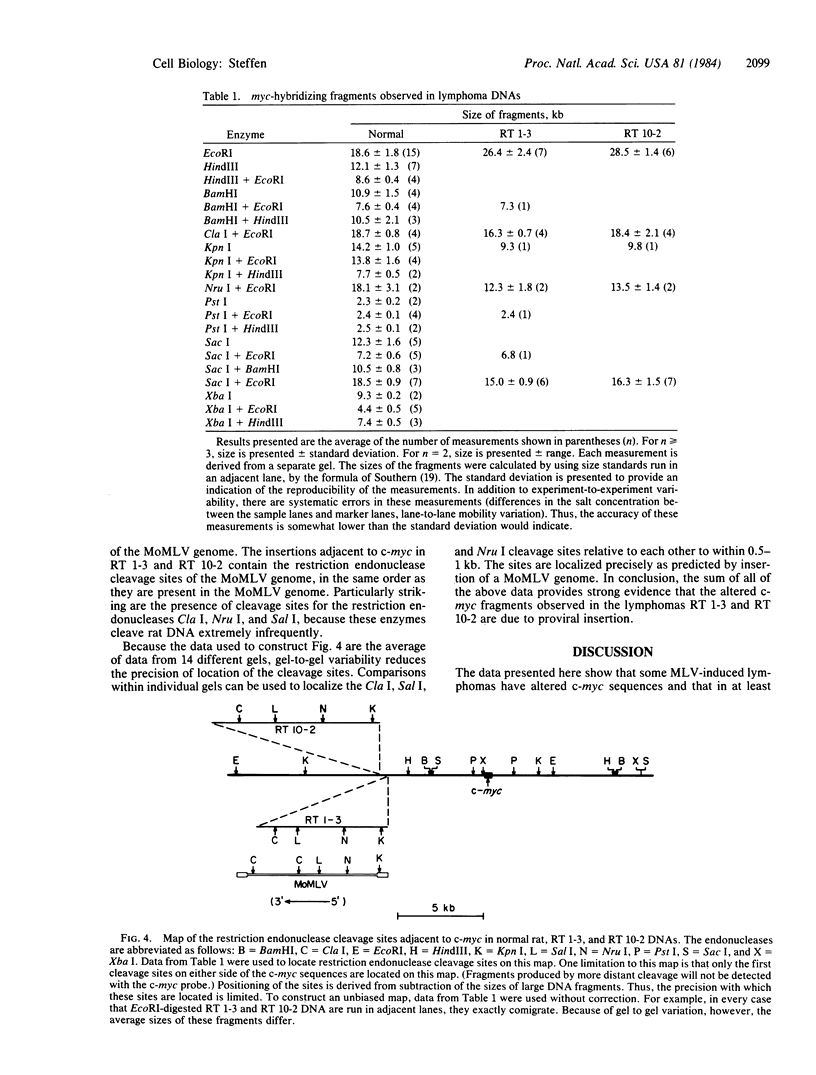
Abstract
Sixty-one murine leukemia virus-induced lymphomas were examined for evidence of proviral insertions adjacent to known oncogenes. Five of these lymphomas were found to have alterations adjacent to c-myc. Two of the lymphomas with altered c-myc sequences were examined in detail. Evidence was obtained showing that the alterations in the c-myc sequences in these two lymphomas were the result of proviral integrations. This result suggests that lymphomagenesis by murine leukemia viruses can result from insertional mutagenesis of cellular oncogenes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Gerondakis S., Webb E., Corcoran L. M., Cory S. Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1982–1986. doi: 10.1073/pnas.80.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Lander M. R., Gupta S., Rands E., Lowy D. R. Origin of mink cytopathic focus-forming (MCF) viruses:comparison with ecotropic and xenotropic murine leukemia virus genomes. Virology. 1981 Sep;113(2):465–483. doi: 10.1016/0042-6822(81)90175-6. [DOI] [PubMed] [Google Scholar]
- Fan H., Paskind M. Measurement of the sequence complexity of cloned Moloney murine leukemia virus 60 to 70S RNA: evidence for a haploid genome. J Virol. 1974 Sep;14(3):421–429. doi: 10.1128/jvi.14.3.421-429.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischinger P. J., Nomura S., Bolognesi D. P. A novel murine oncornavirus with dual eco- and xenotropic properties. Proc Natl Acad Sci U S A. 1975 Dec;72(12):5150–5155. doi: 10.1073/pnas.72.12.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung Y. K., Lewis W. G., Crittenden L. B., Kung H. J. Activation of the cellular oncogene c-erbB by LTR insertion: molecular basis for induction of erythroblastosis by avian leukosis virus. Cell. 1983 Jun;33(2):357–368. doi: 10.1016/0092-8674(83)90417-8. [DOI] [PubMed] [Google Scholar]
- Hartley J. W., Wolford N. K., Old L. J., Rowe W. P. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci U S A. 1977 Feb;74(2):789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Ihle J. N., Lee J. C. Possible immunological mechanisms in C-type viral leukemogenesis in mice. Curr Top Microbiol Immunol. 1982;98:85–101. doi: 10.1007/978-3-642-68369-5_7. [DOI] [PubMed] [Google Scholar]
- Marcu K. B., Harris L. J., Stanton L. W., Erikson J., Watt R., Croce C. M. Transcriptionally active c-myc oncogene is contained within NIARD, a DNA sequence associated with chromosome translocations in B-cell neoplasia. Proc Natl Acad Sci U S A. 1983 Jan;80(2):519–523. doi: 10.1073/pnas.80.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushinski J. F., Bauer S. R., Potter M., Reddy E. P. Increased expression of myc-related oncogene mRNA characterizes most BALB/c plasmacytomas induced by pristane or Abelson murine leukemia virus. Proc Natl Acad Sci U S A. 1983 Feb;80(4):1073–1077. doi: 10.1073/pnas.80.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel B. G., Hayward W. S., Robinson H. L., Fang J., Astrin S. M. Avian leukosis virus-induced tumors have common proviral integration sites and synthesize discrete new RNAs: oncogenesis by promoter insertion. Cell. 1981 Feb;23(2):323–334. doi: 10.1016/0092-8674(81)90128-8. [DOI] [PubMed] [Google Scholar]
- Noori-Daloii M. R., Swift R. A., Kung H. J., Crittenden L. B., Witter R. L. Specific integration of REV proviruses in avian bursal lymphomas. Nature. 1981 Dec 10;294(5841):574–576. doi: 10.1038/294574a0. [DOI] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982 Nov;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Bishop J. M., Varmus H. E. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982 Jan 21;295(5846):209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- Peters G., Brookes S., Smith R., Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983 Jun;33(2):369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Shen-Ong G. L., Keath E. J., Piccoli S. P., Cole M. D. Novel myc oncogene RNA from abortive immunoglobulin-gene recombination in mouse plasmacytomas. Cell. 1982 Dec;31(2 Pt 1):443–452. doi: 10.1016/0092-8674(82)90137-4. [DOI] [PubMed] [Google Scholar]
- Shields A., Witte W. N., Rothenberg E., Baltimore D. High frequency of aberrant expression of Moloney murine leukemia virus in clonal infections. Cell. 1978 Jul;14(3):601–609. doi: 10.1016/0092-8674(78)90245-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Measurement of DNA length by gel electrophoresis. Anal Biochem. 1979 Dec;100(2):319–323. doi: 10.1016/0003-2697(79)90235-5. [DOI] [PubMed] [Google Scholar]
- Steffen D., Bird S., Rowe W. P., Weinberg R. A. Identification of DNA fragments carrying ecotropic proviruses of AKR mice. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4554–4558. doi: 10.1073/pnas.76.9.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen D., Weinberg R. A. The integrated genome of murine leukemia virus. Cell. 1978 Nov;15(3):1003–1010. doi: 10.1016/0092-8674(78)90284-2. [DOI] [PubMed] [Google Scholar]
- Taub R., Kirsch I., Morton C., Lenoir G., Swan D., Tronick S., Aaronson S., Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichlis P. N., Strauss P. G., Hu L. F. A common region for proviral DNA integration in MoMuLV-induced rat thymic lymphomas. 1983 Mar 31-Apr 6Nature. 302(5907):445–449. doi: 10.1038/302445a0. [DOI] [PubMed] [Google Scholar]
- Vennström B., Moscovici C., Goodman H. M., Bishop J. M. Molecular cloning of the avian myelocytomatosis virus genome and recovery of infectious virus by transfection of chicken cells. J Virol. 1981 Aug;39(2):625–631. doi: 10.1128/jvi.39.2.625-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman I. L., McGrath M. S. Retrovirus lymphomagenesis: relationship of normal immune receptors to malignant cell proliferation. Curr Top Microbiol Immunol. 1982;98:103–112. doi: 10.1007/978-3-642-68369-5_8. [DOI] [PubMed] [Google Scholar]
- van der Putten H., Quint W., van Raaij J., Maandag E. R., Verma I. M., Berns A. M-MuLV-induced leukemogenesis: integration and structure of recombinant proviruses in tumors. Cell. 1981 Jun;24(3):729–739. doi: 10.1016/0092-8674(81)90099-4. [DOI] [PubMed] [Google Scholar]