Abstract
Kinase cascades and the modification of proteins by phosphorylation are major mechanisms for cell signaling and communication, and evolution of these signaling pathways can contribute to new developmental or environmental response pathways. The Saccharomyces cerevisiae kinase Ime2 has been well characterized for its role in meiosis. However, recent studies have revealed alternative functions for Ime2 in both S. cerevisiae and other fungi. In the filamentous fungus Neurospora crassa, the IME2 homolog (ime-2) is not required for meiosis. Here we determine that ime-2 interacts genetically with a transcription factor vib-1 during nonself recognition and programmed cell death (PCD). Mutations in vib-1 (Δvib-1) suppress PCD due to nonself recognition events; however, a Δvib-1 Δime-2 mutant restored wild-type levels of cell death. A role for ime-2 in the post-translational processing and localization of a mitochondrial matrix protein was identified, which may implicate mitochondria in N. crassa nonself recognition and PCD. Further, Δvib-1 strains do not produce extracellular proteases, but protease secretion reverted to near wild-type levels in a Δvib-1 Δime-2 strain. Mass spectrometry analysis revealed that the VIB-1 protein is phosphorylated at several sites, including a site that matches the IME-2 consensus. The genetic and biochemical data for ime-2 and vib-1 indicate that IME-2 is a negative regulator of VIB-1 and suggest parallel negative regulation by IME-2 of a cell death pathway in N. crassa that functions in concert with the VIB-1 cell death pathway. Thus, IME2 kinase function has evolved following the divergence of S. cerevisiae and N. crassa and provides insight into the evolution of kinases and their regulatory targets.
Keywords: Neurospora; heterokaryon incompatibility (HI); cell death, kinase; mitochondria
CELL–CELL signaling via kinase cascades is an essential mechanism for communication within and between organisms. Protein kinases are one of the largest protein families in eukaryotes and as much as 30% of any given eukaryotic proteome is phosphorylated (Deshmukh et al. 2010; Moses and Landry 2010). Although kinases have constrained target specificities, these proteins are often structured in a modular way, such that they can evolve new functions via interaction with scaffolds, adapters, or docking proteins (Bhattacharyya et al. 2006). In addition, duplication of kinase targets can result in reciprocal loss of phosphorylation sites and subfunctionalization of these targets and/or a gain of new phosphorylation sites, resulting in neo-functionalization (Amoutzias et al. 2010). Thus, changes in kinase structure, along with target duplication and divergence, can affect the structure and signaling output of kinase pathways. Although the major classes of kinases are conserved across fungal species, there is evidence for duplication, family expansion, and differences in domain organization, suggesting that fungi can change their kinase signaling pathways to accommodate changes in environment or developmental processes (Kosti et al. 2010).
In Saccharomyces cerevisiae, Ime2 is a serine/threonine protein kinase involved in the induction of meiosis and sporulation (Smith and Mitchell 1989). Ime2 has both early and late roles in meiosis, including the initiation of meiosis, meiotic DNA replication, meiotic divisions I and II, and spore formation (Benjamin et al. 2003; Honigberg 2004; Brush et al. 2012). Nutritional signals for meiosis converge at Ime1, a transcriptional regulator of Ime2, as well as at Ime2 itself, to coordinate meiotic initiation (Honigberg and Purnapatre 2003; Kassir et al. 2003). One of the major roles of Ime2 is to contribute to activation of the major middle meiotic transcription factor NDT80 (Pak and Segall 2002a; Sopko et al. 2002; Shin et al. 2010), in part through phosphorylation of the repressor Sum1 (Pak and Segall 2002b; Ahmed et al. 2009; Winter 2012). In addition, Ime2 directly phosphorylates Ndt80, which is associated with an increased ability to activate transcription of Ndt80 target genes (Sopko et al. 2002; Shubassi et al. 2003). Ndt80 binds to the middle sporulation element (MSE) and activates expression of middle meiotic genes (Chu and Herskowitz 1998; Chu et al. 1998); cells lacking Ndt80 arrest at pachytene, prior to nuclear division in meiosis I (Xu et al. 1995).
In the filamentous fungus Neurospora crassa, there is one IME2 homolog (ime-2), but three NDT80 homologs (female sexual development, fsd-1; vegetative incompatibility blocked, vib-1; and NCU04729) (Borkovich et al. 2004; Hutchison and Glass 2010). Homologs to IME1 and SUM1 are lacking in the N. crassa genome. Recently, we showed neither ime-2 nor the NDT80 homologs fsd-1, vib-1, or NCU04729 are involved in meiotic functions in N. crassa (Hutchison and Glass 2010). Mutations in ime-2 did not affect the transcription or activity of fsd-1, the homolog most closely related to S. cerevisiae NDT80. However, ime-2, vib-1, and fsd-1 mutants were affected in the production of female reproductive structures, termed protoperithecia. The development of protoperithecia in N. crassa is induced under conditions of nitrogen starvation (Westergaard and Mitchell 1947; Hirsh 1954). The Δfsd-1 and Δvib-1 mutants formed few protoperithecia under nitrogen starvation and an Δfsd-1 Δvib-1 mutant was female sterile. In contrast, a Δime-2 strain produced protoperithecia under conditions where development of these structures is normally suppressed (nitrogen sufficiency) and significantly more protoperithecia are under nitrogen starvation conditions (Hutchison and Glass 2010). A deletion of ime-2 restored protoperithecial development in a Δfsd-1 mutant, while a Δime-2 Δvib-1 mutant showed a Δvib-1 phenotype (few protoperithecia). These observations indicate a network of regulatory interactions between ime-2 and the NDT80 homologs fsd-1 and vib-1 during development of female reproductive structures in N. crassa.
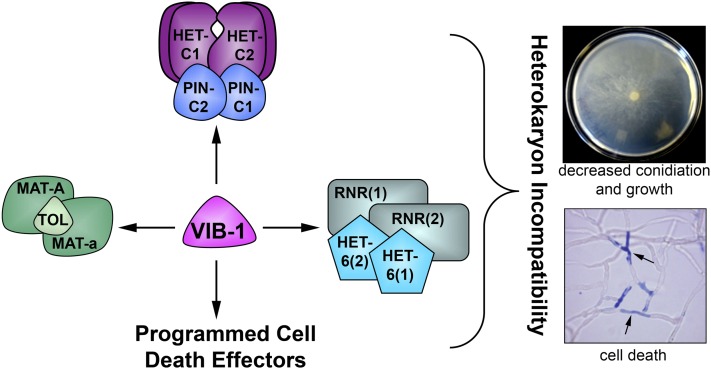
The vib-1 mutant was first identified in a search for mutations that alleviate heterokaryon incompatibility (HI) in N. crassa (Xiang and Glass 2002). Heterokaryon incompatibility mediates nonself recognition and is a ubiquitous phenomenon in filamentous fungi (Aanen et al. 2010; Choi et al. 2012). Within a single filamentous fungal colony, individual hyphae can fuse and form interconnected networks (Fleissner et al. 2008; Read et al. 2010). However, if fusion occurs between strains that contain alternative specificities at heterokaryon incompatibility (het) loci, either the fusion cell is walled off and rapidly killed or the growth of the heterokaryon is inhibited (Glass and Kaneko 2003; Aanen et al. 2010) (Figure 1). In N. crassa, there are 11 het loci and genetic differences at any one of these 11 loci are sufficient to restrict heterokaryon formation (Glass and Dementhon 2006). Strains carrying genetic differences at het loci, but also loss-of-function mutations in vib-1, will form vigorous heterokaryons and do not show HI-associated programmed cell death (PCD) (Xiang and Glass 2002, 2004; Lafontaine and Smith 2012). vib-1 is also necessary for the expression of several genes known to be involved in cell death due to HI (Figure 1) (Dementhon et al. 2006). In Aspergillus nidulans, deletion of a vib-1 homolog, xprG, prevents secretion of extracellular proteases upon nitrogen or carbon starvation (Katz et al. 2006) and vib-1 mutants exhibit this phenotype as well.
Figure 1 .
Schematic for VIB-1 regulation of HI and cell death. VIB-1 is required for HI mediated by genetic differences at mating type, het-6, and het-c pin-c (Xiang and Glass 2002, 2004; Lafontaine and Smith 2012) in addition to activating PCD through additional unknown downstream effectors (Dementhon et al. 2006). het-6 incompatibility is mediated by un-24 (ribonucleotide reductase) and het-6 interactions (Micali and Smith 2006; Lafontaine and Smith 2012); mating-type incompatibility is mediated by mating type A-1, mating type a-1, and tol (Pittenger 1957; Newmeyer 1970; Glass et al. 1990; Shiu and Glass 1999); while het-c incompatibility is mediated by het-c pin-c interactions (Glass and Kaneko 2003; Kaneko et al. 2006); alternative HET-C polypeptides have been shown to physically interact (Sarkar et al. 2002). Phenotypic consequences of HI in N. crassa include growth inhibition and suppression of conidiation (Perkins 1988). Hyphal compartments within heterokaryons that carry alternate het haplotypes (such as het-c/pin-c, rnr/het-6, or mat-a/A/tol) undergo compartmentation and rapid cell death (Glass and Kaneko 2003) and thus stain positive for vital dyes such as methylene blue (arrows).
In this study, we show that in N. crassa, a deletion of ime-2 restores HI-induced PCD in a Δvib-1 strain and also restores the production of extracellular proteases. We further investigated the ime-2 phenotype using transcriptional profiling to assess physiological differences between ime-2 mutants and a wild-type strain and identified a possible role for IME-2 in mitochondrial homeostasis. Our data suggest that IME-2 is a negative regulator of a cell death pathway that functions in parallel to the VIB-1 HI pathway to specifically regulate nonself recognition and cell death when strains carry incompatible specificities at het loci.
Materials and Methods
Strains and growth conditions
All strains used in this study are listed in Table 1. Deletion strains (FGSC 11308, FGSC 11309, FGSC 17936, and FGSC 17937) were constructed by the Neurospora program project grant (Colot et al. 2006) and obtained from the Fungal Genetics Stock Center (FGSC) (McCluskey 2003). Strains were grown on Vogel’s minimal media (MM) (Vogel 1956) unless otherwise specified, and crosses were performed on Westergaard’s media (Westergaard and Mitchell 1947). Transformations were performed as previously described (Margolin et al. 1997). To obtain forced heterokaryons, conidial suspensions from strains of complementary auxotrophic markers were mixed and plated on minimal media. Growth rates were determined by growing strains in race tubes. Protoperithecial development was assessed over a 9-day time period of growth on water agar (Hutchison and Glass 2010).
Table 1 . List of strains used in this study.
| Strain name | Genotypea | Origin or referenceb |
|---|---|---|
| FGSC 2489 | A | FGSC |
| FGSC 11308, FGSC 11309 | Δvib-1::hph a; Δvib-1::hph A | FGSC |
| FGSC 17936, FGSC 17937 | Δime-2::hph a; Δime-2::hph A | FGSC |
| FGSC 4564 | ad-3B cyh-1 am1 | FGSC |
| R15-7 | his-3; a | Dementhon et al. (2006) |
| C9-15 | het-c2 pin-c2 thr-2 A | Smith et al. (2000) |
| C9-2 | het-c2 pin-c2 thr-2 a | Smith et al. (2000) |
| Xa-3 | het-c2 pin-c2 arg-5; pan-2 A | Xiang and Glass (2002) |
| JH3 | het-c2 pin-c2 arg-5; a | C9-2 × Xa-3 |
| Dementhon et al. (2006) | ||
| R14-42 | his-3 rid-1 Δsad-1::hph A | Gift from P. K. T. Shiu; Rasmussen et al. (2008) |
| KD02-10 | his-3; pyr-4; pan-2 a | Dementhon et al. (2006) |
| DVI.4 | Δime-2::hph; Δvib-1::hph A | Hutchison and Glass (2010) |
| D49.10 | his-3; ΔNCU09915::hph; ΔNCU04729::hph A | Hutchison and Glass (2010) |
| KD13-21 | his-3; Δvib-1::hph A | Dementhon et al. (2006) |
| KD13-51 | Δvib-1::hph; pan-2 A | Dementhon et al. (2006) |
| KD13-33 | Δvib-1::hph; pan-2 a | Dementhon et al. (2006) |
| KD13-01 | het-c2 pin-c2 thr-2; Δvib-1::hph a | Dementhon et al. (2006) |
| KD13-23 | his-3; het-c2 pin-c2; Δvib-1::hph; pan-2 A | Dementhon et al. (2006) |
| DVI.HIS.40 | his-3; Δime-2; Δvib-1 a | DVI.4 × KD02-10 |
| SV1 | his-3::pccg1-vib-1-gfp; pyr-4; Δvib-1; pan-2 A | Gift from J. Sun, Glass laboratory |
| BH13c | his-3::pccg1-vib-1-gfp; Δvib-1::hph | SV1 × DVI.HIS.40 |
| DI.PYR.4 | Δime-2::hph pyr-4 a | DVI.4 × KD02-10 |
| DI.HIS.10 | his-3; Δime-2::hph a | DVI.4 × KD02-10 |
| DI.A.22 | Δime-2::hph arg-5 het-c2 pin-c2 a | DVI.A.78 × D49.10 |
| DVI.PYR.63 | Δime-2::hph pyr-4; Δvib-1::hph a | DVI.4 × KD02-10 |
| DVI.HIS.48 | his-3; Δime-2::hph; Δvib-1::hph a | DVI.4 × KD02-10 |
| DVI.A.101 | Δime-2::hph arg-5; Δvib-1::hph a | DVI.4 × JH3 |
| DV.80 | het-c2 pin-c2 arg-5; Δvib-1 A | KD02-10 × DVI.4 |
| R14-42arg4gfp | his-3::pccg1-arg-4-gfp rid-1 Δsad-1::hph A | R14-42 [pccg1-arg-4-gfp] |
| Δvib-1arg4gfp | his-3::pccg1-arg4-gfp; Δvib-1::hph A | KD13-21 [pccg1-arg-4-gfp] |
| Δime2arg4gfp | his-3::pccg1-arg4-gfp; Δime-2::hph a | DI.HIS.10 [pccg1-arg-4-gfp] |
| 2XKOarg4gfp | his-3::pccg1-arg4-gfp; Δime-2; Δvib-1::hph | DVI.HIS.48 [pccg1-arg-4-gfp] |
| DVI.A.78 | Δime-2::hph arg-5; Δvib-1::hph a | DVI.4 × JH3 |
| D49VI.HIS.1 | his-3; Δime-2::hph; ΔNCU09915::hph; Δvib-1::hph; ΔNCU04729::hph a | D49.10 × DVI.A.78 |
| 1XA | his-3::pccg1-vib-1(S60A)-gfp; het-c2 pin-c2 Δvib-1::hph; pan-2 A | KD13-23 [pccg1-vib-11 × PMA-gfp] |
| 5XA | his-3::pccg1-vib-1(S60A, S413A, S537A, S542A, S545A)-gfp; het-c2 pin-c2 Δvib-1::hph; pan-2 A | KD13-23 [pccg1-vib-14 × PMA-gfp] |
| 1XD | his-3::pccg1-vib-1(S60D)-gfp; het-c2 pin-c2 Δvib-1::hph; pan-2 A | KD13-23 [pccg1-vib-11 × PMD-gfp] |
| 5XD | his-3::pccg1-vib-1(S60D, S413D, S537D, S542D, S545D)-gfp; het-c2 pin-c2 Δvib-1::hph; pan-2 A | KD13-23 [pccg1-vib-14 × PMD-gfp] |
Strains are of het-c1 pin-c1 genotype unless otherwise indicated.
“x”, strains derived from crosses.
Protease and cell death assays
For extracellular protease assays, strains were grown (in triplicate) in MM (Vogel 1956) overnight. Protease assays were performed as described previously (Dementhon et al. 2006). Cell death frequency was measured by staining with methylene blue (Hutchison et al. 2009). Heterokaryons were inoculated onto MM overlaid with cellophane, grown for 2–3 days, and stained for 1–2 min with 0.003% methylene blue. Approximately 20 random images were taken and the percentage of dead (blue) hyphal compartments was determined.
RNA extraction and quantitative RT-PCR
RNA extraction was performed on mycelia ground in liquid nitrogen or on sections of mycelia grown on cellophane. Mycelia were mixed with 0.3 g of 0.5-mm silica beads and 1 mL of TRIzol (Invitrogen, Carlsbad, CA) and disrupted using a bead beater (Mini-BeadBeater-8; Biospec Products). RNA was extracted according to the manufacturer’s protocol for TRIzol (Invitrogen). Samples were purified using an RNAeasy kit (QIAGEN, Valencia, CA) and DNA was removed with QIAGEN DNase (no. 79254) or Ambion Turbo DNase (no. AM2238). RNA concentration and quality were assessed using a Nanodrop (Thermo Scientific) and gel electrophoresis. Quantitative RT-PCR (Q-RT-PCR) was performed using an EXPRESS One-Step SYBR GreenER kit (Invitrogen) according to the manufacturer’s protocol, run on an ABI 7300 machine, and analyzed with ABI 7300 system software. Actin mRNA was used as the endogenous control, and reactions were performed in triplicate.
Microarray analysis
Microarray slide production, hybridization, and analysis were performed as described in Tian et al. (2007). Neurospora microarray slides are available from the FGSC (http://www.fgsc.net/). Approximately 10 μg of DNase-treated RNA was used as a template for cDNA synthesis (ChipShot Indirect cDNA Synthesis kit; Promega, Madison, WI), and hybridizations were performed using ProntoPlus kits (Promega), according to manufacturer instructions. Slides were scanned using an Axon GenePix 4000B scanner and analyzed using GenePix Pro 6 software (Molecular Devices). Three independent hybridizations pooled from three biological replicates were performed. Data were analyzed using Bayesian analysis of gene expression levels (BAGEL) (Townsend and Hartl 2002). Microarray data were verified by Q-RT-PCR, using template RNA from an independent experiment. All microarray data were deposited at the Filamentous Fungal Gene Expression Database (Zhang and Townsend 2010) and the Gene Expression Omnibus (GEO) database (ID GSE35905). Functional category enrichment analysis was carried out through the Munich Information Center for Protein Sequences (MIPS) database (http://www.helmholtz-muenchen.de/en/mips/projects/funcat) (Ruepp et al. 2004), which uses a hypergeometric distribution to calculate P-values.
Mitochondrial staining
Mitochondria were visualized using 10 μM MitoTracker Red FM (Invitrogen; no. M22425) (Hickey et al. 2004). Approximately 105 conidia were inoculated into a 30-ml flask of MM and shaken at 30° for ∼6 hr. MitoTracker Red FM was added to 1-ml aliquots of the conidia, followed by shaking at 30° for an additional 15–20 min. Conidia were pelleted by centrifugation and washed once with MM. Conidia were then spread on a MM plate and incubated at 30° for 5–10 min. Mitochondria were imaged using a Deltavision Spectris DV4 deconvolution microscope (Applied Precision Instruments). A stack of ∼20 images was taken 0.2 μm apart, deconvolved using SVI Huygens, and visualized using Bitplane Imaris software.
Protein extraction, immunoprecipitation, and Western blotting
Protein was extracted from mycelia for immunoprecipitation (IP), using a method adapted from the FGSC Neurospora protocol page (http://www.fgsc.net/neurosporaprotocols/Immunoprecipitation%20final.pdf). Briefly, 20–30 g of mycelia was ground in liquid nitrogen and homogenized using a 6770 Freezer/Mill from the SPEX CertiPrep Group, using three cycles of 1 min precool, 1 min run time, and 1 min cool time, at a speed of 15 cycles per second (CPS). Homogenized mycelia were added to HEPES IP extraction buffer [50 mM HEPES (pH 7.4), 137 mM NaCl, 10% glycerol] containing complete mini-EDTA–free protease inhibitor and PhosSTOP phosphatase inhibitor (Roche) and vortexed to homogenization. Samples were centrifuged at 3400 rpm for 10 min. Supernatants were concentrated via centrifugation, using Vivaspin 15R protein concentrators [10,000 molecular weight cut off (MWCO); Sartorius Stedium Biotech]. Four milliliters of each sample was immunoprecipitated using Protein G Dynabeads (Invitrogen), according to manufacturer’s instructions, with the following exceptions: mouse anti-GFP antibody (Roche) was incubated with the beads for 1 hr, and the sample was immunoprecipitated for 2 hr at 4°. Protein was removed from the beads by boiling for 5 min, and samples were run on a 4–15% Criterion Tris-HCl gel (Bio-Rad, Hercules, CA). Protein gels were either subjected to Western blot analysis using standard methods or stained with SimplyBlue SafeStain Coomassie G-250 stain (Invitrogen) to visualize protein, and gel bands of interest were extracted using a razor blade.
Mass spectrometry
Gel bands of interest from Coomassie-stained gels were cut out and minced into <1-mm2 pieces. Protein bands were extracted following a protocol adapted from the University of California (Berkeley) QB3 Proteomics/Mass Spectrometry Laboratory (http://qb3.berkeley.edu/pmsl/protocols.htm). Briefly, gel pieces were washed in 500 μl NH4HCO3 for 20 min. After discarding the NH4HCO3 wash, 150 μl of NH4HCO3 (100 mM) and 10 μl of DTT (45 mM) were added and samples were incubated for 15 min at 50°. After cooling to room temperature, 10 μl of iodoacetamide (100 mM) was added and samples were incubated at room temperature for 15 min in the dark. The solvent was discarded and gel pieces were washed in 500 μl of a 1:1 mixture of acetonitrile and NH4HCO3 (100 mM) for 20 min. Then, gel pieces were incubated for 10–15 min in 50 μl of acetonitrile and dried in a speedvac. Gel pieces were rehydrated in 10 μl of NH4HCO3 (25 mM) and digested overnight with either trypsin or endoproteinase C, according to the manufacturer’s instructions. Proteins were extracted twice with a mix of 60% acetonitrile and 0.1% formic acid and once with 100% acetonitrile. Extracted proteins were dried using a speedvac and two-dimensional mass spectrometry analysis was performed at the QB3 Proteomics/Mass Spectrometry facility.
Results
A deletion of ime-2 restores HI-mediated cell death in a Δvib-1 mutant
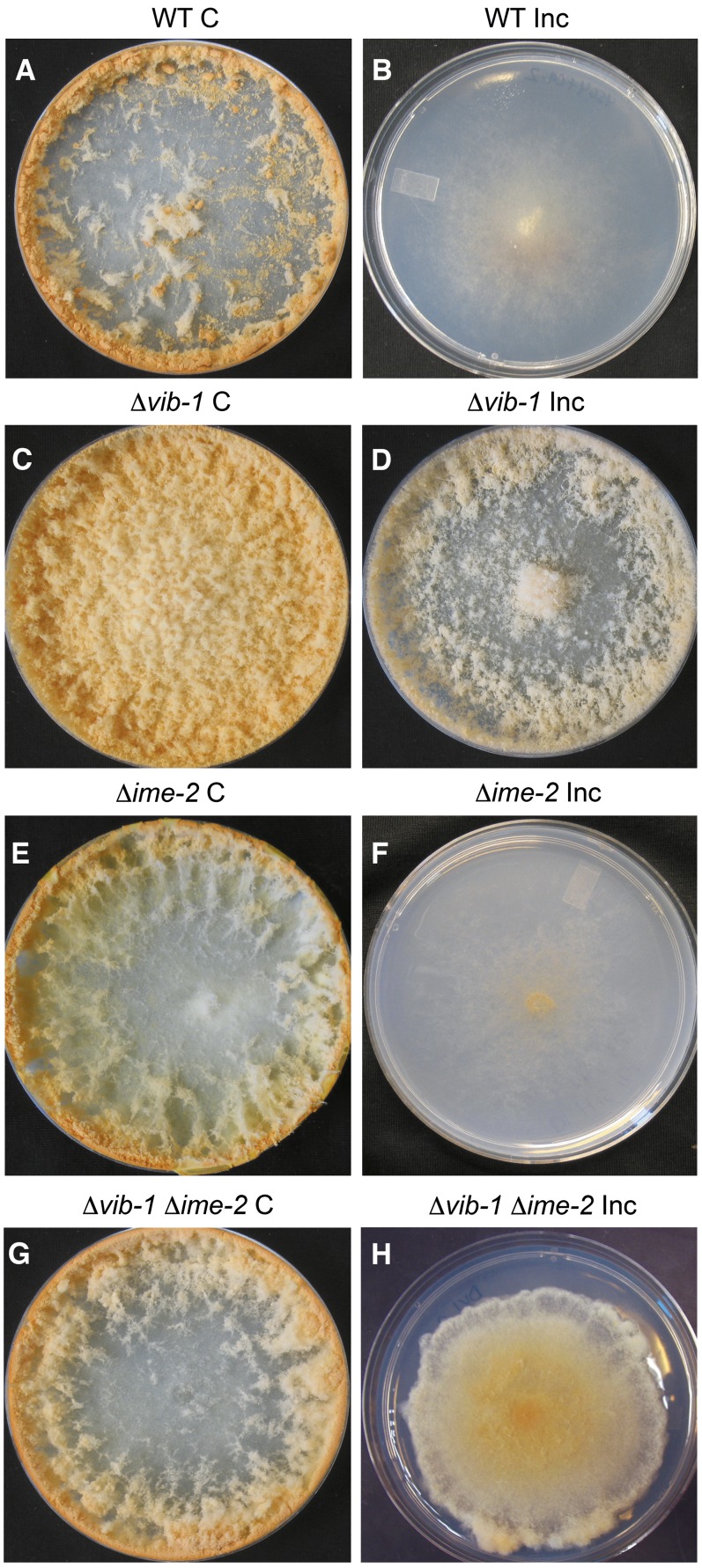
In N. crassa, if two individuals are genetically identical at all nonself recognition loci (termed het), they can undergo cell fusion to form a “compatible” heterokaryon that looks identical to a homokaryotic wild-type strain. However, if individuals are genetically different at any 1 of 11 het loci, hyphal fusion results in compartmentalization of the fusion cell and subsequent death; these individuals are referred to as “incompatible” (Glass and Kaneko 2003) (Figure 1). In addition to cell death, incompatible heterokaryons have a reduced growth rate and do not conidiate. Strains that contain loss-of-function mutations in vib-1 form vigorous heterokaryons even if they differ in het allelic specificity (Xiang and Glass 2002, 2004) (Figure 2, C and D). Whereas compatible wild-type heterokaryons conidiate upon contact with plate edges, Δvib-1 heterokaryons of identical or alternate het-c pin-c haplotype display deregulated conidiation (Figure 2, C and D).
Figure 2 .
A deletion of ime-2 partially restores HI in Δvib-1 mutants. Heterokaryons of identical het-c pin-c haplotype (C, compatible) show a growth and conidiation pattern indistinguishable from that of homokaryotic strains, including WT (A) (FGSC 4564 + R15-7), Δvib-1 (C) (KD13-21 + KD13-51), Δime-2 (E) (DI.PYR.4 + DI.HIS.10), and Δvib-1 Δime-2 (G) (DVI.HIS.48 + DVI.PYR.63) heterokaryons (Table 1). Strains carrying incompatible het-c pin-c haplotypes (Inc, incompatible) in a WT background show growth inhibition and suppression of conidiation (B) (FGSC 4564 + C9-15). Heterokaryons of incompatible het-c pin-c haplotypes, but carrying homozygous Δvib-1 mutations are similar in phenotype to compatible Δvib-1 heterokaryons (D) (KD13-33 + KD13-1). Heterokaryons carrying incompatible het-c pin-c haplotypes and homozygous Δime-2 mutations show typical HI (F) (DI.HIS.10 + DI.A.22), while heterokaryons carrying incompatible het-c pin-c haplotypes and homozygous for Δvib-1 Δime-2 mutations show partial restoration of HI (decreased growth rate and suppression of conidiation) (H) (DVI.PYR.63 + DVI.A.101).
Due to genetic interactions identified between ime-2 and vib-1 with regard to formation of female reproductive structures (Hutchison and Glass 2010), we evaluated whether IME-2 plays a role in HI by assessing the incompatibility phenotype of Δime-2 and Δvib-1 Δime-2 mutants compared to wild-type and Δvib-1 mutants. To evaluate HI, we used a forced heterokaryon approach, using strains with the same het-c pin-c specificity (het-c1 pin-c1 and thus compatible) vs. using strains containing alternate het-c pin-c haplotypes (het-c1 pin-c1 + het-c2 pin-c2 and thus incompatible) (Kaneko et al. 2006; Hall et al. 2010) (Figure 1 and Table 1). Further, these strains have different, complementary auxotrophic markers; when isolates carrying different auxotrophic markers are placed on minimal media, only the heterokaryotic strain is able to grow.
Compatible heterokaryons of identical het-c pin-c haplotype of wild-type, Δvib-1, Δime-2, or Δvib-1 Δime-2 strains were phenotypically identical to a homokaryotic wild-type or mutant strain by itself (Figure 2, A, C, E, and G). Wild-type heterokaryons with incompatible combinations of het-c pin-c haplotypes exhibited a severely decreased growth rate and lack of conidiation (compare Figure 2B with 2A). Heterokaryons carrying incompatible het-c pin-c haplotype and homozygous deletions of ime-2 also showed an identical incompatible phenotype to wild type (compare Figure 2F with 2B), indicating that ime-2 is not required for HI. In contrast, homozygous Δvib-1 heterokaryons of incompatible het-c pin-c haplotype showed a phenotype more similar to compatible Δvib-1 heterokaryons (compare Figure 2D to 2C); a deletion of vib-1 suppresses HI. However, a heterokaryon carrying incompatible het-c pin-c haplotypes, plus homozygous Δvib-1 Δime-2 mutations showed significantly poorer growth and conidiation than a Δvib-1 suppressed incompatible heterokaryon (compare Figure 2H to 2D). The Δvib-1 Δime-2 het-c pin-c incompatible heterokaryon had a growth rate of 2.80 ± 0.55 cm/day (Figure 2H), compared to a wild-type incompatible heterokaryon (1.16 ± 0.53 cm/day; Figure 2B) and a Δvib-1 incompatible heterokaryon (6.7 ± 0.40 cm/day; Figure 2D). In addition, the Δvib-1 Δime-2 het-c pin-c incompatible heterokaryon produced fewer conidia compared to a Δvib-1 suppressed incompatible heterokaryon (compare Figure 2H to 2D) and exhibited an altered, dense growth pattern that did not resemble either single mutants (Δvib-1 or Δime-2; Figure 2, C and E) or the Δvib-1 Δime-2 compatible strain (Figure 2G). Thus, in a Δvib-1 mutant, a deletion of ime-2 partially restored the inhibited growth and aconidial phenotype associated with nonself recognition and HI.
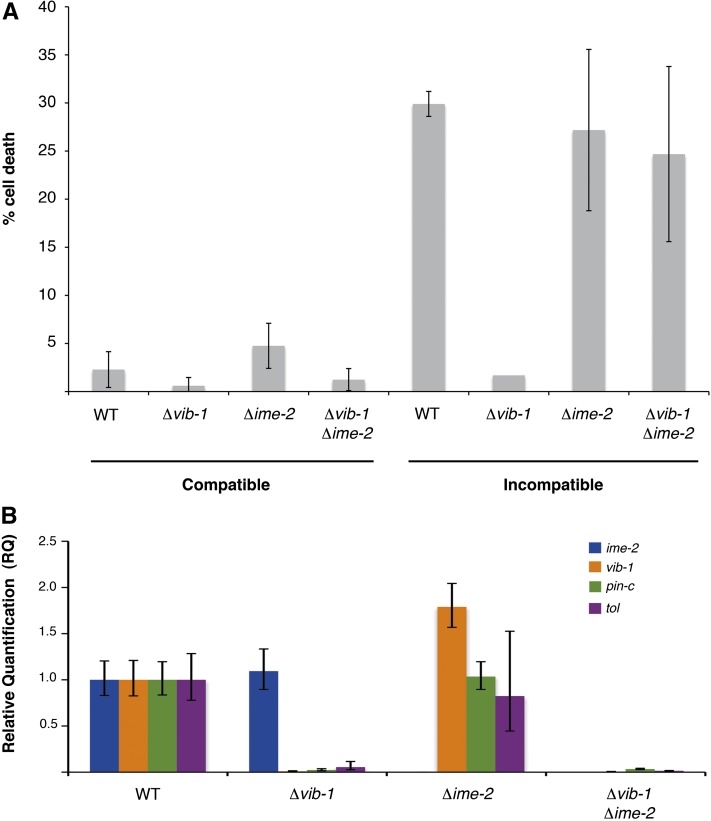
In addition to growth inhibition and absence of conidiation, wild-type heterokaryons carrying incompatible het-c pin-c haplotypes show substantial PCD, with ∼30% of hyphal segments showing compartmentalization and death, which is assessed by staining with vital dyes such as methylene blue (Figure 1) (Glass and Kaneko 2003). The Δvib-1, Δime-2, and Δvib-1 Δime-2 heterokaryons of identical het-c pin-c haplotype exhibited very little cell death, ∼2–5%, which was similar to that of a wild-type compatible heterokaryon (Figure 3A). Wild-type heterokaryons carrying incompatible het-c pin-c haplotypes showed ∼30% hyphal compartmentation and death (Xiang and Glass 2002). Similar to wild-type incompatible heterokaryons and consistent with the HI phenotype (Figure 2F), the Δime-2 heterokaryon carrying incompatible het-c pin-c haplotypes showed ∼27% hyphal compartmentation and death (Figure 3A). As observed previously (Xiang and Glass 2002), Δvib-1 heterokaryons carrying incompatible het-c pin-c haplotypes showed a substantially reduced cell death frequency that was similar to that of either Δvib-1 or wild-type compatible heterokaryons (∼2–5%). In contrast, the Δvib-1 Δime-2 heterokaryon carrying incompatible het-c pin-c haplotypes showed a cell death frequency similar to that of wild-type incompatible heterokaryons (∼25%) (Figure 3A). These data indicate that loss-of-function mutations in ime-2 restored hyphal compartmentation and death to Δvib-1 heterokaryons carrying incompatible het-c pin-c haplotypes.
Figure 3 .
A deletion of ime-2 restores wild-type levels of cell death in Δvib-1 incompatible heterokaryons. (A) Heterokaryons of compatible and incompatible het-c pin-c haplotype from Figure 2 were plated on minimal media overlaid with cellophane and grown for 1–3 days. The percentage of dead cell compartments was evaluated using microscopy and methylene blue staining (Xiang and Glass 2002; Hutchison et al. 2009). Heterokaryons of incompatible het-c pin-c haplotype but carrying homozygous vib-1 deletions were suppressed for cell death (Δvib-1), while the addition of Δime-2 mutation in these strains restored wild-type levels of cell death (Δvib-1 Δime-2). (B) Expression of ime-2, vib-1, and two HET domain genes (pin-c, tol) was assessed using quantitative RT-PCR. Deletion of vib-1 abolishes expression of HET domain genes during vegetative growth. Expression of pin-c and tol was not restored in a Δvib-1 Δime-2 mutant.
Many proteins involved in HI, like PIN-C, contain a conserved protein domain of unknown function, termed HET (PF06985) (Espagne et al. 2002). HET domains are filamentous fungal-specific protein domains that can cause an HI-like cell death when overexpressed (Paoletti and Clave 2007). Previously, it was shown that vib-1 is necessary for the expression of HET domain genes pin-c, tol, and het-6, which are required for HI in N. crassa (Dementhon et al. 2006) (Figure 1). To test whether restoration of cell death in Δvib-1 Δime-2 strains correlated with HET domain gene expression, we performed quantitative RT-PCR for pin-c and tol in wild-type, Δvib-1, Δime-2, and Δvib-1 Δime-2 strains (Figure 3B). As expected, expression of pin-c and tol was not detected in a Δvib-1 mutant. Although expression of pin-c and tol in Δime-2 strains was not significantly different from that in wild type, expression of vib-1 was significantly increased, suggesting that IME-2 negatively regulates vib-1 expression levels. In the Δvib-1 Δime-2 strain, however, expression of pin-c and tol was similar to expression levels observed in the Δvib-1 strain (Figure 3B), indicating that restoration of cell death by Δime-2 mutations in a Δvib-1 strain carrying incompatible het-c pin-c haplotypes is not due to induced expression of pin-c.
Microarray analysis reveals that mutations in ime-2 affect mitochondrial function
Because we identified a genetic interaction between vib-1 and ime-2 during HI, we evaluated what physiological processes were affected in the Δime-2 mutant by performing gene expression profiling of wild type vs. a Δime-2 strain under nitrogen starvation conditions. The initiation of female reproductive structures (protoperithecia) in N. crassa is regulated by the availability of nitrogen (Westergaard and Mitchell 1947; Hirsh 1954), and we hypothesized that differences in gene expression between wild-type and Δime-2 deletion strains may be more pronounced under nitrogen starvation conditions (Hutchison and Glass 2010). The wild-type strain (FGSC 2489) and a Δime-2 strain (FGSC 17937) were grown overnight (∼16 hr) in minimal media. The mycelia were then washed and subsequently transferred to a flask containing minimal media without nitrogen and grown for an additional 4 hr (see Materials and Methods). Mycelia from both strains were harvested for RNA extraction and microarray analysis. Three replicate microarrays, including dye swaps, were performed.
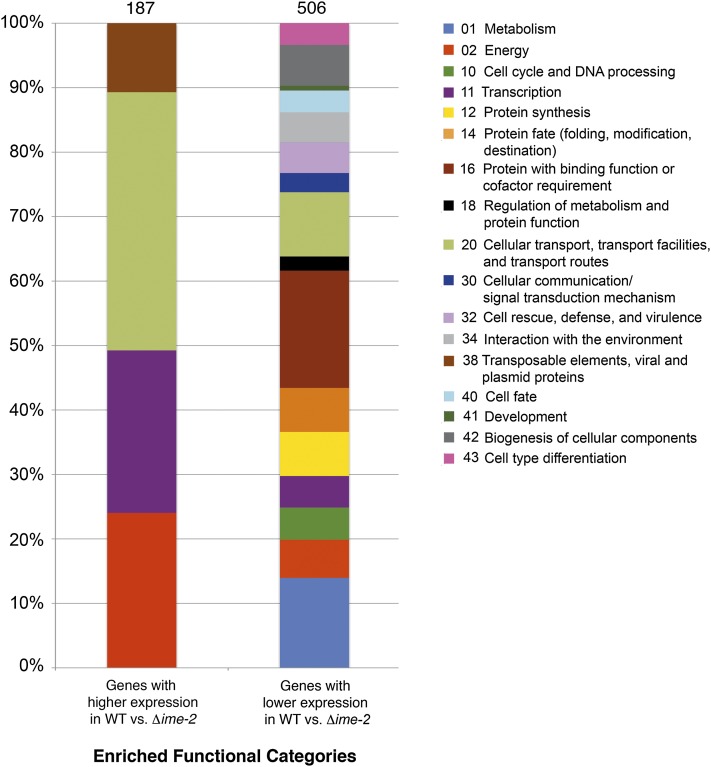
In the Δime-2 strain, a total of 187 genes showed a significant decrease in expression level of at least 1.5-fold. Functional category analysis (Ruepp et al. 2004) of this gene set showed enrichment for energy (P < 0.0001); transcription (P < 0.05); cellular transport (P < 0.005); and transposable elements, viral and plasmid proteins (P < 1e-11) (Figure 4). Many of the genes in the energy functional category belonged to pathways involving electron transport and respiration. Furthermore, the enrichment of genes belonging to the “transposable elements, viral and plasmid proteins” functional category was due almost exclusively to genes belonging to the mitochondrial genome. In fact, 13 of the 29 genes composing the mitochondrial genome showed decreased expression levels in the Δime-2 strain compared to wild type (Supporting Information, Table S1), a significant enrichment (P < 0.006). These observations suggest mitochondrial impairment in the Δime-2 mutant and are consistent with the fact that Δime-2 strains exhibit a slower growth rate (3.5–4 cm/day) compared to wild-type strains (∼7 cm/day).
Figure 4 .
Functional category analysis of gene expression differences in wild type vs. the Δime-2 mutant. Shown is the distribution of significantly enriched MIPS functional categories (http://www.helmholtz-muenchen.de/en/mips/projects/funcat) (Ruepp et al. 2004) for the microarray data set of wild type compared to a Δime-2 deletion strain. A total of 187 genes showed a reduction in expression in Δime-2 relative to wild type, while 506 genes showed an increase in expression level in the Δime-2 strain relative to wild type.
A total of 506 genes showed increased expression levels in the Δime-2 strain compared to wild type (Figure 4). These genes were enriched in a variety of functional categories (Ruepp et al. 2004), including metabolism (P < 1e-11); energy (P < 1e-17); cell-cycle and DNA processing (P < 0.0001); transcription (P < 0.005); protein synthesis (P < 1e-24); protein fate (P < 1e-5); protein with binding function or cofactor requirement (P < 1e-44); regulation of metabolism and protein function (P < 0.0005); cellular transport (P < 1e-14); cellular communication and signal transduction (P < 1e-4); cell rescue, defense, and virulence (P < 0.0005); interaction with the environment (P < 1e-6); cell fate (P < 1e-8); development (P < 0.005); diogenesis of cellular components (P < 1e-12); and cell-type differentiation (P < 1e-6). Although we hypothesized that the constitutive production of protoperithecia in Δime-2 strains (Hutchison and Glass 2010) may be due to a defect in nitrogen sensing, we did not observe significant differences in gene expression with respect to nitrogen metabolism genes or genes involved in the metabolism of amino acids. Thus, the array data instead suggest that Δime-2 mutants are not deficient in nitrogen sensing specifically, but that these strains may have a more general nutrient-sensing defect.
Interestingly, genes involved in mitochondrial biogenesis (within the biogenesis of cellular components category) were also significantly enriched among genes that showed increased expression levels in the Δime-2 mutant (P < 0.0001). A recent study by Keeping et al. (2010) used mass spectrometry as well as computational methods to compile a comprehensive list of 738 genes that compose the N. crassa mitochondrial proteome. Using this data set, we asked whether nuclear-encoded mitochondrial genes were expressed differently between wild-type and Δime-2 strains. In fact, these genes were significantly enriched (P < 1e-18) in the set of 506 genes that showed increased expression levels in the Δime-2 mutant. As previously mentioned, 13 of the 29 genes composing the mitochondrial genome showed lower expression levels in the Δime-2 strain (Table S1). These data suggest that the Δime-2 mutant may have impaired mitochondria (evidenced by decreased expression of genes within the mitochondrial genome), resulting in a regulatory feedback loop such that Δime-2 strains increase expression of nuclear-encoded mitochondrial genes to compensate for this defect. Overall, the microarray data suggest that IME-2 plays a role in mitochondrial function in N. crassa. We therefore assessed, via microscopy, whether mitochondrial morphology or protein localization was altered in Δime-2 mutants compared to wild type and the Δvib-1 mutants.
ime-2 mutants affect post-translational processing of the mitochondrial protein ARG-4
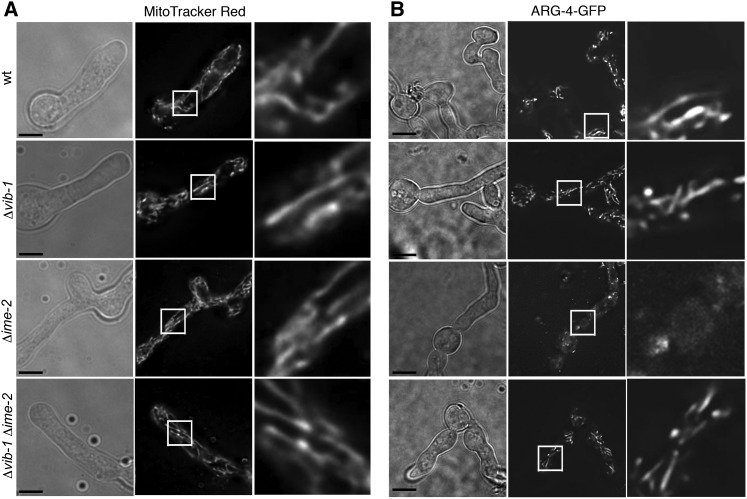
To evaluate the mitochondrial phenotype in wild-type, Δvib-1, Δime-2, and Δvib-1 Δime-2 strains, we stained conidial germlings with MitoTracker Red FM and also transformed each strain with the nuclear-encoded GFP-tagged mitochondrial marker gene encoding ARG-4 (Bowman et al. 2009). The nuclear arg-4 locus encodes acetylornithine-glutamate acetyltransferase (arginine biosynthetic pathway), which is imported into the mitochondrial matrix (Cybis and Davis 1975). In wild-type hyphae, mitochondria appear as long tubules in the apical regions of hyphae and as more punctate structures farther back from the hyphal tip (Bowman et al. 2009). When stained with MitoTracker Red FM, all strains (Δvib-1, Δime-2, and Δvib-1 Δime-2) looked identical to wild type (Figure 5A) and had brightly stained, long, tubular mitochondrial networks. Similarly, when mitochondria were visualized via ARG-4-GFP localization, wild type and the Δvib-1 mutant exhibited long, tubular mitochondria that looked identical to the MitoTracker Red FM-stained mitochondria (Figure 5B). However, Δime-2 strains exhibited an altered localization pattern. Instead of long, tubular structures, ARG-4-GFP localized either to vesicles or to punctate structures or was diffuse in the cytoplasm (Figure 5B). MitoTracker Red FM can permeate the cell membrane and accumulates in mitochondria based on membrane potential (Macho et al. 1996; Poot et al. 1996), and thus it is likely that the mitochondria observed in all strains, including the Δime-2 strains, were active and have a functioning membrane potential. However, the lack of ARG-4-GFP localization in the Δime-2 strain suggested that IME-2 may play a role in protein targeting to the mitochondria. Interestingly, the Δvib-1 Δime-2 strain restored localization of ARG-4-GFP to tubular mitochondria (Figure 5B).
Figure 5 .
Mutations in ime-2 affect localization of the mitochondrial protein ARG-4. (A) Mitochondria stained with MitoTracker Red FM in FGSC 2489 (WT), FGSC 11308 (Δvib-1), FGSC 17937 (Δime-2), and DVI.4 (Δvib-1 Δime-2) (Table 1). The right column is an enlargement of the region highlighted by a white box in the center column. (B) Localization of ARG-4-GFP to mitochondria in wild-type and deletion strains transformed with pccg1-arg-4-gfp (Table 1). Localization of ARG-4-GFP to mitochondria in wild type is identical to mitochondria stained by MitoTracker Red FM and to that previously reported (Bowman et al. 2009). ARG-4-GFP localization in the Δvib-1 and the Δvib-1 Δime-2 mutants was identical to that in WT. However, in the Δime-2 strain, mitochondrial tubule structures were not observed and instead ARG-4-GFP localized to either vesicles or punctae or was diffuse in the cytoplasm. Bar, 5 μm.
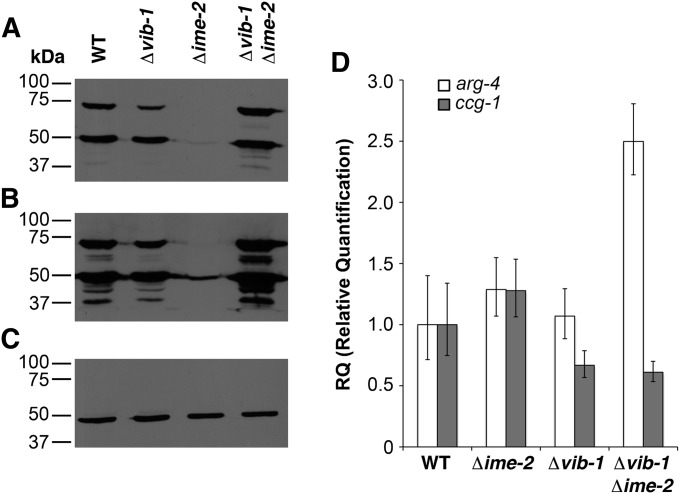
In yeast, the ARG-4 homolog Arg7 undergoes a post-translational autoproteolytic processing step that results in the formation of two smaller subunits, each of which localizes to the mitochondrial matrix, where they associate in a complex (Abadjieva et al. 2000). The autoproteolytic activity of yeast Arg7 is dependent on a threonine residue, and this residue is conserved in N. crassa ARG-4. Therefore, we determined whether the defect in ARG-4-GFP localization in Δime-2 strains was due to a defect in protein processing. Two distinct bands for N. crassa ARG-4-GFP (∼75 kDa and 50 kDa) have previously been reported (Bowman et al. 2009), which is expected if ARG-4 is proteolytically cleaved at the conserved threonine residue. In the wild-type and Δvib-1 strains, two ARG-4-GFP bands at ∼75 kDa and 50 kDa were detected (Figure 6, A and B). However, in Δime-2 strains much less ARG-4-GFP protein was present and only the 50-kDa ARG-4-GFP band was detected. Despite the lack of detectable full-length ARG-4 in the Δime-2 mutants, these mutants were not arginine auxotrophs. In S. cerevisiae, arg7 mutants exhibit a leaky Arg phenotype (Crabeel et al. 1997). Consistent with microscopy results, the Δvib-1 Δime-2 strain showed a wild-type pattern for ARG-4 processing (Figure 6, A and B). In fact, Δvib-1 Δime-2 strains appeared to produce more ARG-4-GFP than wild-type or Δvib-1 deletion strains.
Figure 6 .
Strains carrying a deletion in ime-2 affect the post-transcriptional regulation of ARG-4. (A) Western blot for ARG-4-GFP in wild-type, Δvib-1, Δime-2, and Δvib-1 Δime-2 strains, with molecular weight ladder (kDa) on the left. ARG-4-GFP is detected as two distinct bands, ∼75 kDa and 50 kDa in WT, as previously reported (Bowman et al. 2009). (B) Longer exposure of the blot from A, more clearly showing the 50-kDa ARG-4-GFP in the Δime-2 strain. (C) Western blot of β-tubulin showing that equal amounts of protein were loaded in each well. (D) Quantitative RT-PCR of arg-4 and ccg-1 transcript levels in wild-type, Δvib-1, Δime-2, and Δvib-1 Δime-2 strains.
The arg-4-gfp construct used to visualize ARG-4-GFP localization is under the regulation of the ccg-1 promoter (Bowman et al. 2009), which is commonly used for constitutive gene expression in N. crassa. The wild-type and the Δvib-1, Δime-2, and Δvib-1 Δime-2 strains transformed with the pccg-arg-4-gfp construct also contain a native copy of arg-4. Thus, we quantified the transcription of arg-4 in comparison to ccg-1, using Q-RT-PCR in all strains to assess expression levels of arg-4 (Figure 6D). Expression levels for arg-4 (a readout for both the ccg-1–regulated and the resident arg-4 genes) were very similar between wild type and Δvib-1 and Δime-2 mutants. These data indicate that the differences observed in ARG-4 protein levels between wild-type and Δime-2 strains were not due to decreased expression levels of arg-4 in the Δime-2 mutant (Figure 6, A, B, and D). However, the expression level of arg-4 was significantly elevated in the Δvib-1 Δime-2 strain, consistent with increased ARG-4 protein levels in this strain detected via Western blot (Figure 6B). Wild type and the Δime-2 mutant also showed similar levels of ccg-1 expression, while the Δvib-1 and Δvib-1 Δime-2 strains showed slightly lower levels of ccg-1 expression (Figure 6D). Because the ccg-1 promoter was not downregulated in the Δime-2 strain or upregulated in the Δvib-1 Δime-2 strain, it is likely that differences in proteins levels of ARG-4 are not due to the regulation of the arg-4 transgene. Either the increased arg-4 transcription in the Δvib-1 Δime-2 strain originated from the native arg-4 locus or arg-4 transcripts were stabilized in this strain. Thus, ime-2 and vib-1 affect the post-translational modification of ARG-4 and, to some degree in the Δvib-1 Δime-2 strain, transcriptional regulation of arg-4.
Mutations in ime-2 revert the protease secretion phenotype of vib-1 mutants
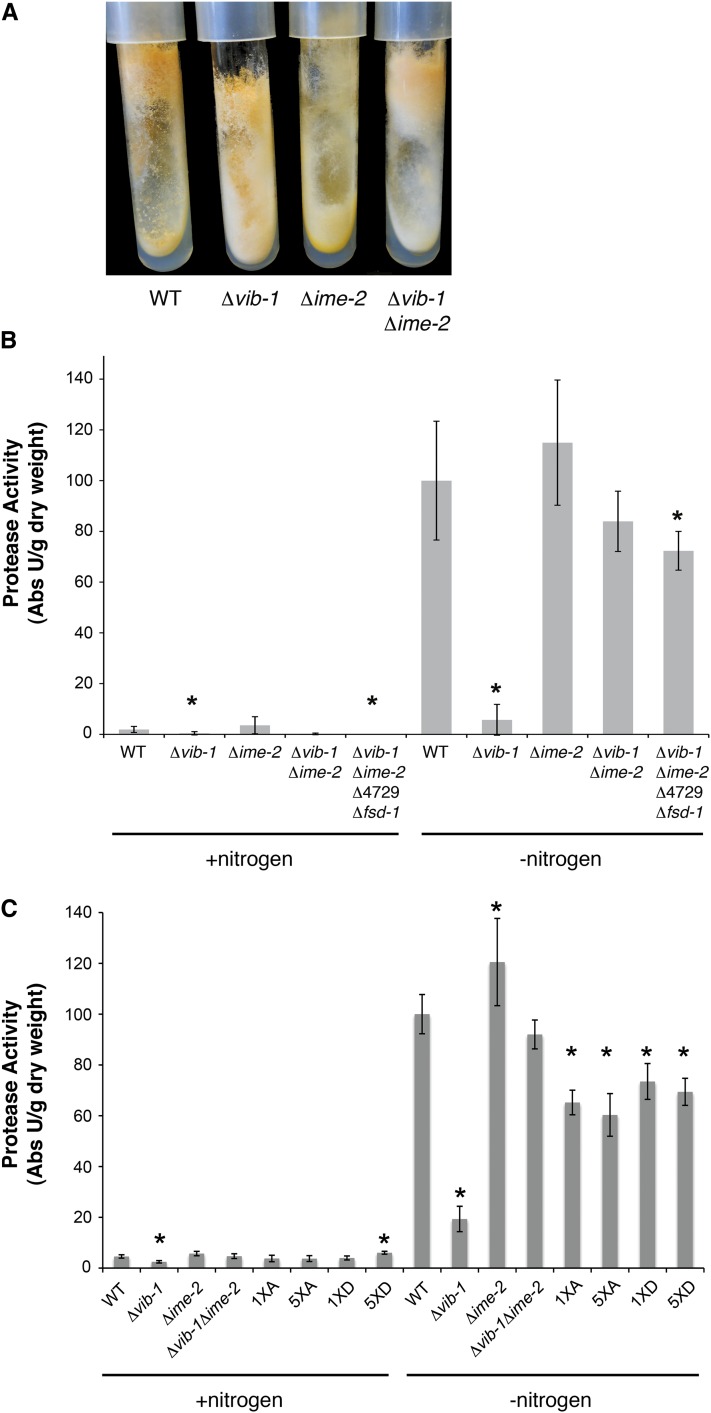
Mutations in the transcription factor vib-1 cause a visible phenotype during vegetative growth, such that Δvib-1 mutants show pinkish (rather than orange) conidial pigmentation, deregulated conidiation, decreased aerial hyphae formation (Figures 2C and 7A), and a slight decrease in growth rate compared to wild type (Xiang and Glass 2002; Dementhon et al. 2006; Hutchison et al. 2009). In contrast, wild type and the Δime-2 mutant show robust aerial hyphae formation and conidiation at the top of the tube or edge of the plate and were nearly indistinguishable, with the exception of the slightly yellow pigmentation observed in the Δime-2 mutant (Figure 7A). In the Δvib-1 Δime-2 mutant, aerial hyphae formation was restored and conidiation occurred only at the top of the slants, a phenotype similar to wild type and the Δime-2 mutant (Figure 7A). However, conidia of the Δime-2 Δvib-1 mutant are pinkish in color, like those of the Δvib-1 single mutants.
Figure 7 .
An ime-2 deletion restores wild-type conidiation patterns and protease production to a Δvib-1 mutant. (A) Wild-type (FGSC 2489), Δvib-1 (FGSC 11308), Δime-2 (FGSC 17936), and Δvib-1 Δime-2 (DVI.4) strains grown on minimal media slants. (B) Extracellular protease activity of wild-type and deletion strains (from A), as well as that of the Δvib-1 Δime-2 Δfsd-1 ΔNCU04729 strain (D49VI.HIS.1), was assessed on media with and without nitrogen. Only the vib-1 deletion strain showed inability to secrete proteases. Protease activity units are normalized to WT in nitrogen starvation (−nitrogen) media. (C) Extracellular protease activity of vib-1 phospho-mutants (vib-1S60A 1XA, vib-1S60D 1XD, vib-1S60A;S413A;S537A;S542A;S545A 5XA, and vib-1S60D;S413D;S537D;S542D;S545D 5XD) (Table 1). Data are shown as the fold increase of protease secretion compared to that in the strains grown on media containing nitrogen. Asterisks (*) in B and C indicate strains with protease production significantly different from that in WT (P < 0.05).
In addition to the vegetative conidiation phenotype, Δvib-1 mutants do not secrete extracellular proteases in response to nitrogen or carbon starvation (Dementhon et al. 2006), a phenotype similar to the vib-1 homolog in A. nidulans, xprG (Katz et al. 2006). We therefore evaluated whether mutations in ime-2 restored protease activity in the Δvib-1 mutant. When nitrogen was provided, extracellular proteases were not induced in the wild-type, Δvib-1, Δime-2, or Δvib-1 Δime-2 strains (Figure 7B). When the wild-type strain was transferred to nitrogen starvation medium, extracellular protease activity was induced, while no activity was detected in the Δvib-1 mutant. A strain carrying a deletion of Δime-2 produced slightly elevated levels of proteases in response to nitrogen starvation (Figure 7, B and C). However, unlike the Δvib-1 mutant, the Δvib-1 Δime-2 mutant showed wild-type protease activity in response to nitrogen starvation (Figure 7, B and C), indicating that loss-of-function mutations in ime-2 suppressed the defect in protease secretion in Δvib-1 mutants.
Based on the Δvib-1 Δime-2 phenotype, we hypothesized that additional regulators may be functioning redundantly to vib-1 to restore protease production. Two obvious candidate genes that may have redundant functions with vib-1 are the vib-1 paralogs fsd-1 and NCU04729. Previously, we determined that strains containing a deletion of NCU04729 were indistinguishable from wild type under all conditions, while Δfsd-1 mutants shows defects in protoperithecial formation and ascospore maturation (Hutchison and Glass 2010); neither fsd-1 nor NCU04729 affect cell death due to HI. We therefore tested the ability of a Δvib-1 Δime-2 Δfsd-1 ΔNCU04729 deletion strain (Table 1, strain D49VI.HIS.1) to produce extracellular proteases. As shown in Figure 7, the quadruple mutant (Δvib-1 Δime-2 Δfsd-1 ΔNCU04729) produced near wild-type levels of extracellular proteases (Figure 7B), indicating that neither fsd-1 or NCU04729 were responsible for the restoration of protease secretion in the Δvib-1 Δime-2 mutant.
VIB-1 is phosphorylated at a predicted IME-2 consensus site
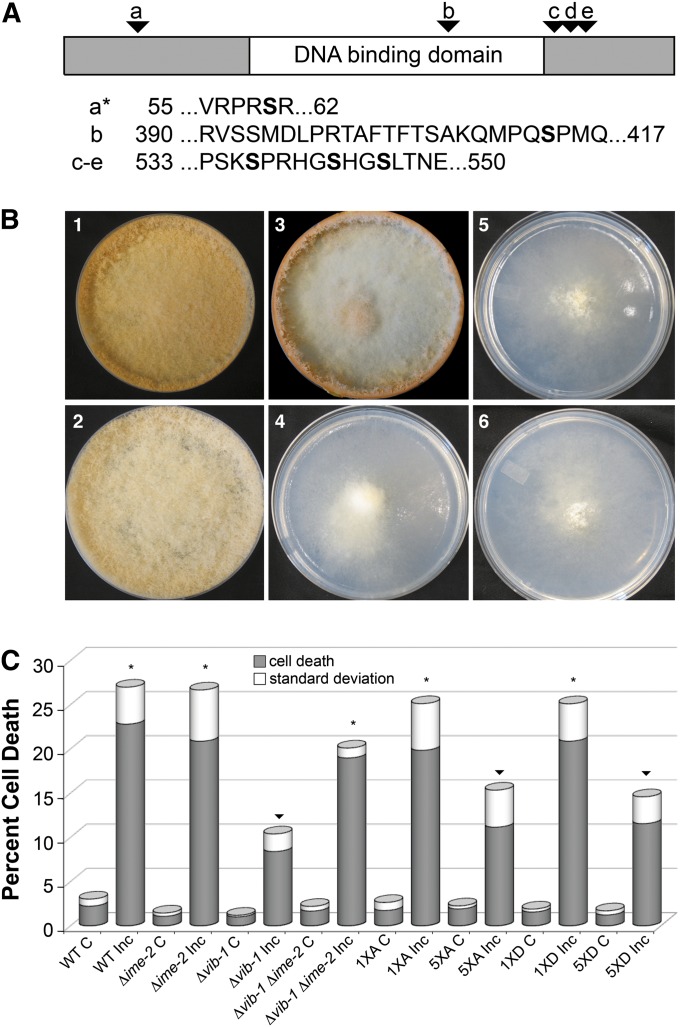
In S. cerevisiae, the consensus sequence for Ime2 phosphorylation (R-P-x-S/T-A/R-G) has been well characterized (Holt et al. 2007; Moore et al. 2007). We analyzed the N. crassa genome for matches to the yeast Ime2 phosphorylation consensus sequence, using the Scansite program (http://scansite.mit.edu/) (Obenauer et al. 2003) with a slightly modified phosphorylation consensus identified for N. crassa (R-P-x-S/T-P/A/R-G) (L. Holt Laboratory, unpublished data). There are 30 total predicted phosphorylation targets of IME-2 in the N. crassa genome (Table S2). Consistent with the role of ime-2 in protoperithecial formation, one of the predicted IME-2 phosphorylation targets present in the Scansite data set is NIT-2 (NCU09068), a major regulator of nitrogen utilization. In addition, both AL-1 (albino-1; NCU00552), a phytoene dehydrogenase involved in carotenoid biosynthesis (Schmidhauser et al. 1990), and NRC-2 (NCU01797), a serine-threonine kinase involved in regulation of entry into the conidiation pathway and conidial development (Kothe and Free 1998), were present in the Scansite data set. Δime-2 strains appear to have a slightly different conidiation phenotype from that of wild type (WT), including less pigmentation and fewer conidia (Figures 2E and 7A). The N. crassa homolog of the yeast protein kinase Ste20 (NCU03894) is also a predicted IME-2 phosphorylation target. Ste20 and its homologs in mammals (Mst1 and Mst2) have been previously shown to have a role in the apoptotic signaling cascade (Madeo et al. 2009; Radu and Chernoff 2009). Additionally, the VIB-1 protein contains a match for the Ime2 consensus, while neither of the other two NDT80 homologs in N. crassa (fsd-1 or NCU04729) have an Ime2 consensus site. From a phosphoproteomics study (A. Leeder and N. L. Glass, unpublished results), we identified a phosphopeptide for VIB-1 at the predicted IME-2 consensus site (RPRS*60), as well as four additional phosphorylation sites (MPQS*413, PSKS*537, and RHGS*542HGS*545) (Figure 8A).
Figure 8 .
Phosphorylation of VIB-1 and phenotype of phospho-site mutants. (A) VIB-1 is phosphorylated on multiple sites (determined by mass spectrometry), including a predicted IME-2 consensus site. The asterisk indicates the phosphorylation site that matches the consensus for yeast Ime2. (B) Mutations of the IME-2 consensus phosphorylation site in VIB-1 do not affect growth inhibition and suppression of conidiation associated with HI. Δvib-1 compatible (1, KD13-21 + KD13-51) and incompatible (2, KD13-21 + DV.80) heterokaryons (suppressed for HI) are shown. A heterokaryon with only one functional copy of vib-1 and carrying incompatible het-c pin-c haplotypes shows typical HI (Δvib-1 mutations are recessive) (4, KD13-51 + JH1) compared to a heterokaryon carrying one functional copy of vib-1, but of compatible het-c pin-c haplotype (3, KD13-21 + FGSC 6103). Heterokaryons with a phospho-null mutation (S60 to A) (5, 1XA + KD13-21) or a phospho-mimetic mutation (S60 to D) (6, 1XD + KD13-21) at the predicted IME-2 consensus site in VIB-1 exhibited a typical wild-type HI phenotype. (C) Mutations of the IME-2 consensus phosphorylation site in VIB-1 do not affect cell-death percentages associated with HI. However, mutations at all five VIB-1 phosphorylation sites reduce cell-death percentages, although growth inhibition is still observed. All incompatible (Inc) heterokaryons exhibit significantly higher cell death than compatible (C) heterokaryons. Cell death percentages for incompatible heterokaryons labeled with an asterisk (*) were significantly different (P < 0.05) from those for heterokaryons labeled with an arrowhead (▼).
To test whether the predicted IME-2 consensus site was necessary for VIB-1 function, we constructed mutant vib-1 alleles such that the IME-2 site was mutated to alanine (predicted to be phospho-null; S60-to-A mutation) (1XA, Table 1) or mutated to aspartate (phospho-mimetic; S60-to-D mutation) (1XD, Table 1). The growth rates of strains carrying the vib-1S60A and vib-1S60D alleles were identical to those of wild type as was nuclear localization of VIB-1S60A-GFP and VIB-1S60D -GFP (Figure S1 and Figure S2). Although identical in phenotype to wild type, both the vib-1S60A-gfp and the vib-1S60D -gfp strains showed slightly lower protease levels, suggesting that phosphorylation of the S60 site contributes to VIB-1 activity (Figure 7C). Similarly, mutations of the IME-2 consensus sequence in VIB-1 significantly reduced the numbers of protoperithecia produced in vib-1S60A-gfp and vib-1S60D -gfp strains under conditions of nitrogen starvation (Figure S3). These data indicate that mutations in the Ime2 consensus site on VIB-1 negatively affect protoperithecial development.
To assess the role of the Ime2 phosphorylation sites on HI, the vib-1S60A-gfp and vib-1S60D-gfp alleles were transformed into Δvib-1 strains of het-c2 pin-c2 haplotype (Table 1). Each strain was then forced in a heterokaryon with a Δvib-1 strain of het-c1 pin-c1 haplotype. The Δvib-1 mutation is recessive, such that a single functional copy of vib-1 in a heterokaryon is sufficient to trigger HI (Xiang and Glass 2002; Dementhon et al. 2006) (Figure 8B, compare panels 3 and 4). When strains containing vib-1S60A-gfp or vib-1S60D-gfp were forced in a heterokaryon with a Δvib-1 strain of incompatible het-c pin-c haplotype, a HI response was triggered that was indistinguishable from a wild-type HI phenotype (Figure 8B, panels 5 and 6). In addition, the vib-1S60A-gfp and vib-1S60D-gfp incompatible heterokaryons displayed wild-type levels of cell death (Figure 8C).
We also constructed strains where all five identified phosphorylation sites were mutated to alanine (S60A, S413A, S537A, S542A, S545A, and 5XA) or aspartate (S60D, S413D, S537D, S542D, S545D, and 5XD) (Table 1). Both the 5XA and 5XD strains grew significantly slower than WT (Figure S2) and were also impaired in cell death during HI (Figure 8C). The 5XA and 5XD mutants also showed a significant decrease in protoperithecial formation compared to WT (Figure S2), with the 5XD strain showing the most significant reduction, with values similar to those of the Δvib-1 and Δvib-1Δ ime-2 mutants. Consistent with our data that five amino acid substitutions in VIB-1 negatively affect its function, the 5XA and 5XD strains showed less protease activity than WT (P = 0.05), with values similar to those of the 1XA and 1XD strains.
Discussion
Diversification of kinase cascades may provide a mechanism for eukaryotes to evolve new developmental pathways or adapt to new environments (Bhattacharyya et al. 2006). In this study, we showed that N. crassa ime-2 regulates cell death due to HI in the absence of vib-1 and also regulates post-translational processing of the mitochondrial matrix protein ARG-4. IME2 homologs have not been previously implicated in programmed cell death, but recent studies in yeast and other fungi have provided evidence that Ime2 and its homologs can function in cellular processes other than meiosis. For instance, Strudwick et al. (2010) described a role for Ime2 in yeast pseudohyphal formation, and studies in other fungal species showed that although Ime2 homologs often function in sexual differentiation or nutrient sensing, they are not generally meiotic regulators (Irniger 2011). Our results provide additional evidence that the function of the Ime2 pathway differs among fungal species and implicate N. crassa ime-2 in nonself recognition and programmed cell death.
Data from this study as well as from a previous study (Hutchison and Glass 2010) indicate that ime-2 interacts genetically with the transcription factor vib-1. For some phenotypes, such as HI, protease production, and conidiation, ime-2 is epistatic to vib-1. However, for other phenotypes, including protoperithecial formation, HET domain gene expression, and ARG-4 localization and protein processing, vib-1 is epistatic to ime-2. These data suggest that the ime-2/vib-1 signaling pathway is not a simple, linear interaction, but that there are other genetic interactors present depending on the cellular process. We propose that the overall structure of the pathway (with the IME2 homolog functioning upstream of the NDT80 homolog) is likely conserved, that IME-2 negatively regulates VIB-1 (likely at the protein level), and that IME-2 regulates a parallel pathway that functions redundantly with VIB-1 (Figure 9) to regulate HI and protease production. In this scenario, protease production and cell death in a Δvib-1 Δime-2 strain is restored. Mutation of components in the parallel cell-death induction pathway in addition to vib-1 mutations would ameliorate cell death and HI completely, regardless of the presence or absence of ime-2. Further experiments will be needed to identify IME-2 targets and additional members of this pathway.
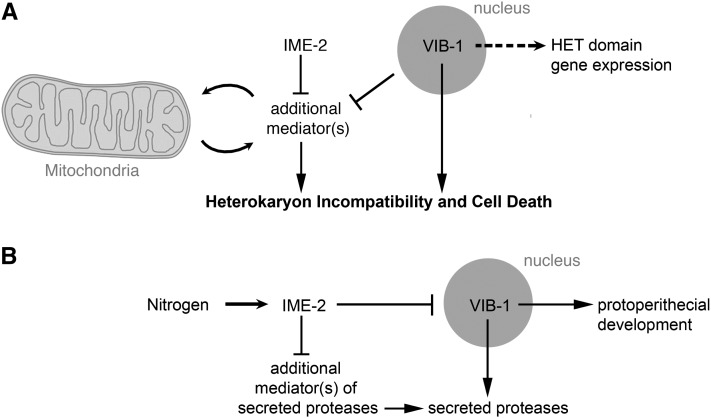
Figure 9 .
Model for the ime-2 and vib-1 genetic pathway. (A) VIB-1 positively regulates HI and cell death, as well as HET domain gene expression (via a separate mechanism). IME-2 negatively regulates additional HI cell death mediators that function in parallel to the VIB-1 pathway. Due to the role of IME-2 in the processing of a mitochondrial protein, we reasoned that the alternate cell-death effectors and/or pathway could be acting through the mitochondria. Further, since deletions in ime-2 do not exhibit increased cell death during HI, we suggest that VIB-1 negatively regulates these alternate death effectors. (B) VIB-1 also positively regulates secreted proteases and, similar to the mechanism for HI cell-death regulation, IME-2 negatively regulates a parallel pathway for secreted proteases. Deletions in ime-2 cause a significant increase in secreted proteases, and thus in this pathway, VIB-1 does not regulate the parallel pathway for secreted proteases. Further, IME-2 appears to negatively regulate VIB-1 with respect to protoperithecial development via a separate pathway. For both pathways, it is likely that IME-2 is regulated by cellular signals of nitrogen availability.
Loss-of-function mutations in vib-1 result in an inability to secrete proteases in response to nitrogen starvation and presence of extracellular protein, a phenotype that is suppressed in Δime-2 Δvib-1 mutants. In eukaryotes, including S. cerevisiae, caspase proteases (metacaspases in S. cerevisiae) are integral in the activation of the apoptotic cell-death cascade (Madeo et al. 2004; Tait and Green 2010; Abdelwahid et al. 2011). In N. crassa, metacaspases are not required for HI-induced cell death (Hutchison et al. 2009). However, it is possible that a link between HI-induced cell death and VIB-1/IME-2–regulated proteases occurs in N. crassa by the utilization of nonmetacaspase proteases to induce cell death.
Mitochondria are key players in apoptosis and cell-death pathways (Tait and Green 2010). During apoptotic cell death, the pro-apoptotic Bcl-2 proteins BAX and BAK can cause the mitochondrial outer membrane to permeabilize, disrupting mitochondrial function, energy production, and redox potential and promoting the release of pro-apoptotic factors such as cytochrome c (Degterev and Yuan 2008; Tait and Green 2010; Abdelwahid et al. 2011). In fungi, mitochondria have also been implicated in apoptotic cell death and have important roles in life span and senescence (Madeo et al. 2004; Maheshwari and Navaraj 2008; Sharon et al. 2009). Mutations in ime-2 affected localization and post-transcriptional processing of ARG-4, a phenotype that was restored to a wild-type pattern in the Δvib-1 Δime-2 mutant. The observation that IME-2 affects post-translational processing of a mitochondrial matrix protein suggests that the parallel cell-death pathway could be acting through the mitochondria or a mitochondria-related pathway. S. cerevisiae IME2 and its homolog in Schizosaccharomyces pombe, mde3/pit1, have a role in meiosis (Abe and Shimoda 2000; Kassir et al. 2003; Honigberg 2004) as well as in pseudohyphal growth (Strudwick et al. 2010). Both of these developmental processes are associated with nitrogen starvation, suggesting an additional role for Ime2 in nutrient sensing, similar to that proposed for N. crassa (Hutchison and Glass 2010). It will be of interest to assess whether ime2Δ mutants in S. cerevisiae also have in common a defect in the localization and processing of Arg7 (ortholog of N. crassa arg-4), as observed in N. crassa Δime-2 mutants. Further experiments on the relationship between ime-2/IME2 and mitochondrial function are warranted.
We identified five phosphorylation sites on VIB-1, including a site that matches the predicted consensus site for Ime2. However, mutations in the Ime2 consensus site in VIB-1 that are predicted to result in phospho-null mutations (S to A) or activating mutations (S to D) resulted in no to only subtle phenotypic differences from WT. In particular, no role for the Ime2 phosphorylation site was observable for HI. Strains containing vib-1 alleles containing five S-to-A or five S-to-D mutations showed more severe defects, particularly reduced vegetative growth and protoperithecial development, as well as a reduced percentage of cell death during HI. These observations suggest that altering these five sites results in a VIB-1 protein that is not fully functional (hypomorphic allele). Our genetic and phenotypic data show a genetic interaction between ime-2 and vib-1 during protoperithecial development and during HI and protease secretion in response to nitrogen starvation and for the localization and processing of a mitochondrial matrix protein. These observations suggest a complex regulatory interaction between these two proteins in a number of cellular functions. Further experiments will unravel the interconnection of these two proteins and the signaling and transcriptional regulatory pathways they regulate in different cellular contexts.
Supplementary Material
Acknowledgments
We thank the Glass laboratory for helpful comments and discussions. Additionally, we thank Abby Leeder for help with confocal microscopy and for sharing unpublished results on VIB-1 phosphorylation and Jianping Sun for her expertise with protein and mass spectrometry experiments. We also thank Lori Kohlstaedt at the University of California (Berkeley) Mass Spectrometry facility. Finally, we thank Liam Holt for helpful discussions regarding Ime2, yeast meiosis, and phosphorylation site prediction methods. This work was supported by a National Institutes of Health (NIH) grant to N.L.G. (GM60468). We acknowledge use of materials generated by NIH grant P01 GM068087, “Functional analysis of a model filamentous fungus.”
Footnotes
Communicating editor: A. P. Mitchell
Literature Cited
- Aanen D. K., Debets A. J., Glass N. L., Saupe S. J., 2010. Biology and genetics of vegetative incompability in fungi, pp. 274–288 in Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. ASM Press, Washington, DC. [Google Scholar]
- Abadjieva A., Hilven P., Pauwels K., Crabeel M., 2000. The yeast ARG7 gene product is autoproteolyzed to two subunit peptides, yielding active ornithine acetyltransferase. J. Biol. Chem. 275: 11361–11367 [DOI] [PubMed] [Google Scholar]
- Abdelwahid E., Rolland S., Teng X., Conradt B., Hardwick J. M., et al. , 2011. Mitochondrial involvement in cell death of non-mammalian eukaryotes. Biochim. Biophys. Acta 1813: 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H., Shimoda C., 2000. Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4 and a genome-wide search for its target genes. Genetics 154: 1497–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N. T., Bungard D., Shin M. E., Moore M., Winter E., 2009. The Ime2 protein kinase enhances the disassociation of the Sum1 repressor from middle meiotic promoters. Mol. Cell. Biol. 29: 4352–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias G. D., He Y., Gordon J., Mossialos D., Oliver S. G., et al. , 2010. Posttranslational regulation impacts the fate of duplicated genes. Proc. Natl. Acad. Sci. USA 107: 2967–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin K. R., Zhang C., Shokat K. M., Herskowitz I., 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17: 1524–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya R. P., Remenyi A., Yeh B. J., Lim W. A., 2006. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 75: 655–680 [DOI] [PubMed] [Google Scholar]
- Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., et al. , 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68: 1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman B. J., Draskovic M., Freitag M., Bowman E. J., 2009. Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa. Eukaryot. Cell 8: 1845–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush G. S., Najor N. A., Dombkowski A. A., Cukovic D., Sawarynski K. E., 2012. Yeast IME2 functions early in meiosis upstream of cell cycle-regulated SBF and MBF targets. PLoS ONE 7: e31575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G. H., Dawe A. L., Churbanov A., Smith M. L., Milgroom M. G., et al. , 2012. Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 190: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S., Herskowitz I., 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1: 685–696 [DOI] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., et al. , 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705 [DOI] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M., Abadjieva A., Hilven P., Desimpelaere J., Soetens O., 1997. Characterization of the Saccharomyces cerevisiae ARG7 gene encoding ornithine acetyltransferase, an enzyme also endowed with acetylglutamate synthase activity. Eur. J. Biochem. 250: 232–241 [DOI] [PubMed] [Google Scholar]
- Cybis J., Davis R. H., 1975. Organization and control in the arginine biosynthetic pathway of Neurospora. J. Bacteriol. 123: 196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A., Yuan J., 2008. Expansion and evolution of cell death programmes. Nat. Rev. Mol. Cell Biol. 9: 378–390 [DOI] [PubMed] [Google Scholar]
- Dementhon K., Iyer G., Glass N. L., 2006. VIB-1 is required for expression of genes necessary for programmed cell death in Neurospora crassa. Eukaryot. Cell 5: 2161–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh K., Anamika K., Srinivasan N., 2010. Evolution of domain combinations in protein kinases and its implications for functional diversity. Prog. Biophys. Mol. Biol. 102: 1–15 [DOI] [PubMed] [Google Scholar]
- Espagne E., Balhadere P., Penin M. L., Barreau C., Turcq B., 2002. HET-E and HET-D belong to a new subfamily of WD40 proteins involved in vegetative incompatibility specificity in the fungus Podospora anserina. Genetics 161: 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner A., Simonin A. R., Glass N. L., 2008. Cell fusion in the filamentous fungus, Neurospora crassa. Methods Mol. Biol. 475: 21–38 [DOI] [PubMed] [Google Scholar]
- Glass N. L., Dementhon K., 2006. Non-self recognition and programmed cell death in filamentous fungi. Curr. Opin. Microbiol. 9: 553–558 [DOI] [PubMed] [Google Scholar]
- Glass N. L., Kaneko I., 2003. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot. Cell 2: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N. L., Grotelueschen J., Metzenberg R. L., 1990. Neurospora crassa A mating-type region. Proc. Natl. Acad. Sci. USA 87: 4912–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C., Welch J., Kowbel D. J., Glass N. L., 2010. Evolution and diversity of a fungal self/nonself recognition locus. PLoS ONE 5: e14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey P. C., Swift S. R., Roca M. G., Read N. D., 2004. Live-cell imaging of filamentous fungi using vital flourescent dyes and confocal microscopy. Methods Microbiol. 34: 63–87 [Google Scholar]
- Hirsh H. M., 1954. Environmental factors influencing the differentiation of protoperithecia and their relation to tyrosinase and melanin formation in Neurospora crassa. Physiol. Plant. 7: 72–92 [Google Scholar]
- Holt L. J., Hutti J. E., Cantley L. C., Morgan D. O., 2007. Evolution of Ime2 phosphorylation sites on Cdk1 substrates provides a mechanism to limit the effects of the phosphatase Cdc14 in meiosis. Mol. Cell 25: 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg S. M., 2004. Ime2p and Cdc28p: co-pilots driving meiotic development. J. Cell. Biochem. 92: 1025–1033 [DOI] [PubMed] [Google Scholar]
- Honigberg S. M., Purnapatre K., 2003. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J. Cell Sci. 116: 2137–2147 [DOI] [PubMed] [Google Scholar]
- Hutchison E., Brown S., Tian C., Glass N. L., 2009. Transcriptional profiling and functional analysis of heterokaryon incompatibility in Neurospora crassa reveals that reactive oxygen species, but not metacaspases, are associated with programmed cell death. Microbiology 155: 3957–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison E. A., Glass N. L., 2010. Meiotic regulators Ndt80 and Ime2 have different roles in Saccharomyces and Neurospora. Genetics 185: 1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S., 2011. The Ime2 protein kinase family in fungi: more duties than just meiosis. Mol. Microbiol. 80: 1–13 [DOI] [PubMed] [Google Scholar]
- Kaneko I., Dementhon K., Xiang Q., Glass N. L., 2006. Nonallelic interactions between het-c and a polymorphic locus, pin-c, are essential for nonself recognition and programmed cell death in Neurospora crassa. Genetics 172: 1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y., Adir N., Boger-Nadjar E., Raviv N. G., Rubin-Bejerano I., et al. , 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224: 111–171 [DOI] [PubMed] [Google Scholar]
- Katz M. E., Gray K. A., Cheetham B. F., 2006. The Aspergillus nidulans xprG (phoG) gene encodes a putative transcriptional activator involved in the response to nutrient limitation. Fungal Genet. Biol. 43: 190–199 [DOI] [PubMed] [Google Scholar]
- Keeping A., Deabreu D., Dibernardo M., Collins R. A., 2010. Gel-based mass spectrometric and computational approaches to the mitochondrial proteome of Neurospora. Fungal Genet. Biol. 48: 526–536 [DOI] [PubMed] [Google Scholar]
- Kosti I., Mandel-Gutfreund Y., Glaser F., Horwitz B. A., 2010. Comparative analysis of fungal protein kinases and associated domains. BMC Genomics 11: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe G. O., Free S. J., 1998. The isolation and characterization of nrc-1 and nrc-2, two genes encoding protein kinases that control growth and development in Neurospora crassa. Genetics 149: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D. L., Smith M. L., 2012. Diverse interactions mediate asymmetric incompatibility by the het-6 supergene complex in Neurospora crassa. Fungal Genet. Biol. 49: 65–73 [DOI] [PubMed] [Google Scholar]
- Macho A., Decaudin D., Castedo M., Hirsh T., Susin S. A., et al. , 1996. Chloromethyl-X-Rosamine is an aldehyde-fixable potential-sensitive fluorochrome for the detection of early apoptosis. Cytometry 25: 333–340 [DOI] [PubMed] [Google Scholar]
- Madeo F., Herker E., Wissing S., Jungwirth H., Eisenberg T., et al. , 2004. Apoptosis in yeast. Curr. Opin. Microbiol. 7: 655–660 [DOI] [PubMed] [Google Scholar]
- Madeo F., Carmona-Gutierrez D., Ring J., Buettner S., Eisenberg T., et al. , 2009. Caspase-dependent and caspase-independent cell death pathways in yeast. Biochem. Biophys. Res. Commun. 382: 227–231 [DOI] [PubMed] [Google Scholar]
- Maheshwari R., Navaraj A., 2008. Senescence in fungi: the view from Neurospora. FEMS Microbiol. Lett. 280: 135–143 [DOI] [PubMed] [Google Scholar]
- Margolin B. S., Freitag M., Selker E. U., 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44: 34–36 [Google Scholar]
- McCluskey K., 2003. The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52: 246–262 [DOI] [PubMed] [Google Scholar]
- Micali C. O., Smith M. L., 2006. A nonself recognition gene complex in Neurospora crassa. Genetics 173: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M., Shin M. E., Bruning A., Schindler K., Vershon A., et al. , 2007. Arg-Pro-X-Ser/Thr is a consensus phosphoacceptor sequence for the meiosis-specific Ime2 protein kinase in Saccharomyces cerevisiae. Biochemistry 46: 271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses A. M., Landry C. R., 2010. Moving from transcriptional to phospho-evolution: generalizing regulatory evolution? Trends Genet. 26: 462–467 [DOI] [PubMed] [Google Scholar]
- Newmeyer D., 1970. A suppressor of the heterokaryon-incompatibility associated with mating type in Neurospora crassa. Can. J. Genet. Cytol. 12: 914–926 [DOI] [PubMed] [Google Scholar]
- Obenauer J. C., Cantley L. C., Yaffe M. B., 2003. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31: 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Segall J., 2002a. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 6417–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Segall J., 2002b. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 6430–6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M., Clave C., 2007. The fungus-specific HET domain mediates programmed cell death in Podospora anserina. Eukaryot. Cell 6: 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D., 1988. Main features of vegetative incompatibility in Neurospora crassa. Fungal Genet. Newsl. 35: 44–46 [Google Scholar]
- Pittenger T. H., 1957. The mating type alleles and heterokaryon formation in Neurospora crassa. Microbiol. Genet. Bull. 15: 21–22 [Google Scholar]
- Poot M., Zhang Y. Z., Kramer J. A., Wells K. S., Jones L. J., et al. , 1996. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J. Histochem. Cytochem. 44: 1363–1372 [DOI] [PubMed] [Google Scholar]
- Read N. D., Fleissner A., Roca M. G., Glass N. L., 2010. Hyphal fusion, pp. 260–273 in Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. ASM Press, Washington, DC. [Google Scholar]
- Radu M., Chernoff J., 2009. The DeMSTification of mammalian Ste20 kinases. Curr. Biol. 19: R421–R425 [DOI] [PubMed] [Google Scholar]
- Ruepp A., Zollner A., Maier D., Albermann K., Hani J., et al. , 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32: 5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Iyer G., Wu J., Glass N. L., 2002. Nonself recognition is mediated by HET-C heterocomplex formation during vegetative incompatibility. EMBO J. 21: 4841–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser T. J., Lauter F. R., Russo V. E., Yanofsky C., 1990. Cloning, sequence, and photoregulation of al-1, a carotenoid biosynthetic gene of Neurospora crassa. Mol. Cell. Biol. 10: 5064–5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon A., Finkelstein A., Shlezinger N., Hatam I., 2009. Fungal apoptosis: function, genes and gene function. FEMS Microbiol. Rev. 33: 833–854 [DOI] [PubMed] [Google Scholar]
- Shin M. E., Skokotas A., Winter E., 2010. The Cdk1 and Ime2 protein kinases trigger exit from meiotic prophase in Saccharomyces cerevisiae by inhibiting the Sum1 transcriptional repressor. Mol. Cell. Biol. 30: 2996–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu P. K., Glass N. L., 1999. Molecular characterization of tol, a mediator of mating-type-associated vegetative incompatibility in Neurospora crassa. Genetics 151: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubassi G., Luca N., Pak J., Segall J., 2003. Activity of phosphoforms and truncated versions of Ndt80, a checkpoint-regulated sporulation-specific transcription factor of Saccharomyces cerevisiae. Mol. Genet. Genomics 270: 324–336 [DOI] [PubMed] [Google Scholar]
- Smith H. E., Mitchell A. P., 1989. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 2142–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Raithatha S., Stuart D., 2002. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 22: 7024–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strudwick N., Brown M., Parmar V. M., Schroder M., 2010. Ime1 and Ime2 are required for pseudohyphal growth of Saccharomyces cerevisiae on nonfermentable carbon sources. Mol. Cell. Biol. 30: 5514–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait S. W., Green D. R., 2010. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11: 621–632 [DOI] [PubMed] [Google Scholar]
- Tian C., Kasuga T., Sachs M. S., Glass N. L., 2007. Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot. Cell 6: 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend J. P., Hartl D. L., 2002. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 3: RESEARCH0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H. J., 1956. A convenient growth medium for Neurospora. Microbiol. Genet. Bull. 13: 42–46 [Google Scholar]
- Westergaard M., Mitchell H. K., 1947. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573–577 [Google Scholar]
- Winter E., 2012. The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 76: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q., Glass N. L., 2002. Identification of vib-1, a locus involved in vegetative incompatibility mediated by het-c in Neurospora crassa. Genetics 162: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q., Glass N. L., 2004. The control of mating type heterokaryon incompatibility by vib-1, a locus involved in het-c heterokaryon incompatibility in Neurospora crassa. Fungal Genet. Biol. 41: 1063. [DOI] [PubMed] [Google Scholar]
- Xu L., Ajimura M., Padmore R., Klein C., Kleckner N., 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6572–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Townsend J. P., 2010. The filamentous fungal gene expression database (FFGED). Fungal Genet. Biol. 47: 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.