Abstract
Catalytic Atom Maps (CAMs) are minimal models of enzyme active sites. The structures in the Protein Data Bank (PDB) were examined to determine if proteins with CAM-like geometries in their active sites all share the same catalytic function. We combined the CAM-based search protocol with a filter based on the weighted contact number (WCN) of the catalytic residues, a measure of the “crowdedness” of the microenvironment around a protein residue. Using this technique, a CAM based on the Ser-His-Asp catalytic triad of trypsin was able to correctly identify catalytic triads in other enzymes within 0.5 Å RMSD of the Catalytic Atom Map with 96% accuracy. A CAM based on the Cys-Arg-(Asp/Glu) active site residues from the tyrosine phosphatase active site achieved 89% accuracy in identifying this type of catalytic functionality. Both of these Catalytic Atom Maps were able to identify active sites across different fold types. Finally, the PDB was searched to locate proteins with catalytic functionality similar to that present in the active site of orotidine 5′-monophosphate decarboxylase (ODCase), whose mechanism is not known with certainty. A CAM, based on the conserved Lys-Asp-Lys-Asp tetrad in the ODCase active site, was used to search the PDB for enzymes with similar active sites. The ODCase active site has a geometry similar to that of Schiff base-forming Class I aldolases, with lowest aldolase RMSD to the ODCase CAM at 0.48 Å. The similarity between this CAM and the aldolase active site suggests that ODCase has the correct catalytic functionality present in its active site for the generation of a nucleophilic lysine.
Introduction
The development of general methods for analysis of the chemical functionality present in active sites is an ongoing challenge for biochemists, given the rapid growth in protein structural data. In the last 3 years alone (2009–2011), more than 23,000 structures have been deposited in the Protein Data Bank. (1) In order to keep pace with the wealth of structural data being generated, there exists a need for high-throughput methods to analyze and identify the types of chemical reactions that an enzyme is likely to catalyze. We previously reported SABER (Selection of Active/Binding sites for Enzyme Redesign), a program used to search large sets of protein structures for specific arrangements of atoms that match the geometry described by a minimal model of an enzyme active site, known as a Catalytic Atom Map (CAM). (2) Here we introduce a method, using SABER and a Catalytic Atom Map, to identify the mechanisms available to an enzyme. We show how the precise geometric arrangement of catalytic groups such as nucleophiles, electrophiles, and general acids/bases can be used to predict the type of reaction catalyzed by a specific, preorganized active site. This is consistent with the recognition that much of the enormous proficiency of enzymes results from the precise organization of catalytic groups. (3–5)
To support our claim, we have explored two central questions. The first is whether a Catalytic Atom Map based on the conserved residues in an active site is sufficient to correctly identify the chemistry that occurs in that active site. We have created Catalytic Atom Maps for two enzyme families, one based on a Ser-His-Asp catalytic triad, and the other from a tyrosine phosphatase active site. These CAMs were then used to search the Protein Data Bank for similar active sites. Proteins with close geometric matches to the Catalytic Atom Map were analyzed to determine if they catalyzed reactions with similar mechanisms. We also assessed the effectiveness of using a filter based on a residue’s weighted contact number (WCN) value, a measure of the microenvironment occupied by a residue in the protein. This filter removes matches to the CAM that are unlikely to be part of an active site, because they are surface exposed. (6, 7)
In the second part of this study, we explored whether this technique could be used to identify the type of catalysis that is likely to operate in the active site of an enzyme that is known to catalyze a specific reaction, but for which the mechanism is unknown. We chose to study the enzyme orotidine 5′-monophosphate decarboxylase (ODCase), and created a Catalytic Atom Map based on the conserved residues in its active site. We then used this CAM to search the Protein Data Bank for other enzymes that have the catalytic atoms in their active sites placed in a geometry similar to that of ODCase. Based on this analysis, we gained evidence that ODCase has the correct catalytic functionality to generate a nucleophilic lysine residue present in its active site.
Computational Details
SABER procedure and parameters
Catalytic Atom Map Generation
Coordinates for each of the Catalytic Atom Maps used in the SABER searches are provided in the Supporting Information. These CAMs were generated based on high-resolution X-ray structures from the Protein Data Bank (PDB codes: 1A0J, 1ZC0, and 3LTP).
SABER data collection
The radius of the sphere representing each atom in a CAM was set to 2.0 Å. A cutoff of 0.7 Å RMSD to each CAM was applied, and matches with RMSD values higher than this were discarded. Matches to each CAM were binned according to their geometric similarity to the Catalytic Atom Map in 0.1 Å increments, resulting in seven bins for each dataset: 0 – 0.10 Å, 0.11 – 0.2 Å, 0.21 – 0.30 Å, 0.31 – 0.40 Å, 0.41 – 0.50 Å, 0.51 – 0.60 Å, and 0.61 – 0.70 Å.
SABER scoring
After the data were binned, each bin was analyzed using the program’s scoring functions. (2) First, each match was assigned an RMSD value to the Catalytic Atom Map. Second, using the ActiveSiteFinder module, matches were scored for overlap with known active site residues according to annotation in the Catalytic Site Atlas. (8) Matches with positive scores had at least one residue that was part of a known or putative active site. Third, each match was analyzed using the BindingSiteFinder module, an alternative method for active/binding site identification based on detection of non-water PDB heteroatoms within 5 Å of the residues in the match. Finally, the SABER-WCN module was used to determine the weighted contact number (WCN) and WCN z-score for each residue. The WCN calculations are described below.
Weighted contact number (WCN) analysis in SABER
A new module was developed for SABER, called WCNcalc, to determine the weighted contact number values and z-scores for the residues identified in the SABER search. The WCN value of a residue is a measure of the crowdedness of the local protein environment around a residue.
The weighted contact number values for the individual residues were generated based on the WCN of the Cα atom for that residue. The WCN for atom i in a protein with N non-hydrogen atoms is defined as
as described in (9).
In order to normalize the WCN values used in this research, the z-score for each WCN was calculated using the formula
where wcni is the WCN of the Cα atom of the residue of interest, is the average WCN for Cα atoms in the protein being analyzed, and σwcn is the standard deviation of the WCN of Cα atoms in the protein. The zwcn value will often be referred to as the z-score for a residue in the text.
Analysis of matches to Catalytic Atom Maps
The SABER results, including WCNcalc results, were analyzed for all matches with an RMSD to the CAM of ≤ 0.7 Å. For the Ser-His-Asp triad CAM and the tyrosine phosphatase CAM, matches were identified as either true positives or false positives based on either their ActiveSiteScore or on literature analysis. Any match with an ActiveSiteScore equal to the number of residues in the Catalytic Atom Map was deemed a true positive. If a match had an ActiveSiteScore less than this number, the literature reference associated with the PDB code of the match was used to determine whether or not the residues identified by the Catalytic Atom Map were part of its active site and had the proposed chemical function in the protein. After this, the WCN z-score values of the residues contained in the true positive and false positive data sets were calculated individually.
For the analysis of ODCase-like active sites, it was not possible to bin these matches into true and false positives, as the mechanism of ODCase has not been conclusively proven. As such, we analyzed only the matches that had at least partial overlap with a known active site. Except where otherwise noted, these were defined as any hit within the 0.7 Å RMSD of the CAM and having an ActiveSiteScore > 0.
Results and Discussion
Analysis of Ser-His-Asp catalytic triads using Catalytic Atom Maps
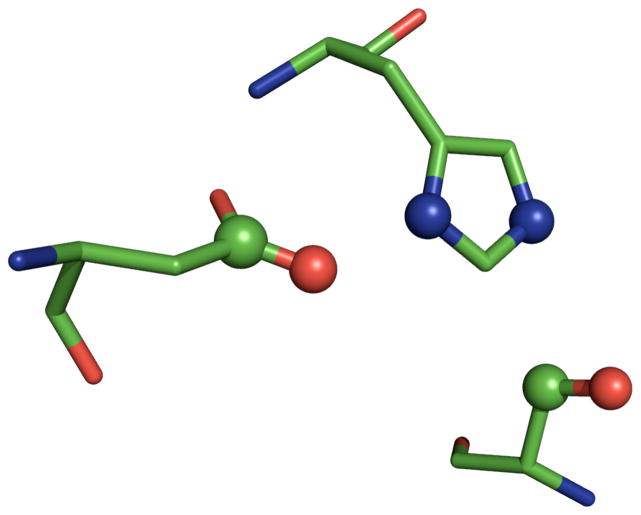
In order to determine whether or not a Catalytic Atom Map could be used to predict the presence of an active site that uses a catalytic triad to generate a nucleophilic serine hydroxyl group, we constructed at CAM based on a high-resolution structure of the serine protease trypsin (PDB code: 1A0J). We chose six atoms to represent the catalytic triad present in this enzyme, as shown in Figure 2. The Catalytic Atom Map parameters allowed the following atom types for each residue: serine (CB/OG), histidine (ND1/NE2), and aspartic acid (CG/OD1, OD2). The 1A0J CAM and the SABER program were used to search a subset of the Protein Data Bank for other catalytic triads. The PDB data set we used contained all X-ray structures of proteins with resolution ≤2.0 Å, with sequences that had ≥ 90% homology removed. We abbreviate this as the PDB90(2Å) dataset; this contained ~11,700 structures.
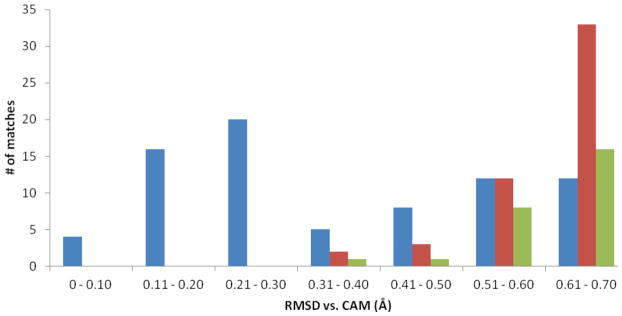
Figure 2.
Results of the analysis of using the 1A0J Catalytic Atom Map to search the PDB90(2Å) data set. The blue bars indicate the number of correctly identified catalytic triads, while the red bars indicate groups of atoms that matched the CAM geometry but were not part of a catalytic triad. The green bars are incorrectly identified triads that remained after the WCN filter was applied.
The SABER program was used to search the PDB90(2Å) dataset using the CAM described above. SABER uses the Jess geometric hashing algorithm to identify matches to the Catalytic Atom Map. (10) The program then uses a series of scoring functions to analyze each match to the CAM. For each match, it (a) provides the RMSD to the CAM, (b) scores for overlap with known active site residues annotated in the Catalytic Site Atlas, and (c) determines if any non-water heteroatoms are within 5 Å of any of the match residues. SABER is described more fully in the Computational Details section. A new module was added to the SABER that performs weighted contact number (WCN) analysis on the match residues, described below.
The weighted contact number (WCN) analysis was used to identify residues that were more likely to be part of an active site, and thus less likely to be either deeply buried in the protein structure or completely solvent exposed. The WCN is a measure of the average “crowdedness” of the environment occupied by a given residue or atom in a protein’s structure. This type of analysis has been used previously to characterize WCN values of catalytic residues vs. WCN values of non-catalytic residues, and the two classes have been show to have distinctly different distributions. (7) The z-scores of the WCN values were used to filter the geometric matches identified during the SABER search, so as to remove matches that were unlikely to be part of an active site.
After performing the SABER search, we analyzed the 127 structures identified during the search that had ≤ 0.7 Å RMSD to the Catalytic Atom Map. Each potential catalytic triad was categorized as either a true positive (part of a known triad) or a false positive (a match with a similar geometry to the CAM, but not a functioning catalytic triad). The results were sorted into bins by RMSD to the CAM in 0.1 Å increments, as shown in Figure 2. Within the 0.0 – 0.6 Å range, 79% of the geometric matches to the CAM were part of known catalytic triads. Beyond this range, there are still true positives, but the number of false positives rises dramatically. In the 0.51 – 60 Å bin, there is a 50% chance of being a false positive, and this probability rises to 73% in the 0.61 – 0.70 Å bin. We then used the SABER WCNcalc module to further analyze these matches, as shown in Table 1.
Table 1.
Weighted contact number z-score analysis for matches to the 1A0J trypsin-based catalytic triad CAM.
| Category | Ser z-score | His z-score | Asp z-score | Avg. z-score for triad |
|---|---|---|---|---|
| True positive | 1.47 ± 0.34 | 0.65 ± 0.42 | 1.11 ± 0.37 | 1.08 ± 0.32 |
| False positive | 0.50 ± 1.16 | 0.58 ± 0.89 | 0.55 ± 1.01 | 0.55 ± 0.91 |
Based on the WCN analysis, it is clear that the catalytic residues in the true positive hits have significantly higher WCN values than the false positive hits, on average. This reflects the fact that catalytic triads, like other active site residues, tend to be in regions of a protein that are more crowded, have restricted motion, and are pre-organized for catalysis. (6, 7) We used the WCN z-score data to remove the false positive matches to the Catalytic Atom Map that were unlikely to be in a catalytic site. All of the matches that had an average z-score lower than the average true positive z-score minus one standard deviation (z-score < 0.76) were removed. The unfiltered false positives are shown in Figure 2 as red bars. We then eliminated the false positives with WCN z-scores < 0.76. These data are shown as green bars in Figure 2.
After application of the WCN filter, 48% of the false positives were eliminated, as these matches were in regions of the protein unlikely to support a functional catalytic triad. Within 0.5 Å of the CAM, the triads were identified with 96% accuracy, and within 0.6 Å, 87% accuracy. Of the remaining false positive matches, the best was in a leukotriene A4 hydrolase enzyme (PDB code: 3B7S), with a 0.37 Å RMSD to the CAM. The Asp residue identified by the CAM, D375, is part of the substrate binding site of this enzyme. (11) The other two residues identified, Ser113 and His139, are not part of the active site. The WCN z-scores for these residues are 0.84 and 1.52, respectively, indicating a serine that is in a less crowded environment and a histidine that is in a much more crowded environment than those in the correctly identified triads. These residues are well-positioned to function as a catalytic triad, but as the Ser and His are not part of the enzyme active site, the triad is inactive. The false positive with the next lowest RMSD, 0.44 Å, was an epoxide hydrolase (PDB code: 3KDA). (12) The potential triad again has reasonable geometric agreement with the CAM, but the residues are in a very crowded region of the protein, with an average WCN z-score of 1.52. The individual residues in the 3KDA match had WCN z-scores of 1.66, 1.62, and 1.27 for Ser, His, and Asp, respectively. The His and Asp residues are in a significantly more crowded protein environment than is typical for a catalytic triad. This makes catalytic function unlikely, as substrate access would be extremely limited, as would water access to the histidine imidazole ring for the release of the acyl-enzyme intermediate.
The correctly identified catalytic triads were not exclusively from trypsin-like enzymes. Catalytic triads were identified in the subtilisin fold and the α/β hydrolase fold as well, indicating that the CAM used is not fold-specific. In addition, the triads identified also included matches in ester hydrolases. The lack of reaction specificity shows that this CAM can be used to identify the catalytic machinery present in the active site, but not the specific reaction that takes place. This is not surprising, since catalytic triads are used to catalyze a number of different reactions in addition to peptide hydrolysis. In the α/β hydrolase fold alone, Ser-His-Asp triads are used for catalysis of peptide hydrolysis, ester hydrolysis, lipid hydrolysis, and haloperoxidase reactions. In every case, the catalytic triad activates the serine nucleophile for attack on an electrophilic carbon atom. (13)
Although the CAM used was able to identify Ser-His-Asp catalytic triads with high accuracy within the 0 – 0.60 Å RMSD range, there were true catalytic triads that fell outside this range. Some enzyme-catalyzed reactions associated with these triads, such as lipid hydrolysis and haloperoxidation, were not identified using the 1A0J Catalytic Atom map within the RMSD limits specified. The enzymes do not appear among the matches in Figure 2, as lipases first appear in the 0.71 – 0.80 Å RMSD bin, and haloperoxidases in the 0.81 – 0.90 Å RMSD bin. This result is not surprising, as we used a Catalytic Atom Map based on a single enzyme structure. While this CAM was able to correctly identify Ser-His-Asp catalytic triads with high accuracy, it does not locate every possible true catalytic triad in the PDB90(2Å) dataset.
Our search identified the enzymes that contain similar catalytic functionality in their active sites prearranged in a specific geometry. The combination of the geometry of the catalytic atoms and WCN scoring was sufficient to locate other active sites that are known to contain this serine nucleophile-generating functionality. We have found that is the RMSD to the CAM is ≤ 0.5 Å, the catalytic mechanism is very likely (> 90%) to operate in the same way. For enzymes with unknown mechanisms, this method has the potential to give insight into the catalytic mechanisms available to a given active site.
Analysis of tyrosine phosphatase active sites using Catalytic Atom Maps
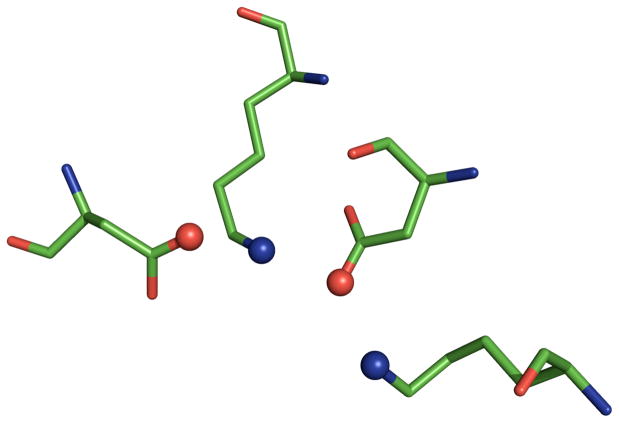
Next, we sought to verify that a Catalytic Atom Map and the SABER program could be used to identify the chemical functionality of other types of active sites. The tyrosine phosphatases (TyrPs) are also known to have a highly conserved active site that exists in more than one fold type. (14) These enzymes catalyze the hydrolysis of of the phosphate group from phosporylated tyrosine residues, using a nucleophilic cysteine residue in the active site. This type of active site contains a highly conserved Cys-Arg-(Asp/Glu) triad, which we used the basis for the Catalytic Atom Map. The atoms in the CAM were constrained to match only the following atom types for each residue: Cys (CB/SG), Arg (CZ/NH1, NH2), and Asp/Glu (CG, CD/OD1, OD2, OE1, OE2).
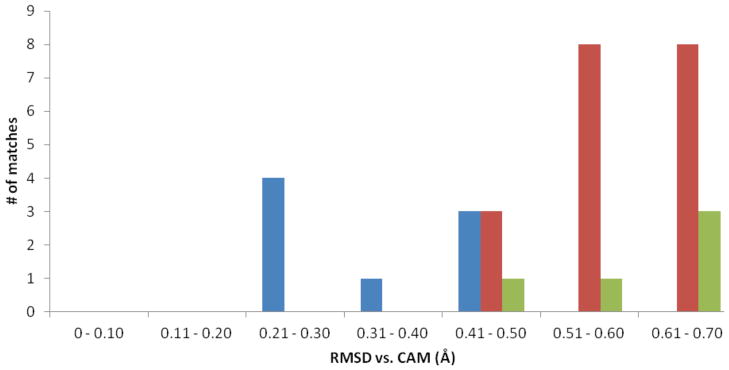
Using SABER, we performed the same procedure as employed previously for the catalytic triad-based CAM search of the PDB90(2Å) dataset with the tyrosine phosphatase CAM in Figure 3. This search identified a total of 27 proteins with tyrosine phosphatase-like triads with RMSD values ≤ 0.7 Å vs. the CAM. There were 11 hits in the 0.0 – 0.5 Å range, 73% of which were true TyrP catalytic triads. The WCN analysis of the catalytic residues was also performed, shown in Table 2.
Figure 3.
The Catalytic Atom Map for the Cys-Arg-(Asp/Glu) triad of tyrosine phosphatase, based on the structure of the human hematopoietic tyrosine phosphatase (PDB code: 1ZC0). Catalytic atoms are rendered as spheres.
Table 2.
Weighted contact number z-score analysis for matches to the 1ZC0 TyrP-based catalytic triad CAM.
| Category | Cys z-score | Arg z-score | Asp/Glu z-score | Avg. z-score for Cys/Arg |
|---|---|---|---|---|
| True positive | 1.51 ± 0.39 | 1.51 ± 0.31 | 0.21 ± 0.43 | 1.51 ± 0.34 |
| False positive | 0.75 ± 0.68 | 0.40 ± 0.87 | 0.26 ± 1.14 | 0.58 ± 0.74 |
Unlike the Ser-His-Asp catalytic triads, the TyrP active site has the ability to tolerate significant variation in the packing around the Asp/Glu residue, and this residue is in a much less crowded environment than is typical for catalytic residues. Without an analysis of WCN z-scores for many different types of enzyme active sites, it is impossible to know if it is unusual for an active site residue to have such wide variation in its WCN z-score. The WCN z-score values for the Asp/Glu residue do not distinguish at all between catalytic and non-catalytic arrangements, but the WCN z-score from the Cys and Arg residues are excellent indicators. Only these z-scores from Cys and Arg were used in our analysis, shown in Figure 4.
Figure 4.
Results of the analysis of using the 1ZC0 Catalytic Atom Map to search the PDB90(2Å) data set. The blue bars indicate the number of correctly identified TyrP-like catalytic triads, while the red bars indicate groups of atoms that matched the CAM geometry but were not part of a TyrP catalytic triad. The green bars are incorrectly identified triads that remained after the WCN filter was applied.
As was the case with the Ser-His-Asp catalytic triads, the WCN z-score filter removed a significant number of false positives, reducing them by 74%. Within 0 – 0.5 Å RMSD to the CAM, true Cys-Arg-(Asp/Glu) triads were identified with 89% accuracy, and within 0 – 0.6 Å they were identified with 80% accuracy. The only false positive within the 0.0 – 0.5 Å range was the YdcF protein from E. coli, at 0.47 Å RMSD to the TyrP CAM. The function of this protein is not known, but it does bind S-adenosyl methionine. (15) All the residues identified by the TyrP CAM in this structure are in a very crowded region of the protein. The WCN z-scores for Cys, Arg, and Glu are 1.76, 1.52, and 0.93, respectively. This indicated a region of the protein that is likely to be completely inaccessible. The acid residue in particular is deeply buried compared to a true tyrosine phosphatase. Beyond 0.5 Å, all of the matches were false positives. Also like the Ser-His-Asp catalytic triad CAM, the TyrP CAM was also to identify active sites across different fold types. TyrP enzymes with the Class I fold and the low molecular weight fold were both correctly identified.
Analysis of the ODCase active site using Catalytic Atom Maps
Based on the analysis of Ser-His-Asp catalytic triads (96% accuracy within 0.5 Å of the CAM) and the Cys-Arg-(Asp/Glu) residues of the tyrosine phosphatase active site (89% accuracy within 0.5 Å of the CAM), we concluded that this methodology could be used to identify enzymes with a common active site chemistry, in this case the deprotonation of serine or cysteine to activate them as nucleophiles. Next, we turned to investigating orotidine 5′-monophosphate decarboxylase (ODCase) in order to identify other enzymes with active sites having similar atomic geometries, and to use this information to predict the possible cataltyic function of the conserved groups present in the ODCase active site.
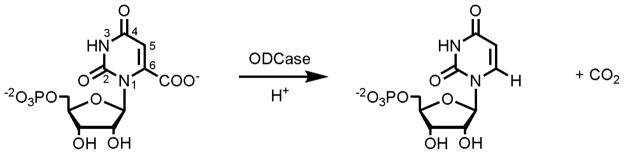
Despite more than two decades of research, the mechanism of ODCase has not been definitively established. The overall reaction catalyzed by this enzyme is shown in Figure 5. As described by Wolfenden in 1995, ODCase has an unusually high catalytic proficiency (kcat/KM/kuncat) of 4.8 × 1022 M−1 and a kcat/kuncat of 7.1 × 1016. At the time, ODCase was the most proficient enzyme yet discovered. (16) While there have been discoveries of enzymes with higher catalytic proficiencies, such as uroporphyrinogen decarboxylase (17) and the S-O cleaving sulfatases (18), ODCase still represents a fascinating challenge to scientists because the source of its extreme catalytic proficiency has yet to be definitively established.
Figure 5.
The reaction catalyzed by orotidine 5′-monophosphate decarboxylase (ODCase).
ODCase provides its enormous rate acceleration despite the fact that its active site does not utilize any cofactors, such as pyridoxal, or metal ions. (19–21) Several cofactor and ion-free mechanisms have been proposed, such as protonation at O2, O4, C5, or C6. (22–27) Nucleophilic mechanisms, such as iminium ion formation at C4 or a Michael addition at C5, have also been suggested. These mechanisms are generally discounted due to lack of oxygen exchange with 18O-labeled water and kinetic isotope effects, respectively. (28–30) In addition, both electrostatic stabilization of the transition state and electrostatic destabilization of the substrate have been proposed as the source of the catalytic proficiency. (31, 32) These mechanistic proposals have all been reviewed in detail in reference (33).
Additional information about the ODCase mechanism has been obtained from structural data. The active sites of ODCases from many species has been extensively studied using X-ray crystallography and subjected to a great deal of mutational analysis. (34–37) The catalytic functionality of the enzyme is controlled by a Lys-Asp-Lys-Asp tetrad in the active site. Mutation of any of these residues reduces the activity of ODCase by more than five orders of magnitude. (38) This active site tetrad is highly conserved, and is found across the ODCase enzymes from many species, including E. coli, S. cerevisiae, M. thermoautotrophicum, B. subtilis, and others. Beyond the Lys-Asp-Lys-Asp tetrad, other interactions with the substrate have also been implicated in ODCase catalysis. Binding of the substrate’s phosphate group contributes significantly to catalysis in this enzyme. (39–41) A trio of hydrophobic residues in the active site have been shown to reduce kcat by as much as 400-fold upon mutation to neutral hydrophilic residues, and mutation of a serine residue near the O4 atom of the substrate to a proline reduces kcat/KM by more than a factor of 106, consistent with proton transfer occurring within the vicinity of O4 in the transition state. (42, 43) Even with all of this information, there has been no definitive consensus on the mechanism of this enzyme.
As with the previous two active sites, we constructed a Catalytic Atom Map based on a high-resolution crystal structure of ODCase (PDB code: 3LTP). For this CAM, the nitrogen atoms were allowed to match only the sidechain nitrogen (NZ) atoms of Lys residues, and the oxygen atoms were allowed to match only the carboxylate oxygens of Asp or Glu (OD1, OD2, OE1, OE2).
We set out to look for all functions of the ODCase Lys-Asp-Lys-Asp tetrad, as we had no way to distinguish true positives from false positives. We examined all of the geometric matches to the ODCase CAM that were within 0.7 Å RMSD and had an ActiveSiteScore > 0, indicating that the geometric match had at least partial overlap with a known active site. In addition, we used the BindingSiteFinder module in SABER to identify any potential active site matches with a metal ion within 2.5 Å of the atoms identified by the Catalytic Atom Map. As the ODCase active site is known to be free of metals, any match with a metal ion in such close proximity to the catalytic residues is unlikely to share a similar mechanism. (19) The results of this analysis are shown in Table 3.
Table 3.
Results of the analysis of using the 3LTP Catalytic Atom Map to search the PDB90 data set. Data is shown for matches to the CAM with a GeometryScore ≤ 0.7 Å and an ActiveSiteScore > 0, except where noted.
| PDB | Protein | RMSD (Å) | Avg. WCN z-scorea | Metal within 2.5 Å? | Catalytic Element Identified |
|---|---|---|---|---|---|
| 2FDS | Orotidine 5′-monophosphate decarboxylase | 0.42 | 1.58 | No | ? |
| 3HJZb | Transaldolase | 0.48 | 0.53 | No | One lysine functions as a nucleophile; reaction proceed through a Schiff base intermediate |
| 2PS2 | Mandelate racemase | 0.48 | 1.09 | Yes | Carboxylate residues bind Mg2+, lysines function as general acid/base |
| 2CZD | Orotidine 5′-monophosphate decarboxylase | 0.54 | 1.70 | No | ? |
| 2HXT | L-fuconate dehydratase | 0.61 | 1.39 | Yes | Carboxylate residues bind Mg2+, lysines function as general acid/base |
| 3H12 | Mandelate racemase | 0.64 | 0.94 | Yes | Carboxylate residues bind Mg2+, lysines function as general acid/base |
| 2A4A | Deoxyribose phosphate aldolase | 0.65 | 1.42 | No | One lysine functions as a nucleophile; reaction proceed through a Schiff base intermediate |
| 2ZAD | Muconate cycloisomerase | 0.65 | 1.63 | Yes | Carboxylate residues bind Mg2+, lysines function as general acid/base |
| 1N7K | Deoxyribose phosphate aldolase | 0.69 | 1.13 | No | One lysine functions as a nucleophile; reaction proceed through a Schiff base intermediate |
WCN z-score is the average of the WCN z-scores for each of the residues identified by the Catalytic Atom Map.
3HJZ was the aldolase structure with the lowest RMSD identified, but its ActiveSiteScore was 0, as it had not been annotated in the Catalytic Site Atlas at the time of this analysis.
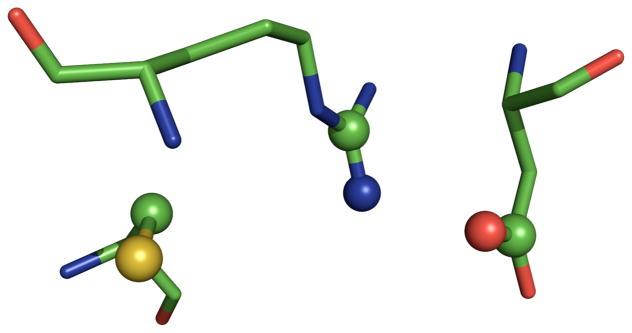
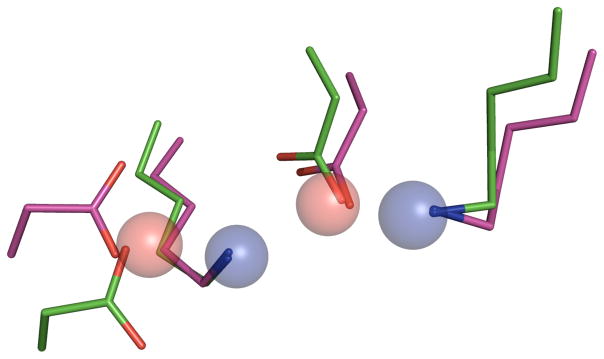
It is immediately apparent that all of the active sites identified conform to the observation that active site residues are likely to be in more crowded regions of an enzyme’s structure, as evidenced by the high WCN z-score values. Even the lowest average WCN z-score of 0.53 is more than one-half of one standard deviation higher in terms of WCN versus an average residue in that protein. Beyond that, the matches can be classified into three categories. The first of these categories was other ODCase enzymes, which we expected would be present, given the highly conserved geometry of this enzyme’s active site. The Lys-Asp-Lys-Asp catalytic tetrads from the 2FDS and 2CZD ODCase matches are shown in Figure 7, superimposed with the Catalytic Atom Map used in the SABER search of the PDB90(2Å) data set.
Figure 7.
Superposition of the ODCase matches from the SABER search on the 3LTP-based ODCase Catalytic Atom Map. The ODCase active site from 2FDS is shown in green, while the active site from 2CZD is shown in magenta. The Catalytic Atom Map is shown as spheres.
The second category consists of members of the enolase superfamily, known to include mandelate racemase, muconate cycloisomerase, and L-fuconate dehydratase. All of these enzymes use an Mg2+ ion bound by three carboxylate-containing sidechains to stabilize an enediolate intermediate, accompanied by other residues, often lysines, that act as general acids/bases. (44–48) Previous work has shown that a search of the PDB based on Catalytic Atom Map of the o-succinyl benzoate synthase (OSBS), another member of the enolase superfamily, does not locate the active site of ODCase, but does locate mandelate racemase and muconate cycloisomerase. (49) This is due to the ODCase active site‘s lack of a third carboxylate sidechain, in a geometry appropriate to bind the Mg2+ ion. Each of the enolase superfamily enzymes located using the ODCase CAM had a metal ion within 2.4 Å of one of the carboxylate sidechain oxygens. Despite the structural similarities of these active sites in terms of residue placement, the existence of the metal ion makes any sort of mechanistic overlap with ODCase extremely unlikely.
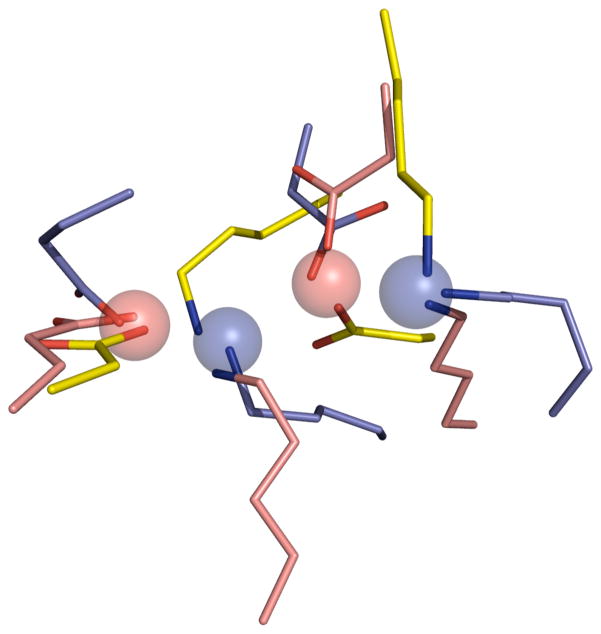
The third category of matches to the ODCase Catalytic Atom Map were the Class I aldolases: transaldolase (PDB code: 3HJZ) and deoxyribose-phosphate aldolase (PDB codes: 2A4A and 1N7K). The Class I aldolases use a metal-free Schiff base mechanism to catalyze aldol condensation reactions and have a highly conserved Lys-Asp-Lys triad in the active site. (50) For example, the enzyme 2-dexoyribose-5-phosphate aldolase (DERA), which catalyzes an aldol condensation between acetaldehyde and glyceraldehyde-3-phosphate, uses the nucleophilic lysine in the Lys-Asp-Lys triad to attack the carbonyl carbon of acetaldehyde and form a Schiff base. This covalent intermediate has been observed via X-ray crystallography. (51) The SABER search revealed that the active sites of these Class I aldolase enzymes have a very similar geometry to the ODCase active site. The superposition of the three Class I aldolase matches with the ODCase Catalytic Atom Map is shown in Figure 8.
Figure 8.
Superposition of the aldolase matches from the SABER search on the 3LTP-based ODCase Catalytic Atom Map. The aldolase active site from 2A4A is shown in yellow, the active site from 1N7K is shown in pink, and the active site from 3HJZ is shown in purple. The Catalytic Atom Map is shown as spheres.
This research does not prove that the ODCase active site functions through a similar mechanism, although the formation of an iminium ion at C4 has been suggested previously. (29) What the geometric similarity of these active sites does suggest, however, is that the active site of ODCase contains catalytic residues placed in a geometry appropriate for the generation of a nucleophilic lysine, in the same manner of the Class I aldolase active sites. This has been demonstrated experimentally, as covalent inhibitors have been developed for ODCase. 6′-iodouridine-5′-monophosphate (6-iodo-UMP) has been shown using X-ray crystallography to form a covalent bond with a Lys residue (M. thermoautotrophicum Lys72) in the ODCase active site. (52) 6′-azidouridine-5′-monophosphate (6-azido-UMP) has also been shown to be a covalent inhibitor of the ODCase enzymes from both M. thermoautotrophicum and P. falciparum. (53) Unlike the iminium ion mechanism that was proposed that requires attack at C4, or the Silverman mechanism where nucleophilic attack would occur at C5, the nucleophilic lysine that has been observed in the ODCase active site reacts with the C6 atom, where the iodide leaving group was attached.
Although it is not possible to establish definitively the reaction mechanism of ODCase using this approach, its similarity to the Class I aldolase active site allows us to narrow the choice of potential mechanisms to investigate. Given the nucleophilic character of one of the lysines on the ODCase active site, this means it must spend a significant amount of time in the unprotonated and uncharged state. This has implications for the charge state, and thus the catalytic function, of the other residues in the ODCase catalytic tetrad. In addition to narrowing the choice of likely reaction mechanisms, this methodology is also useful for identification of additional functionality in an enzyme’s active site that can be utilized for both inhibitor design and for active site redesign. The identification of a functional nucleophile in an enzyme’s active site, whether or not it participates in the reaction mechanism, provides a target for inhibition. Likewise, a nucleophile identified in this manner could be utilized as part of a rational active site redesign process, in which the redesigned active site utilizes the nucleophile identified as part of its catalytic mechanism. This type of enzyme design strategy is discussed in reference (2).
As was the case with Ser-His-Asp catalytic triads, a geometric match to the Catalytic Atom Map does not determine the overall reaction catalyzed by the enzyme. The CAM predicts a mode of catalysis that the functionality in an enzyme’s active site can effect. The geometric similarity of the atoms in the ODCase CAM to the catalytic atoms in the aldolase active site indicates that the ODCase active site has the correct chemical functionality to generate a lysine nucleophile. While this prediction is correct, as demonstrated by the covalent inhibitors described above, it does not establish that a nucleophilic lysine participates in the ODCase reaction mechanism. Given the placement of the lysine nuceophile in the active site of this enzyme, it is difficult to argue in support of the iminium ion mechanism based on these results alone. However, as we have no information regarding the nucleophilicity of the other lysine residue in the ODCase active site, this remains an open question.
Conclusion
We have presented a computational method for analysis of the chemical functionality present in the active site of an enzyme based on comparison to the active site of other enzymes with known mechanisms. As high-throughput structure determination projects such as the Protein Structure Initiative (54) expand our knowledge of the protein structures that exist in nature, this analysis too will aid in the prediction of enzyme functions. This is the second application for which we have used SABER. Previously, we have also demonstrated the utility of this program for enzyme design, and now it has been used successfully for the prediction of catalytic functionality in active sites. (2)
Figure 1.
The Catalytic Atom Map for a Ser-His-Asp catalytic triad, based on the structure of trypsin (PDB code: 1A0J). The atoms used in the CAM are rendered as spheres.
Figure 6.
The Catalytic Atom Map for orotidine 5′-monophosphate decarboyxlase, based on the structure of the M. thermoautotrophicum ODCase (PDB code: 3LTP). Catalytic atoms are rendered as spheres.
Acknowledgments
Funding: This work was supported by the National Institute of General Medical Science, National Institutes of Health (GM 75962 and T32 GM008496-16).
References
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nosrati GR, Houk KN. SABER: A computational method for identifying active sites for new reactions. Protein Sci. 2012;21:697–706. doi: 10.1002/pro.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warshel A. Electrostatic origin of the catalytic power of enzymes and the role of preorganized active sites. J Biol Chem. 1998;273:27035–27038. doi: 10.1074/jbc.273.42.27035. [DOI] [PubMed] [Google Scholar]
- 4.Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson MH. Electrostatic basis for enzyme catalysis. Chem Rev. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 5.Smith AJ, Muller R, Toscano MD, Kast P, Hellinga HW, Hilvert D, Houk KN. Structural reorganization and preorganization in enzyme active sites: comparisons of experimental and theoretically ideal active site geometries in the multistep serine esterase reaction cycle. J Am Chem Soc. 2008;130:15361–15373. doi: 10.1021/ja803213p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin CP, Huang SW, Lai YL, Yen SC, Shih CH, Lu CH, Huang CC, Hwang JK. Deriving protein dynamical properties from weighted protein contact number. Proteins. 2008;72:929–935. doi: 10.1002/prot.21983. [DOI] [PubMed] [Google Scholar]
- 7.Huang SW, Yu SH, Shih CH, Guan HW, Huang TT, Hwang JK. On the Relationship Between Catalytic Residues and their Protein Contact Number. Curr Protein Pept Sc. 2011;12:574–579. doi: 10.2174/138920311796957676. [DOI] [PubMed] [Google Scholar]
- 8.Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Research. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 10.Barker JA, Thornton JM. An algorithm for constraint-based structural template matching: application to 3D templates with statistical analysis. Bioinformatics. 2003;19:1644–1649. doi: 10.1093/bioinformatics/btg226. [DOI] [PubMed] [Google Scholar]
- 11.Tholander F, Muroya A, Roques BP, Fournie-Zaluski MC, Thunnissen MMGM, Haeggstrom JZ. Structure-Based Dissection of the Active Site Chemistry of Leukotriene A4 Hydrolase: Implications for M1 Aminopeptidases and Inhibitor Design. Chemistry & Biology. 2008;15:920–929. doi: 10.1016/j.chembiol.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Bahl CD, Morisseau C, Bomberger JM, Stanton BA, Hammock BD, O’Toole GA, Madden DR. Crystal Structure of the Cystic Fibrosis Transmembrane Conductance Regulator Inhibitory Factor Cif Reveals Novel Active-Site Features of an Epoxide Hydrolase Virulence Factor. J Bacteriol. 2010;192:1785–1795. doi: 10.1128/JB.01348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmquist M. Alpha/Beta-Hydrolase Fold Enzymes: Structures, Functions and Mechanisms. Curr Protein Pept Sc. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- 14.Moorhead GBG, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–409. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- 15.Chao KL, Lim K, Lehmann C, Doseeva V, Howard AJ, Schwarz FP, Herzberg O. The Escherichia coli YdcF binds S-adenosyl-L-methionine and adopts an alpha/beta-fold characteristic of nucleotide-utilizing enzymes. Proteins-Structure Function and Bioinformatics. 2008;72:506–509. doi: 10.1002/prot.22046. [DOI] [PubMed] [Google Scholar]
- 16.Radzicka A, Wolfenden R. A proficient enzyme. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 17.Lewis CA, Wolfenden R. Uroporphyrinogen decarboxylation as a benchmark for the catalytic proficiency of enzymes. P Natl Acad Sci USA. 2008;105:17328–17333. doi: 10.1073/pnas.0809838105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards DR, Lohman DC, Wolfenden RV. Catalytic Proficiency: The Extreme Case of S-O Cleaving Sulfatases. J Am Chem Soc. 2011;134:525–531. doi: 10.1021/ja208827q. [DOI] [PubMed] [Google Scholar]
- 19.Cui W, DeWitt JG, Miller SM, Wu W. No metal cofactor in orotidine 5′-monophosphate decarboxylase. Biochem Biophys Res Commun. 1999;259:133–135. doi: 10.1006/bbrc.1999.0737. [DOI] [PubMed] [Google Scholar]
- 20.Appleby TC, Kinsland C, Begley TP, Ealick SE. The crystal structure and mechanism of orotidine 5′-monophosphate decarboxylase. Proc Natl Acad Sci U S A. 2000;97:2005–2010. doi: 10.1073/pnas.259441296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller BG, Hassell AM, Wolfenden R, Milburn MV, Short SA. Anatomy of a proficient enzyme: the structure of orotidine 5′-monophosphate decarboxylase in the presence and absence of a potential transition state analog. Proc Natl Acad Sci U S A. 2000;97:2011–2016. doi: 10.1073/pnas.030409797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beak P, Siegel B. Mechanism of decarboxylation of 1,3-dimethylorotic acid. A model for orotidine 5′-phosphate decarboxylase. J Am Chem Soc. 1976;98:3601–3606. doi: 10.1021/ja00428a035. [DOI] [PubMed] [Google Scholar]
- 23.Lee JK, Houk KN. A proficient enzyme revisited: the predicted mechanism for orotidine monophosphate decarboxylase. Science. 1997;276:942–945. doi: 10.1126/science.276.5314.942. [DOI] [PubMed] [Google Scholar]
- 24.Begley TP, Appleby TC, Ealick SE. The structural basis for the remarkable catalytic proficiency of orotidine 5′-monophosphate decarboxylase. Curr Opin Struct Biol. 2000;10:711–718. doi: 10.1016/s0959-440x(00)00148-2. [DOI] [PubMed] [Google Scholar]
- 25.Houk KN, Lee JK, Tantillo DJ, Bahmanyar S, Hietbrink BN. Crystal structures of orotidine monophosphate decarboxylase: Does the structure reveal the mechanism of nature’s most proficient enzyme? Chembiochem. 2001;2:113–118. doi: 10.1002/1439-7633(20010202)2:2<113::AID-CBIC113>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 26.Lee TS, Chong LT, Chodera JD, Kollman PA. An alternative explanation for the catalytic proficiency of orotidine 5′-phosphate decarboxylase. J Am Chem Soc. 2001;123:12837–12848. doi: 10.1021/ja011096f. [DOI] [PubMed] [Google Scholar]
- 27.Lundberg M, Blomberg MRA, Siegbahn PEM. Density functional models of the mechanism for decarboxylation in orotidine decarboxylase. J Mol Model. 2002;8:119–130. doi: 10.1007/s00894-002-0080-2. [DOI] [PubMed] [Google Scholar]
- 28.Silverman RB, Groziak MP. Model Chemistry for a Covalent Mechanism of Action of Orotidine 5′-Phosphate Decarboxylase. Journal of the American Chemical Society. 1982;104:6434–6439. [Google Scholar]
- 29.Shostak K, Jones ME. Orotidylate Decarboxylase - Insights into the Catalytic Mechanism from Substrate-Specificity Studies. Biochemistry. 1992;31:12155–12161. doi: 10.1021/bi00163a026. [DOI] [PubMed] [Google Scholar]
- 30.Acheson SA, Bell JB, Jones ME, Wolfenden R. Orotidine-5′-Monophosphate Decarboxylase Catalysis - Kinetic Isotope Effects and the State of Hybridization of a Bound Transition-State Analog. Biochemistry. 1990;29:3198–3202. doi: 10.1021/bi00465a007. [DOI] [PubMed] [Google Scholar]
- 31.Warshel A, Florian J, Strajbl M, Villa J. Circe effect versus enzyme preorganization: what can be learned from the structure of the most proficient enzyme? Chembiochem. 2001;2:109–111. doi: 10.1002/1439-7633(20010202)2:2<109::AID-CBIC109>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 32.Wu N, Mo Y, Gao J, Pai EF. Electrostatic stress in catalysis: structure and mechanism of the enzyme orotidine monophosphate decarboxylase. Proc Natl Acad Sci U S A. 2000;97:2017–2022. doi: 10.1073/pnas.050417797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houk KN, Tantillo DJ, Stanton C, Hu YF. What have theory and crystallography revealed about the mechanism of catalysis by orotidine monophosphate decarboxylase? Top Curr Chem. 2004;238:1–22. [Google Scholar]
- 34.Miller BG, Hassell AM, Wolfenden R, Milburn MV, Short SA. Anatomy of a proficient enzyme: The structure of orotidine 5 ′-monophosphate decarboxylase in the presence and absence of a potential transition state analog. P Natl Acad Sci USA. 2000;97:2011–2016. doi: 10.1073/pnas.030409797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Appleby TC, Kinsland C, Begley TP, Ealick SE. The crystal structure and mechanism of orotidine 5 ′-monophosphate decarboxylase. P Natl Acad Sci USA. 2000;97:2005–2010. doi: 10.1073/pnas.259441296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris P, Poulsen JCN, Jensen KF, Larsen S. Structural basis for the catalytic mechanism of a proficient enzyme: Orotidine 5 ′-monophosphate decarboxylase. Biochemistry. 2000;39:4217–4224. doi: 10.1021/bi992952r. [DOI] [PubMed] [Google Scholar]
- 37.Wu N, Mo YR, Gao JL, Pai EF. Electrostatic stress in catalysis: Structure and mechanism of the enzyme orotidine monophosphate decarboxylase. P Natl Acad Sci USA. 2000;97:2017–2022. doi: 10.1073/pnas.050417797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller BG, Snider MJ, Wolfenden R, Short SA. Dissecting a charged network at the active site of orotidine-5′-phosphate decarboxylase. J Biol Chem. 2001;276:15174–15176. doi: 10.1074/jbc.M011429200. [DOI] [PubMed] [Google Scholar]
- 39.Goryanova B, Amyes TL, Gerlt JA, Richard JP. OMP Decarboxylase: Phosphodianion Binding Energy Is Used To Stabilize a Vinyl Carbanion Intermediate. Journal of the American Chemical Society. 2011;133:6545–6548. doi: 10.1021/ja201734z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnett SA, Amyes TL, Wood BM, Gerlt JA, Richard JP. Dissecting the total transition state stabilization provided by amino acid side chains at orotidine 5 ′-monophosphate decarboxylase: A two-part substrate approach. Biochemistry. 2008;47:7785–7787. doi: 10.1021/bi800939k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amyes TL, Richard JP, Tait JJ. Activation of orotidine 5 ′-monophosphate decarboxylase by phosphite dianion: The whole substrate is the sum of two parts. Journal of the American Chemical Society. 2005;127:15708–15709. doi: 10.1021/ja055493s. [DOI] [PubMed] [Google Scholar]
- 42.Iams V, Desai BJ, Fedorov AA, Fedorov EV, Almo SC, Gerlt JA. Mechanism of the Orotidine 5 ′-Monophosphate Decarboxylase-Catalyzed Reaction: Importance of Residues in the Orotate Binding Site. Biochemistry. 2011;50:8497–8507. doi: 10.1021/bi2012355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood BM, Amyes TL, Fedorov AA, Fedorov EV, Shabila A, Almo SC, Richard JP, Gerlt JA. Conformational Changes in Orotidine 5 ′-Monophosphate Decarboxylase: “Remote” Residues That Stabilize the Active Conformation. Biochemistry. 2010;49:3514–3516. doi: 10.1021/bi100443a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yew WS, Fedorov AA, Fedorov EV, Rakus JF, Pierce RW, Almo SC, Gerlt JA. Evolution of enzymatic activities in the enolase superfamily: L-fuconate dehydratase from Xanthomonas campestris. Biochemistry. 2006;45:14582–14597. doi: 10.1021/bi061687o. [DOI] [PubMed] [Google Scholar]
- 45.Gerlt JA, Babbitt PC, Rayment I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Arch Biochem Biophys. 2005;433:59–70. doi: 10.1016/j.abb.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 46.Hasson MS, Schlichting I, Moulai J, Taylor K, Barrett W, Kenyon GL, Babbitt PC, Gerlt JA, Petsko GA, Ringe D. Evolution of an enzyme active site: The structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase. P Natl Acad Sci USA. 1998;95:10396–10401. doi: 10.1073/pnas.95.18.10396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helin S, Kahn PC, Guha BL, Mallows DG, Goldman A. The Refined X-Ray Structure of Muconate Lactonizing Enzyme from Pseudomonas-Putida Prs2000 at 1.85 Angstrom Resolution. J Mol Biol. 1995;254:918–941. doi: 10.1006/jmbi.1995.0666. [DOI] [PubMed] [Google Scholar]
- 48.Neidhart DJ, Howell PL, Petsko GA, Powers VM, Li RS, Kenyon GL, Gerlt JA. Mechanism of the Reaction Catalyzed by Mandelate Racemase .2. Crystal-Structure of Mandelate Racemase at 2.5-a Resolution - Identification of the Active-Site and Possible Catalytic Residues. Biochemistry. 1991;30:9264–9273. doi: 10.1021/bi00102a019. [DOI] [PubMed] [Google Scholar]
- 49.Nosrati G, Houk KN. SABER: A Computational Method for Identifying Active Sites for New Reactions, Accepted to. Protein Science. 2012 doi: 10.1002/pro.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heine A, Luz JG, Wong CH, Wilson IA. Analysis of the class I aldolase binding site architecture based on the crystal structure of 2-deoxyribose-5-phosphate aldolase at 0.99 angstrom resolution. J Mol Biol. 2004;343:1019–1034. doi: 10.1016/j.jmb.2004.08.066. [DOI] [PubMed] [Google Scholar]
- 51.Heine A, DeSantis G, Luz JG, Mitchell M, Wong CH, Wilson IA. Observation of covalent intermediates in an enzyme mechanism at atomic resolution. Science. 2001;294:369–374. doi: 10.1126/science.1063601. [DOI] [PubMed] [Google Scholar]
- 52.Bello AM, Poduch E, Fujihashi M, Amani M, Li Y, Crandall I, Hui R, Lee PI, Kain KC, Pai EF, Kotra LP. A potent, covalent inhibitor of orotidine 5 ′-monophosphate decarboxylase with antimalarial activity (vol 50, pg 915, 2007) J Med Chem. 2007;50:2541–2541. doi: 10.1021/jm060827p. [DOI] [PubMed] [Google Scholar]
- 53.Bello AM, Poduch E, Liu Y, Wei LH, Crandall I, Wang XY, Dyanand C, Kain KC, Pai EF, Kotra LP. Structure-activity relationships of C6-uridine derivatives targeting Plasmodia orotidine monophosphate decarboxylase. J Med Chem. 2008;51:439–448. doi: 10.1021/jm7010673. [DOI] [PubMed] [Google Scholar]
- 54.Montelione GT. The Protein Structure Initiative: achievements and visions for the future. F1000 Biol Rep. 2012;4:7. doi: 10.3410/B4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]