Abstract
Phenotypes relevant to oxidative phosphorylation (OXPHOS) in eukaryotes are jointly determined by nuclear and mitochondrial DNA (mtDNA). Thus, in humans, the variable clinical presentations of mitochondrial disease patients bearing the same primary mutation, whether in nuclear or mitochondrial DNA, have been attributed to putative genetic determinants carried in the “other” genome, though their identity and the molecular mechanism(s) by which they might act remain elusive. Here we demonstrate cytoplasmic suppression of the mitochondrial disease-like phenotype of the Drosophila melanogaster nuclear mutant tko25t, which includes developmental delay, seizure sensitivity, and defective male courtship. The tko25t strain carries a mutation in a mitoribosomal protein gene, causing OXPHOS deficiency due to defective intramitochondrial protein synthesis. Phenotypic suppression was associated with increased mtDNA copy number and increased mitochondrial biogenesis, as measured by the expression levels of porin voltage dependent anion channel and Spargel (PGC1α). Ubiquitous overexpression of Spargel in tko25t flies phenocopied the suppressor, identifying it as a key mechanistic target thereof. Suppressor-strain mtDNAs differed from related nonsuppressor strain mtDNAs by several coding-region polymorphisms and by length and sequence variation in the noncoding region (NCR), in which the origin of mtDNA replication is located. Cytoplasm from four of five originally Wolbachia-infected strains showed the same suppressor effect, whereas that from neither of two uninfected strains did so, suggesting that the stress of chronic Wolbachia infection may provide evolutionary selection for improved mitochondrial fitness under metabolic stress. Our findings provide a paradigm for understanding the role of mtDNA genotype in human disease.
Keywords: mitochondrial DNA, cybrid, mitochondrial biogenesis, Wolbachia, bang sensitivity
THE oxidative phosphorylation (OXPHOS) system depends upon the cooperative expression of genes in nuclear and mitochondrial DNA (mtDNA). In most metazoans, including humans and Drosophila, the mitochondrial genome encodes 13 essential polypeptides of the OXPHOS complexes of the inner mitochondrial membrane, plus the two rRNAs and 22 tRNAs required for their synthesis inside mitochondria. The remaining polypeptides of the OXPHOS complexes, as well as all of the proteins required for mtDNA maintenance and expression, are nuclear coded.
Mitochondrial disease in humans takes many forms ranging from rare neurological syndromes to cases of common multifactorial disorders (Greaves et al. 2012; Ylikallio and Suomalainen 2012). Pathological OXPHOS mutations can be in nuclear or mitochondrial genes, and these have also been hypothesized to interact to modify disease phenotype (Brown et al. 1997, 2002; Hudson et al. 2007). Variants of mtDNA have also been postulated to be under evolutionary selection, in a complex interaction with the nuclear genome and environmental factors (Willett and Burton 2004; Wallace 2005; Meiklejohn et al. 2007). Although direct evidence for a modifier effect of mtDNA in disease is lacking, naturally occurring variants have been experimentally shown to have effects on OXPHOS biochemistry (Katewa and Ballard 2007), reactive oxygen species (ROS) production (Moreno-Loshuertos et al. 2006), and other aspects of cellular (Kazuno et al. 2006) and organismal physiology (Ballard et al. 2007a; Pravenec et al. 2007).
Drosophila provides an ideal model system in which the effects of mtDNA variation on OXPHOS disease can be directly tested. The tko25t strain, bearing a point mutation in the gene encoding mitoribosomal protein S12 (Toivonen et al. 2001), shows an organismal phenotype of developmental delay, bang sensitivity, impaired sound response, and defective male courtship, associated with severe deficiency of the four OXPHOS complexes (I, III, IV, and V) containing mitochondrial translation products (Shah et al. 1997). The primary effect of the mutation appears to be on the stability or assembly of the mitoribosomal small subunit, based on measurements of rRNA levels (Toivonen et al. 2001, 2003). We previously determined that the mitochondrial disease-like phenotype of tko25t is partially suppressible by duplication of the tko locus (Kemppainen et al. 2009), although mtDNA effects were not analyzed in earlier studies.
In Drosophila, the frequent presence of the endosymbiont Wolbachia, one of the closest extant relatives of the mitochondrial ancestor, provides an additional dimension to nuclear–cytoplasmic genetic interactions, especially since its presence is correlated with mtDNA copy-number variation (Ballard and Melvin 2007). Wolbachia has elsewhere been reported to influence susceptibility to viral pathogens Hedges et al. 2008; Teixeira et al. 2008), manipulate lifespan (Fry and Rand 2002; Reynolds et al. 2003; Toivonen et al. 2007), and increase insulin signaling (Ikeya et al. 2009). We therefore set out to determine whether Wolbachia-containing cytoplasm could modify the mitochondrial phenotype of tko25t and whether any such effect depended on the actual presence of Wolbachia, on coinherited mtDNA variants, or the combination.
Materials and Methods
Drosophila stocks and maintenance
tko25t flies in the original strain background were maintained at room temperature as balanced stock (FM7 balancer), as previously shown (Toivonen et al. 2001). Introgression into other cytoplasmic backgrounds was performed essentially as shown in Supporting Information, Figure S1B, with generally five or more generations of backcrossing before balanced stocks were reconstituted, which were then used to generate tko25t homozygotes and hemizygotes for individual experiments. sesB1 flies, also in the original Oregon R background (Homyk and Sheppard 1977; Zhang et al. 1999), obtained from M. Ashburner, University of Cambridge, Cambridge, UK, were balanced, maintained, and manipulated similarly. The UAS-Spargel (UAS-Srl) line, kindly supplied by C. Frei, ETH, Zurich, contains an insertion on chromosome 2 of the coding region of the Spargel gene under upstream activation sequence (UAS) promoter control (Tiefenböck et al. 2010), balanced with CyO. Wild-type strains BER1, KSA2, Oregon R-C, Reids-1, CO3, QI2, BS1, and M2 were obtained from the Bloomington Stock Center. Standard lab strains Canton-S and Oregon R, and the da-GAL4 driver strain (Wodarz et al. 1995) have been maintained in our own laboratories for >10 yr. Prior to testing for Wolbachia infection, or when flies were found to be infected, flies were maintained in quarantine from all other stocks. Infected flies were cured of Wolbachia infection by growth for two generations at room temperature on medium containing 30 μg/ml tetracycline (Dobson et al. 2002) and then retested (Figure S1A). Flies were maintained in standard medium with supplements as previously shown (Toivonen et al. 2001) and reared for experiments at 25° except where stated.
Developmental time
Groups of eight virgin females and five males of a given strain or genotype were mated 5 days after eclosion for 3 days, being tipped to a fresh vial each day and then discarded. Each experiment used batches of five identical vials, and each was repeated, to generate means ± SD of the eclosion day as shown in the figures. Wild-type (Oregon R) flies were included as a control in every experiment.
Behavioral and lifespan assays
For behavioral analysis, flies were collected under CO2 anesthesia within 2 days of eclosion and then kept for up to a further 5 days in vials with 15 flies of the same sex per vial, and transferred to fresh food vials every 2 days. Bang sensitivity and courtship were measured at room temperature, as previously shown (Toivonen et al. 2001). For bang sensitivity, 20–25 individual flies of each sex and genotype to be tested were used in each experiment. For courtship, at least 15 mating pairs of the genotypes to be tested were used in each experiment, which was conducted in triplicate. Courtship was scored as the proportion of females that mated within 1 hr. Lifespan was measured as previously shown (Sanz et al. 2010a,b), using groups of 150–300 flies of each sex, collected within 24 hr of eclosion, and kept at a density of 20 flies per vial at 25°C in a controlled 12 hr light–dark cycle. Vials were changed every 2–3 days and the number of dead flies was counted. Each independent experiment was repeated twice: data were pooled and analyzed together.
DNA extraction and quantitation
For mtDNA copy-number analysis, total DNA was prepared from batches of 10–30 adult flies, homogenized in 500 μl ice-cold homogenization medium (75 mM NaCl, 25 mM EDTA pH 8.0) in a 1.5-ml Eppendorf tube, using a motor-driven polypropylene pestle (VWR). Following addition of 70 μl 10% SDS, 200 μg proteinase K (Fermentas), and 100 μg DNase-free, proteinase-free RNase A (Fermentas) homogenates were incubated overnight at 37° with gentle shaking, followed by two extractions with phenol-chloroform-isoamyl alcohol (25:24:1) for 1 hr with gentle shaking. Extracts were centrifuged at room temperature for 15 min at 5000 gmax. DNA was recovered from the final aqueous phase by ethanol precipitation and resuspended in 200 μl TE buffer (pH 8.0). Relative mtDNA copy number was measured by real-time qPCR using primers for mitochondrial large subunit (LSU) rRNA and nuclear 18S rRNA (Table S2), in a StepOnePlus instrument (Applied Biosystems) using Fast SYBR Green Master Mix (Applied Biosystems) under the manufacturer’s recommended conditions, with 20 sec of enzyme activation at 95°, followed by 40 cycles of 95° for 3 sec and 60° for 30 sec. For mtDNA sequencing and noncoding A + T region (NCR) repeat analysis, mitochondria were prepared from batches of 50–200 flies, essentially as described by Miwa et al. (2003) (see Supporting Information for full details). The mitochondrial pellet was resuspended in 500 μl ice-cold homogenization medium and processed for DNA isolation as above.
RNA extraction and quantitation
RNA was isolated as previously shown (Fernández-Ayala et al. 2009; Sanz et al. 2010a,b) from three biological replicate samples of each sex and genotype to be tested. Expression of mitochondrial small subunit (SSU) and LSU rRNAs and of various nuclear gene transcripts was measured relative to that of the housekeeping gene RpL32 by qRT–PCR using customized primers (see Table S2 for details), as previously shown (Fernández-Ayala et al. 2009; Kemppainen et al. 2009; Sanz et al. 2010a,b), together with the SYBR Green Master Mix (Applied Biosystems). Following an initial enzyme activation at 95° for 20 sec, 40 reaction cycles of denaturation for 3 sec at 95° followed by annealing and extension for 30 sec at 60° were carried out, using the StepOne real-time PCR instrument (Applied Biosystems).
Protein analysis by Western blotting
Batches of 15–20 adult (2–5 days old) flies were placed in a 1.5-ml Eppendorf tube with 100 μl PBS. An equal volume of 2× SDS–PAGE sample buffer (100 mm Tris-HCl, 4% SDS, 0.2% bromophenol blue, 20% glycerol, 200 mm β-mercaptoethanol, pH 6.8) was added. Samples were homogenized using a pellet pestle (VWR), heated to 100° for 5 min, centrifuged for 5 min at 12,000 gmax, and loaded on Criterion precast Tris-HCl 10–20% polyacrylamide gradient gel (Bio-Rad). Electrophoresis, blotting, and detection were essentially as described previously (Fernández-Ayala et al. 2009) (see Supporting Information for full details), except for the antibodies: primary (from Mitosciences: Porin 1:5000, ATP synthase subunit α, 1:100,000, and complex I subunit NDFUS3, 1:10,000; from Santa Cruz Biotechnology; α-actinin, 1:10,000), secondary (Vector Laboratories, peroxidase linked) antimouse IgG (H + L), or for α-actinin detection, antirabbit IgG (1:10,000). The amount of each protein was measured by densitometry with Image Lab 3.0.1 software (Bio-Rad) and normalized first to the α-actinin signal, then to the value for Oregon R wild-type flies of the given sex.
Polarography, ATP assay, and mitochondrial H2O2 production
Mitochondria were isolated and used to measure substrate oxidation rates at state 3 by polarography as previously described (Fernández-Ayala et al. 2009), using a substrate mix of 20 mM sn-glycerol 3-phosphate, 5 mM sodium pyruvate, and 5 mM proline. Mitochondrial ROS production at state 4 was assayed as previously described (Sanz et al. 2010a,b) by measuring Mitosox fluorescence in the absence of added ADP. ATP was assayed on batches of 15 adult females/genotype, collected at room temperature, as described previously (Fergestad et al. 2006), using a Harta MicrolumXS microplate luminometer with Lumiterm II acquisition software (Harta Instruments, Gaithersburg, MD) and ATP determination kit A-22066 (Invitrogen). Sample values were corrected for protein content using DC protein assays (Bio-Rad Laboratories) and ATP concentrations were normalized as a percentage of wild-type control samples.
Long PCR and mtDNA sequencing
The whole mitochondrial genome was amplified in four overlapping fragments (each ∼5.5 kb in size). Three of these, representing the coding region, were sequenced using primer sets generating overlapping reads on both strands (Table S2). The NCR and its boundaries were sequenced by a strategy combining long PCR and cloning of restriction fragments (for full details see Supporting Information). Sequence analysis used the Phred/Phrap/Consed package (Gordon et al. 1998).
Statistical analysis
Data were analyzed by ANOVA or t-test, as appropriate; see figure legends for details.
Results
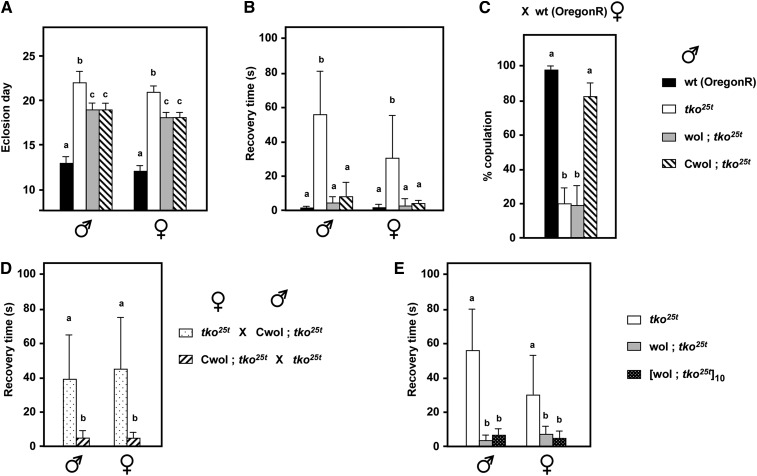
PCR using Wolbachia-specific 16S rRNA gene primers was first used to test for the presence of the endosymbiont in a set of wild-type strains collected from diverse locations and maintained at Bloomington, as well as our own stocks of tko25t. While tko25t was confirmed to be Wolbachia-free (Fernández-Ayala et al. 2010), strains BER1, Oregon R-C, CO3, Reids-1, and QI2 were found to be infected (Figure S1A). One of these stocks, BER1, was selected for detailed studies. By crossing infected BER1 females with tko25t males, using the scheme shown in Figure S1B, we introgressed tko25t into the BER1 cytoplasm (denoted wol; tko25t). After confirming the presence of Wolbachia (Figure S1C, O’Neill et al. 1992), we tested the phenotype of the wol; tko25t flies (Figure 1), finding a partial rescue of developmental delay (Figure 1A) and an almost complete suppression of bang sensitivity (Figure 1B). To test whether the presence of Wolbachia was the cause of this suppression, we treated wol; tko25t flies with tetracycline over two generations, confirmed the elimination of the endosymbiont in the “cured” (Cwol; tko25t) flies (Figure S1D), and then retested for phenotype. Surprisingly, we found an identical degree of suppression in the cured (Cwol; tko25t) as in the infected wol; tko25t flies (Figure 1, A and B), indicating that Wolbachia was not the suppressing determinant of the BER1 cytoplasm.
Figure 1 .
BER1 mtDNA confers partial suppression of the tko25t phenotype. (A) Time to eclosion at room temperature and (B) bang sensitivity (recovery time from mechanical shock) of flies of the sex and genotypes indicated (means ± SD). wol, Wolbachia-infected BER1 cytoplasmic background; Cwol, BER1 cytoplasmic background after Wolbachia removal by tetracycline treatment, in each case crossed into tko25t as shown in Figure S1B. (C) Frequency of successful copulation between wild-type (Oregon R) females and males of the indicated genotypes. (D) Bang sensitivity of progeny from reciprocal cross between tko25t flies in the original and Wolbachia-cured (BER1) backgrounds. (E) Bang sensitivity of tko25t flies in the Wolbachia-infected (BER1) background, before and after backcrossing for 10 generations to tko25t males, alongside bang sensitivity of tko25t flies in the original background, reproduced from A. A, B, and C denote significantly different data classes in each experiment (Newman–Keuls test, P < 0.05, based on ANOVA, P < 0.001, each sex considered separately).
To test the inheritance pattern of the suppressing determinant, we crossed Cwol; tko25t females with tko25t males from our original stock and vice versa (Figure S1E). The suppression of bang sensitivity was transmitted in a strictly maternal fashion (Figure 1D), indicating that it must reside in the mtDNA itself, or else in an unknown, maternally transmitted genetic element. To exclude any effects of nuclear genes, we backcrossed Cwol; tko25t females to tko25t males from the original strain in the Oregon R background over 10 generations, using the scheme of Figure S1F, and then retested the phenotype. The suppression of bang sensitivity was fully retained in the backcrossed flies (Figure 1E). The cured BER1 background also rescued the male courtship defect of tko25t, although the corresponding Wolbachia-infected strain prior to curing was still defective for male courtship (Figure 1C). The original Wolbachia-infected wild-type strain was itself male-courtship defective, when tested in this assay with Oregon R females (giving a copulation frequency of only 57%). Finally, the BER1-derived mtDNA slightly increased the lifespan of tko25t males, but not females (Figure S1G), after homogenization of the nuclear background by backcrossing.
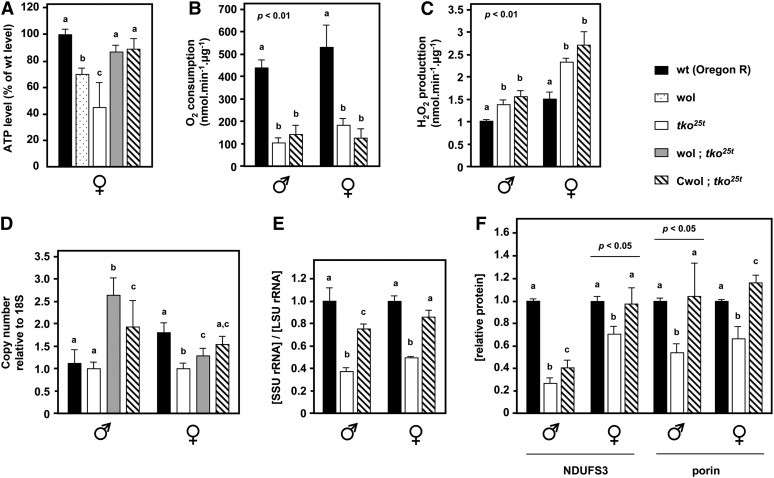
To gain insight into the mechanism of suppression, we assayed total ATP levels in homogenates of adult flies of the different strains and measured mtDNA copy number by qPCR. Both parameters showed a correlation with phenotypic suppression. The total ATP level in the original tko25t strain was significantly lower than in the wild-type Oregon R reference strain, but was restored to almost the wild-type level in wol; tko25t or Cwol; tko25t flies (Figure 2A). Normalized to mitochondrial protein concentration, respiration by isolated mitochondria was much lower in tko25t adults than in controls (Figure 2B), while mitochondrial ROS production was increased (Figure 2C). Neither of these parameters was significantly altered by the BER1 mitochondrial background (Cwol; tko25t). We conclude that BER1 mtDNA does not alter the biochemical phenotype as such. Therefore, we next tested whether it might act simply by increasing the amount of mtDNA and of mitochondria.
Figure 2 .
BER1 mtDNA influences ATP, mtDNA, mtRNA, and protein levels in tko25t flies. (A) ATP levels in whole flies (mean ± SD), plus (B) oxygen consumption, and (C) ROS production of isolated mitochondria from flies of the indicated sex and genotypes, mean ± SE, three or more biological replicates. (D) Copy number of mtDNA in flies of the genotypes indicated, relative to 18S rDNA, means ± SD, normalized to that in tko25t flies of the same sex. (E) Ratio of mitoribosomal SSU to LSU RNA levels in flies of the indicated genotypes, means ± SD wol; tko25t and Cwol; tko25t flies, maintained as balanced stocks, were backcrossed over five or more generations to tko25t in the Oregon R background and rendered homozygous prior to the experiment. (F) Levels of a representative subunit of complex I (NDUFS3) and of a global mitochondrial protein marker (porin) in flies of the sex and genotype indicated, based on densitometry of Western blots as shown in Figure S2E, normalized against a cytosolic loading control (α-actinin). A, B, and C denote significantly different data classes (Newman–Keuls test, P < 0.05, based on ANOVA, P < 0.001 except where indicated, each sex and, where appropriate, each gene considered separately).
The copy number of mtDNA was significantly higher in wol; tko25t or Cwol; tko25t than in tko25t in the original (Oregon R) background, with the differences more pronounced in males (Figure 2D). There was also a substantial recovery in the ratio of small subunit to large subunit mitoribosomal RNA (Figure 2E, Figure S2D). Furthermore, as measured by the levels of the outer mitochondrial membrane marker protein porin and of the nuclear-coded complex I subunit NDUFS3 (Figure 2F, Figure S2E), relative to a cytosolic marker, there was a significant increase in mitochondrial protein from the depressed levels seen in the original tko25t line, although the increase in some other mitochondrial proteins, such as ATP synthase subunit α, was much more modest (Figure S2F), though still significant. Importantly, mtDNA copy number was not elevated in otherwise wild-type flies carrying the BER1 cytoplasm (Figure S2C).
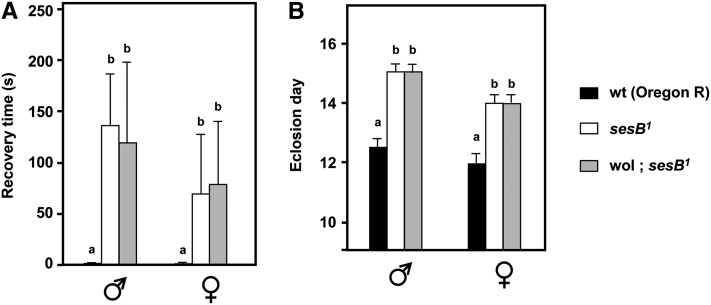
Mitochondrially inherited suppression of a mitochondrial protein synthesis defect by a mechanism that generates additional capacity for ATP synthesis logically should not affect mutants with defects in ATP supply downstream of OXPHOS, such as sesB1 (encoding the major isoform of the adenine nucleotide translocase, Zhang et al. 1999). To test this, we crossed the Wolbachia-containing cytoplasm from BER1 into sesB1, using the same strategy as for tko25t (Figure S1D), and then tested the phenotype. In this case there was no rescue of bang sensitivity (Figure 3A), developmental delay (Figure 3B), or female sterility. There was, however, a modest rescue of the short-lifespan phenotype of sesB1 (Figure S3).
Figure 3 .
BER1 strain mtDNA does not rescue the developmental phenotype of sesB1. (A) Bang sensitivity (mean recovery times ± SD) and (B) time to eclosion at room temperature of flies of the sex and genotypes indicated. wol, Wolbachia-infected BER1 cytoplasmic background, crossed into sesB1. A and B denote significantly different data classes (Newman–Keuls test, P < 0.05, based on ANOVA, P < 0.01, each sex considered separately).
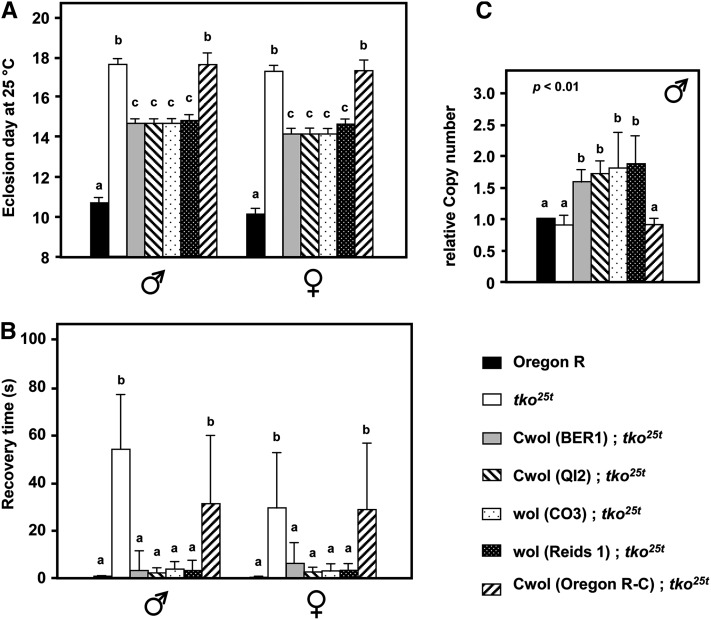
Next we tested the four other Wolbachia-infected strains for suppression of tko25t, using a similar crossing strategy as for BER1, with at least five generations of backcrossing in each case. Two strains (CO3 and Reids-1) were phenotypically tested in the uncured state, whereas the two others (QI2 and Oregon R-C) were tested after curing by tetracycline treatment. The cytoplasm from backgrounds QI2, Reids-1, and CO3 gave a similar degree of rescue of the mutant phenotype as did that of BER1 (Figure 4, A and B), while Oregon R-C cytoplasm gave no rescue. A control crossing strategy using wild-type Oregon R as the cytoplasmic donor also produced no rescue, nor did cytoplasm from wild-type strain Canton-S (Figure S4A). When tested, the nuclear background of all these strains was the same, further supporting the view that the suppression activity is entirely cytoplasmic. Importantly, all four suppressor cytoplasms (after removal of Wolbachia) showed elevated mtDNA copy number in the tko25t background, whereas nonsuppressor Oregon R-C cytoplasm did not (Figure 4C).
Figure 4 .
mtDNA from three other Wolbachia-infected backgrounds partially suppresses tko25t phenotype. (A) Time to eclosion at 25°, (B) bang sensitivity, and (C) mtDNA copy number of flies of the sex and genotypes indicated. wol, strains still infected with Wolbachia; Cwol, strains cured of Wolbachia infection. A, B, and C denote significantly different data classes (Newman–Keuls test, P < 0.05, based on ANOVA, P < 0.001, each sex considered separately).
Logically, a cytoplasmic suppressor acting via an effect on the amount of mtDNA should reside in portions of the mitochondrial genome affecting replication or copy number control. However, the cis-acting signals regulating mtDNA copy number remain to be identified. Moreover, such an effect might also be indirect, e.g., mediated by the properties of the mitochondrially encoded gene products themselves. We therefore sequenced the entire coding region of the mtDNA of the four suppressor strains, plus three nonsuppressor strains (Canton-S, Oregon R-C, and the mtDNA of the original tko25t strain; the coding region of Oregon R is already deposited in the NCBI database, accession no. AF200828). We also characterized the structure and determined the sequence of the NCR (A + T rich and highly repetitive) in representative strains.
The mtDNA coding regions of all strains analyzed contained between 30 and 50 single nucleotide or short indel polymorphisms compared with the reference sequence, NCBI NC_001709 (Table S1). Each strain had a unique sequence, although suppressor strains CO3 and QI2 were very similar, as were nonsuppressor strain Oregon R-C (identical with the previously sequenced Oregon R strain) and the original tko25t stock, which differed by only two polymorphisms. Suppressor strain BER1 and nonsuppressor strain Canton-S shared many polymorphisms not found in other strains. No single coding-region polymorphism was present in all suppressor strains but absent from all nonsuppressor strains (or vice versa), nor was any significant heteroplasmy detected.
The overall structure of the NCR was inferred for several suppressor and nonsuppressor strains using a combination of long PCR (Figure S4B), partial restriction mapping (Figure S4, C–E), and Sanger sequencing, involving both PCR and cloning (Figure S4, F and G). The NCR of BER1 mtDNA, as well as that of most other strains, including both suppressors and nonsuppressors, was the “short” morph, containing only three copies of repeat I, whereas the reference sequence (Lewis et al. 1994) is the “long” morph, containing five (Figure S4H). Complete NCR sequences of suppressor strain BER1 and the original (nonsuppressor) tko25t strain revealed many single-nucleotide and short indel polymorphisms distinguishing the NCRs of these strains from each other and from the reference sequence, many of them affecting the length of oligo(A/T) and oligo(AT/TA) tracts, but also some rare AT-to-GC transitions. There was no evidence in the NCR of BER1 mtDNA for the presence of integrated sequences derived from Wolbachia, nor was there any obvious or systematic difference between the NCRs that might explain the suppressor phenotype.
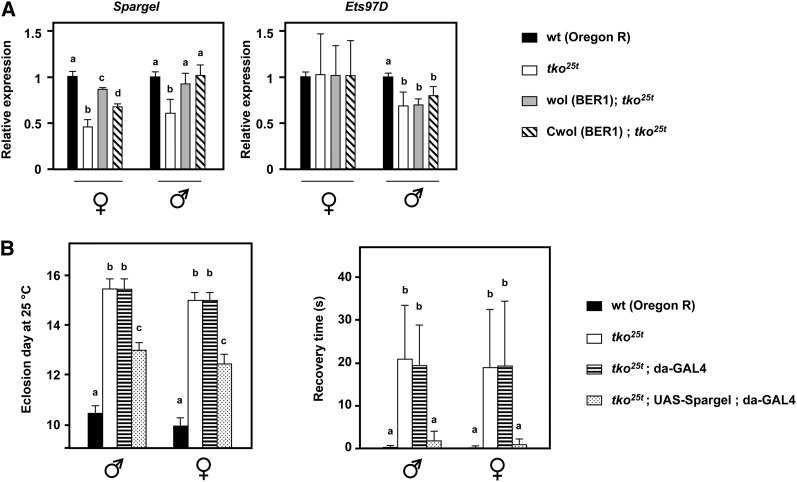
We investigated the expression levels of eight genes with known or hypothesized roles in mtDNA replication or transcription, using qRT–PCR, revealing no significant differences between tko25t in the original or Wolbachia-cured BER1 (Cwol; tko25t) backgrounds (Figure S5A). Next (Figure 5) we investigated two genes proposed to have general roles in the regulation of mitochondrial biogenesis, namely Spargel, homolog of human PPRC, and PPARGC1A (PGC1α), and Ets97D, also known as Delg, ortholog of human GABPA (NRF-2α). Ets97D was slightly downregulated in tko25t males, but was not significantly altered by the mtDNA background in either sex (Figure 5B). However, Spargel was significantly downregulated in tko25t in both sexes, but restored to near wild-type expression in the BER1 mtDNA background, regardless of the presence or absence of Wolbachia (Figure 5A). This is consistent with the BER1 cytoplasm suppressing the phenotype of tko25t by promoting mitochondrial biogenesis and suggests Spargel could be a key target in this process. To confirm this, we drove Spargel overexpression throughout development in tko25t flies (Figure S5B), using the strong and ubiquitous da-GAL4 driver and a UAS-Spargel transgenic line (Tiefenböck et al. 2010), which together phenocopied the suppressor (Figure 5B), both for bang sensitivity and eclosion timing.
Figure 5 .
Expression levels of global regulators of mitochondrial biogenesis in tko25t in different mtDNA backgrounds. (A) qRT–PCR of Spargel and Ets97D mRNAs normalized to that of RpL32 and then to the values for wild-type Oregon R flies of the given sex and genotype. a, b, c, and d denote significantly different data classes (Newman–Keuls test, P < 0.05, based on ANOVA, P < 0.01, each gene in each sex considered separately). Despite the variation between the sexes, and between flies with and without the physical presence of Wolbachia, both the downregulation of Spargel in tko25t flies and its restoration in the presence of the BER1 cytoplasm are statistically significant. (B) Bang sensitivity (mean recovery times ± SD) and time to eclosion at 25 ° of flies of the sex and genotypes indicated. For the crosses required in this experiment, the UAS-Spargel transgene was first rendered homozygous in the tko25t background, with the original nonsuppressor cytoplasm. a, b, and c denote significantly different data classes (Newman–Keuls test, P < 0.05, based on ANOVA, P < 0.01).
Discussion
Nature of the tko25t suppressor
In this study, we set out to test the role of cytoplasmic genetic determinants on the phenotype of tko25t, a Drosophila nuclear OXPHOS mutant. Cytoplasm from the Wolbachia-infected strain BER1 partially suppressed tko25t, rescuing bang sensitivity and male courtship defect, and alleviating developmental delay. Based on reciprocal crosses and backcrossing over 10 generations, the suppressor determinant was entirely cytoplasmic, but independent of the physical presence of Wolbachia, because the tetracycline-cured BER1 strain retained full suppression. The cytoplasmic determinant providing suppression most likely resides in mtDNA itself, although we cannot completely exclude some other maternally inherited element that has escaped detection, such as a tetracycline-resistant, prokaryotic endosymbiont not detected by PCR using universal primers, or a cytoplasmically transmitted plasmid or virus. Suppression was associated with increased mitochondrial biogenesis and mtDNA copy number, supporting the idea that the suppressor resides in mtDNA, e.g., as a cis-acting element that responds to mitochondrial stress. The most logical location for such an element would be within the NCR, which contains the replication origin and terminus (Goddard and Wolstenholme 1978; Saito et al. 2005). At this time, nothing is known of the physiological mechanisms of mtDNA copy-number control in any metazoan, although many genes are known to be required for mtDNA maintenance (Larsson et al. 1998; Iyengar et al. 1999, 2002; Matsushima et al. 2004; Hance et al. 2005).
Natural length variation in the NCR (Townsend and Rand 2004) has been suggested to have selective value, with longer NCR variants hypothesized as advantageous (Solignac et al. 1987; Kann et al. 1998). However, the overall architecture of the NCR did not correlate with suppressor activity. Nonsuppressor strain Canton-S had a long NCR equivalent to that of the reference sequence, with five copies of repeat I, whereas all other strains tested had a shorter NCR (three copies of repeat I, Figure S4), regardless of whether they were suppressors or nonsuppressors.
The NCR contains many strain-specific and shared polymorphisms, notably in the length of oligo(A/T) and oligo(AT/TA) tracts. Suppressor strain BER1 had ∼80 such polymorphisms compared with the reference sequence or the original tko25t strain, including some transitions at phylogenetically conserved positions (Brehm et al. 2001). However, large-scale sequencing and analysis of many wild strains will be needed to identify any specific NCR polymorphism(s) correlating with suppressor activity.
Coding region polymorphisms may also underlie haplotype-specific copy-number regulation (Suissa et al. 2009), for example, by influencing the binding of specific proteins. However, no single polymorphism distinguished suppressor from nonsuppressor strains, nor did we find any evidence for heteroplasmy. Globally, suppressor strain BER1 was most similar to nonsuppressor strain Canton-S, whereas suppressor strains QI2, CO3, and Reids-1 were more similar to nonsuppressor strains Oregon R and Oregon R-C (Table S1). The suppressor thus appears to have arisen independently on different haplotype backgrounds.
The recent confirmation of the presence of both cytosine methylation and hydroxymethylation in mammalian mtDNA (Shock et al. 2011) suggests the possibility that the suppressor might be an epigenetic mark carried in the mitochondrial genome rather than the mtDNA sequence itself. The increased expression of the mitochondrial isoform of the DNA methyltransferase DNMT1 in human cells under hypoxia (Shock et al. 2011) is accompanied by altered mitochondrial gene expression, raising the possibility that an epigenetic alteration of mtDNA might underlie the resistance to the stress imposed by the tko25t mutation.
Mechanism of the mitochondrial suppressor of tko25t
The degree of phenotypic rescue was similar in four different suppressor strains, indicating a common mechanism. Although respiration and ROS production were unchanged, when normalized against the amount of mitochondrial protein, the amount of mitochondrial protein was itself increased up to twofold, accompanied by an increased mtDNA copy number and a shift in the ratio SSU:LSU rRNA toward the equimolarity typical of wild-type flies, as seen also in a nuclear suppressor (Kemppainen et al. 2009), and indicating enhanced mitochondrial biogenesis as the key mechanism of suppression.
The suppressor was independent of any short-term copy-number increase after tetracycline treatment (Ballard and Melvin 2007), because it persisted for many generations. There was also increased expression of Spargel (PGC1α), one of the proposed global regulators of mitochondrial biogenesis and retrograde signaling (Spiegelman 2007; Jones et al. 2012). Importantly, Spargel expression was found to be low in tko25t flies in the nonsuppressor background, and simply restored to wild-type levels in the presence of the suppressor. This may indicate a programmed downregulation of mitochondrial biogenesis in nonsuppressor backgrounds in response to the metabolic stress generated by tko25t (Fernández-Ayala et al. 2010). Suppressor mtDNAs might be refractory to mitochondrial stress signals, or else the gene products of suppressor mtDNA may minimize the production of these signals. Note that the copy number of suppressor-strain mtDNA was not elevated in a wild-type nuclear background, consistent with the idea that the (sex-specific) changes seen in tko25t are a stress response rather than an invariant property of the suppressor-strain mtDNAs. Crucially, ubiquitous overexpression of Spargel was able to phenocopy the suppressor, showing that the expression level of Spargel is not merely a marker for the effect of the suppressor, but is a key mechanistic target thereof.
The signal to which suppressor- and nonsuppressor-strain mtDNAs may respond differently could be a direct metabolic readout of OXPHOS insufficiency or an indirect effect of defective mitochondrial translation, such as a disturbance in protein homeostasis (Dieteren et al. 2011). Different mtDNA backgrounds proposed as modifiers of Leber Hereditary Optic Neuropathy in humans have been previously shown to modulate OXPHOS complex assembly (Pello et al. 2008). In tko25t, one of the most highly upregulated genes is Hsp22 (Fernández-Ayala et al. 2010), also upregulated under stress and in aging (Morrow et al. 2004; Yang and Tower 2009; Kim et al. 2010). Suppressor- and nonsuppressor-strain mtDNAs may thus differ in their induction or implementation of the mitochondrial unfolded protein response (Haynes and Ron 2010).
The suppressor clearly does not act by establishing a bypass of ATP deficiency, since the developmental phenotype of sesB1, affecting the mitochondrial adenine nucleotide translocator ANT (Zhang et al. 1999), was not rescued by BER1 cytoplasm. However, the adult phenotype of sesB1, similar to that of the cytochrome c oxidase mutant levy1 (Liu et al. 2007) was partially alleviated. The phenotype may reflect enhanced oxidative stress and damage, against which the suppressor mtDNA affords some protection. Some molecular correlates of suppression, including Spargel expression, mtDNA copy number, and the amount of assembled complex I, as judged by the NDUFS3 marker, exhibited quantitative differences between the sexes, even though all markers showed significant improvement in both males and females in the presence of suppressor cytoplasm. Sex differences have been reported previously in mitochondrial bioenergetics and oxidative stress handling (Ballard et al. 2007b), and these might underlie the differences observed here. Isolation and study of additional mutants with compromized OXPHOS and/or defects in mitochondrial biogenesis should help to elucidate further the relationship between mitochondrial stress signaling, mtDNA copy number, and the suppressor determinant.
Role of Wolbachia in selecting suppressor cytoplasms
Cytoplasm from four wild strains of Drosophila naturally infected with Wolbachia conferred suppression after curing of Wolbachia, while that from two Wolbachia-free wild-type strains (Oregon R and Canton-S) did not. However, the correlation between Wolbachia infection and suppressor activity is not absolute, since Wolbachia-infected strain Oregon R-C, collected from the wild at the same time as the closely related strain Oregon R (Table S1), was also a nonsuppressor. Oregon R-C was first reported as infected in 1994 (Bourtzis et al. 1994), but has been maintained in laboratories for over 50 years (Clancy and Beadle 1937), and we cannot exclude the possibility that it became infected in the laboratory. Suppressor activity in the wild may therefore have been selected under the metabolic stress imposed by Wolbachia.
Wolbachia is considered an endosymbiont, with variable effects on host reproduction to favor its own spread in the population (Stouthamer et al. 1999; Champion de Crespigny et al. 2006), although host adaptation may also subvert this process (Turelli 1994). Effects on host fitness are influenced dramatically by mitochondrial genetic background in D. simulans (Dean 2006), since Wolbachia may compete with mitochondria for substrates. Flies of a genetic background able to boost mitochondrial functions in the presence of such an intruder might be expected to have a natural advantage.
Paradigm for understanding nuclear–mitochondrial interactions in human disease
Suppressor and nonsuppressor cytoplasmic backgrounds were isolated in multiple examples from nature, and presumably have a different adaptive value depending on the conditions, as hypothesized for different mtDNA haplotypes in the human population (Wallace 2005). In a wild-type nuclear background, suppressor and nonsuppressor cytoplasms confer no obvious biological advantage or disadvantage, but in the tko25t background, where mitochondrial protein synthesis is limiting, the effects are substantial.
Many human pathologies result directly from defects in mitochondrial protein synthesis (Jacobs and Turnbull 2005; Rötig 2011), including mutations in genes for mitoribosomal proteins (Miller et al. 2004; Saada et al. 2007; Smits et al. 2011) and numerous other translation factors. Our findings support the concept that the variable clinical phenotypes seen in these disorders reflect differences in mtDNA genotype.
The role of mtDNA copy number as a modifying factor in mitochondrial disease has long been hypothesized (Tyynismaa and Suomalainen 2009) and is supported by cell culture studies (Bentlage and Attardi 1996). Copy-number aberrations are features of some mtDNA pathologies and may contribute to the clinical phenotype (Liu et al. 2006; Brinckmann et al. 2010), in addition to those disease entities where mtDNA maintenance is a primary pathological target (Lamperti and Zeviani 2009; Rötig and Poulton 2009). They have also been reported in other diseases, such as breast cancer (Shen et al. 2009).
Artificial manipulation of mtDNA copy number can influence disease phenotype in animal models (Ekstrand et al. 2004; Matsushima et al. 2004; Tyynismaa et al. 2004; Ikeuchi et al. 2005; Hokari et al. 2010; Matsushima et al. 2010; Nishiyama et al. 2010), although high copy number is not always beneficial (Ylikallio et al. 2010).
Although we remain ignorant of the physiological mechanisms regulating mtDNA copy number and its responsiveness to stress, the tko25t suppressor provides a paradigm for mtDNA sequence variation playing a role as a disease modifier in humans. The further exploitation of this model and elucidation of the underlying molecular machinery may reveal new drug targets in what is currently an intractable class of human disease.
Supplementary Material
Acknowledgments
We thank Tea Tuomela, Outi Kurronen, Charlotta Putkonen, and Tess Jeffers for technical assistance and Jaakko Pohjoismäki for useful discussions and advice. This work was supported by funding from the Academy of Finland, Tampere University Hospital Medical Research Fund, the Sigrid Juselius Foundation, European Research Council, European Molecular Biology Organization, and National Institutes of Health (grant GM45295).
Footnotes
Communicating editor: R. Anholt
Literature Cited
- Ballard J. W., Melvin R. G., 2007. Tetracycline treatment influences mitochondrial metabolism and mtDNA density two generations after treatment in Drosophila. Insect Mol. Biol. 16: 799–802. [DOI] [PubMed] [Google Scholar]
- Ballard J. W., Melvin R. G., Katewa S. D., Maas K., 2007a. Mitochondrial DNA variation is associated with measurable differences in life-history traits and mitochondrial metabolism in Drosophila simulans. Evolution 61: 1735–1747. [DOI] [PubMed] [Google Scholar]
- Ballard J. W., Melvin R. G., Miller J. T., Katewa S. D., 2007b. Sex differences in survival and mitochondrial bioenergetics during aging in Drosophila. Aging Cell 6: 699–708. [DOI] [PubMed] [Google Scholar]
- Bentlage H. A., Attardi G., 1996. Relationship of genotype to phenotype in fibroblast-derived transmitochondrial cell lines carrying the 3243 mutation associated with the MELAS encephalomyopathy: shift towards mutant genotype and role of mtDNA copy number. Hum. Mol. Genet. 5: 197–205. [DOI] [PubMed] [Google Scholar]
- Bourtzis K., Nirgianaki A., Onyango P., Savakis C., 1994. A prokaryotic dnaA sequence in Drosophila melanogaster: Wolbachia infection and cytoplasmic incompatibility among laboratory strains. Insect Mol. Biol. 3: 131–142. [DOI] [PubMed] [Google Scholar]
- Brehm A., Harris D. J., Hernández M., Cabrera V. M., Larruga J. M., et al. , 2001. Structure and evolution of the mitochondrial DNA complete control region in the Drosophila subobscura subgroup. Insect Mol. Biol. 10: 573–578. [DOI] [PubMed] [Google Scholar]
- Brinckmann A., Weiss C., Wilbert F., von Moers A., Zwirner A., et al. , 2010. Regionalized pathology correlates with augmentation of mtDNA copy numbers in a patient with myoclonic epilepsy with ragged-red fibers (MERRF-syndrome). PLoS ONE 5: e13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. D., Sun F., Wallace D. C., 1997. Clustering of Caucasian Leber hereditary optic neuropathy patients containing the 11778 or 14484 mutations on an mtDNA lineage. Am. J. Hum. Genet. 60: 381–387. [PMC free article] [PubMed] [Google Scholar]
- Brown M. D., Starikovskaya E., Derbeneva O., Hosseini S., Allen J. C., et al. , 2002. The role of mtDNA background in disease expression: a new primary LHON mutation associated with Western Eurasian haplogroup. J. Hum. Genet. 110: 130–138. [DOI] [PubMed] [Google Scholar]
- Champion de Crespigny F. E., Pitt T. D., Wedell N., 2006. Increased male mating rate in Drosophila is associated with Wolbachia infection. J. Evol. Biol. 19: 1964–1972. [DOI] [PubMed] [Google Scholar]
- Clancy C. W., Beadle G. W., 1937. Ovary transplants in Drosophila melanogaster: studies of the characters singed, fused, and female-sterile. Biol. Bull. 72: 47–56. [Google Scholar]
- Dean M. D., 2006. A Wolbachia-associated fitness benefit depends on genetic background in Drosophila simulans. Proc. Biol. Sci. 273: 1415–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieteren C. E., Willems P. H., Swarts H. G., Fransen J., Smeitink J. A., et al. , 2011. Defective mitochondrial translation differently affects the live cell dynamics of complex I subunits. Biochim. Biophys. Acta 1807: 1624–1633. [DOI] [PubMed] [Google Scholar]
- Dobson S. L., Marsland E. J., Rattanadechakul W., 2002. Mutualistic Wolbachia infection in Aedes albopictus: accelerating cytoplasmic drive. Genetics 160: 1087–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand M. I., Falkenberg M., Rantanen A., Park C. B., Gaspari M., et al. , 2004. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 13: 935–944. [DOI] [PubMed] [Google Scholar]
- Fergestad T., Bostwick B., Ganetzky B., 2006. Metabolic disruption in Drosophila bang-sensitive seizure mutants. Genetics 173: 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Ayala D. J., Sanz A., Vartiainen S., Kemppainen K. K., Babusiak M., et al. , 2009. Expression of the Ciona intestinalis alternative oxidase (AOX) in Drosophila complements defects in mitochondrial oxidative phosphorylation. Cell Metab. 9: 449–460. [DOI] [PubMed] [Google Scholar]
- Fernández-Ayala D. J. M., Chen S., Kemppainen E., O’Dell K. M. C., Jacobs H. T., 2010. Gene expression in a Drosophila model of mitochondrial disease. PLoS ONE 5: e8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A. J., Rand D. M., 2002. Wolbachia interactions that determine Drosophila melanogaster survival. Evolution 56: 1976–1981. [DOI] [PubMed] [Google Scholar]
- Goddard J. M., Wolstenholme D. R., 1978. Origin and direction of replication in mitochondrial DNA molecules from Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 75: 3886–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Abajian C., Green P., 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Greaves L. C., Reeve A. K., Taylor R. W., Turnbull D. M., 2012. Mitochondrial DNA and disease. J. Pathol. 226: 274–286. [DOI] [PubMed] [Google Scholar]
- Hance N., Ekstrand M. I., Trifunovic A., 2005. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum. Mol. Genet. 14: 1775–1783. [DOI] [PubMed] [Google Scholar]
- Haynes C. M., Ron D., 2010. The mitochondrial UPR: protecting organelle protein homeostasis. J. Cell Sci. 123: 3849–3855. [DOI] [PubMed] [Google Scholar]
- Hedges L. M., Brownlie J. C., O’Neill S. L., Johnson K. N., 2008. Wolbachia and virus protection in insects. Science 322: 702. [DOI] [PubMed] [Google Scholar]
- Hokari M., Kuroda S., Kinugawa S., Ide T., Tsutsui H., et al. , 2010. Overexpression of mitochondrial transcription factor A (TFAM) ameliorates delayed neuronal death due to transient forebrain ischemia in mice. Neuropathology 30: 401–407. [DOI] [PubMed] [Google Scholar]
- Homyk T., Sheppard D. E., 1977. Behavioral mutants of Drosophila melanogaster. I. Isolation and mapping of mutations which decrease flight ability. Genetics 87: 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G., Carelli V., Spruijt L., Gerards M., Mowbray C., et al. , 2007. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am. J. Hum. Genet. 81: 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Matsusaka H., Kang D., Matsushima S., Ide T., et al. , 2005. Overexpression of mitochondrial transcription factor A ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation 112: 683–690. [DOI] [PubMed] [Google Scholar]
- Ikeya T., Broughton S., Alic N., Grandison R., Partridge L., 2009. The endosymbiont Wolbachia increases insulin/IGF-like signalling in Drosophila. Proc. Biol. Sci. 276: 3799–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar B., Roote J., Campos A. R., 1999. The tamas gene, identified as a mutation that disrupts larval behavior in Drosophila melanogaster, codes for the mitochondrial DNA polymerase catalytic subunit (DNApol-gamma125). Genetics 153: 1809–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar B., Luo N., Farr C. L., Kaguni L. S., Campos A. R., 2002. The accessory subunit of DNA polymerase gamma is essential for mitochondrial DNA maintenance and development in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99: 4483–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H. T., Turnbull D. M., 2005. Nuclear genes and mitochondrial translation: a new class of genetic disease. Trends Genet. 21: 312–314. [DOI] [PubMed] [Google Scholar]
- Jones A. W., Yao Z., Vicencio J. M., Karkucinska-Wieckowska A., Szabadkai G., 2012. PGC-1 family coactivators and cell fate: roles in cancer, neurodegeneration, cardiovascular disease and retrograde mitochondria-nucleus signalling. Mitochondrion 12: 86–99. [DOI] [PubMed] [Google Scholar]
- Kann L. M., Rosenblum E. B., Rand D. M., 1998. Aging, mating, and the evolution of mtDNA heteroplasmy in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 95: 2372–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa S. D., Ballard J. W., 2007. Sympatric Drosophila simulans flies with distinct mtDNA show difference in mitochondrial respiration and electron transport. Insect Biochem. Mol. Biol. 37: 213–222. [DOI] [PubMed] [Google Scholar]
- Kazuno A. A., Munakata K., Nagai T., Shimozono S., Tanaka M., et al. , 2006. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemppainen E., Fernández-Ayala D. J., Galbraith L. C., O’Dell K. M., Jacobs H. T., 2009. Phenotypic suppression of the Drosophila mitochondrial disease-like mutant tko25t by duplication of the mutant gene in its natural chromosomal context. Mitochondrion 9: 353–363. [DOI] [PubMed] [Google Scholar]
- Kim H. J., Morrow G., Westwood J. T., Michaud S., Tanguay R. M., 2010. Gene expression profiling implicates OXPHOS complexes in lifespan extension of flies over-expressing a small mitochondrial chaperone, Hsp22. Exp. Gerontol. 45: 611–620. [DOI] [PubMed] [Google Scholar]
- Lamperti C., Zeviani M., 2009. Encephalomyopathies caused by abnormal nuclear-mitochondrial intergenomic cross-talk. Acta Myol. 28: 2–11. [PMC free article] [PubMed] [Google Scholar]
- Larsson N. G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., et al. , 1998. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18: 231–236. [DOI] [PubMed] [Google Scholar]
- Lewis D. L., Farr C. L., Farquhar A. L., Kaguni L. S., 1994. Sequence, organization, and evolution of the A + T region of Drosophila melanogaster mitochondrial DNA. Mol. Biol. Evol. 11: 532–538. [DOI] [PubMed] [Google Scholar]
- Liu C. S., Cheng W. L., Lee C. F., Ma Y. S., Lin C. Y., et al. , 2006. Alteration in the copy number of mitochondrial DNA in leukocytes of patients with mitochondrial encephalomyopathies. Acta Neurol. Scand. 113: 334–341. [DOI] [PubMed] [Google Scholar]
- Liu W., Gnanasambandam R., Benjamin J., Kaur G., Getman P. B., et al. , 2007. Mutations in cytochrome c oxidase subunit VIa cause neurodegeneration and motor dysfunction in Drosophila. Genetics 176: 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima Y., Garesse R., Kaguni L. S., 2004. Drosophila mitochondrial transcription factor B2 regulates mitochondrial DNA copy number and transcription in Schneider cells. J. Biol. Chem. 279: 26900–26905. [DOI] [PubMed] [Google Scholar]
- Matsushima Y., Goto Y., Kaguni L. S., 2010. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM). Proc. Natl. Acad. Sci. USA 107: 18410–18415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Montooth K. L., Rand D. M., 2007. Positive and negative selection on the mitochondrial genome. Trends Genet. 23: 259–263. [DOI] [PubMed] [Google Scholar]
- Miller C., Saada A., Shaul N., Shabtai N., Ben-Shalom E., et al. , 2004. Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann. Neurol. 56: 734–738. [DOI] [PubMed] [Google Scholar]
- Miwa S., St-Pierre J., Partridge L., Brand M. D., 2003. Superoxide and hydrogen peroxide production by Drosophila mitochondria. Free Radic. Biol. Med. 35: 938–948. [DOI] [PubMed] [Google Scholar]
- Moreno-Loshuertos R., Acin-Perez R., Fernandez-Silva P., Movilla N., Perez-Martos A., et al. , 2006. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet. 38: 1261–1268. [DOI] [PubMed] [Google Scholar]
- Morrow G., Samson M., Michaud S., Tanguay R. M., 2004. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 18: 598–599. [DOI] [PubMed] [Google Scholar]
- Nishiyama S., Shitara H., Nakada K., Ono T., Sato A., et al. , 2010. Over-expression of Tfam improves the mitochondrial disease phenotypes in a mouse model system. Biochem. Biophys. Res. Commun. 401: 26–31. [DOI] [PubMed] [Google Scholar]
- O’Neill S. L., Giordano R., Colbert A. M., Karr T. L., Robertson H. M., 1992. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. USA 89: 2699–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pello R., Martín M. A., Carelli V., Nijtmans L. G., Achilli A., et al. , 2008. Mitochondrial DNA background modulates the assembly kinetics of OXPHOS complexes in a cellular model of mitochondrial disease. Hum. Mol. Genet. 17: 4001–4011. [DOI] [PubMed] [Google Scholar]
- Pravenec M., Hyakukoku M., Houstek J., Zidek V., Landa V., et al. , 2007. Direct linkage of mitochondrial genome variation to risk factors for type 2 diabetes in conplastic strains. Genome Res. 17: 1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds K. T., Thomson L. J., Hoffmann A. A., 2003. The effects of host age, host nuclear background and temperature on phenotypic effects of the virulent Wolbachia strain popcorn in Drosophila melanogaster. Genetics 164: 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rötig A., 2011. Human diseases with impaired mitochondrial protein synthesis. Biochim. Biophys. Acta 1807: 1198–1205. [DOI] [PubMed] [Google Scholar]
- Rötig A., Poulton J., 2009. Genetic causes of mitochondrial DNA depletion in humans. Biochim. Biophys. Acta 1792: 1103–1108. [DOI] [PubMed] [Google Scholar]
- Saada A., Shaag A., Arnon S., Dolfin T., Miller C., et al. , 2007. Antenatal mitochondrial disease caused by mitochondrial ribosomal protein (MRPS22) mutation. J. Med. Genet. 44: 784–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Tamura K., Aotsuka T., 2005. Replication origin of mitochondrial DNA in insects. Genetics 171: 1695–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz A., Fernández-Ayala D. J., Stefanatos R. K., Jacobs H. T., 2010a. Mitochondrial ROS production correlates with, but does not directly regulate lifespan in Drosophila. Aging 2: 200–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz A., Soikkeli M., Portero-Otín M., Wilson A., Kemppainen E., et al. , 2010b. Expression of the yeast NADH dehydrogenase Ndi1 in Drosophila confers increased lifespan independently of dietary restriction. Proc. Natl. Acad. Sci. USA 107: 9105–9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah Z. H., O’Dell K. M., Miller S. C., An X., Jacobs H. T., 1997. Metazoan nuclear genes for mitoribosomal protein S12. Gene 204: 55–62. [DOI] [PubMed] [Google Scholar]
- Shen J., Platek M., Mahasneh A., Ambrosone C. B., Zhao H., 2009. Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion 10: 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock L. S., Thakkar P. V., Peterson E. J., Moran R. G., Taylor S. M., 2011. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. USA 108: 3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P., Saada A., Wortmann S. B., Heister A. J., Brink M., et al. , 2011. Mutation in mitochondrial ribosomal protein MRPS22 leads to Cornelia de Lange-like phenotype, brain abnormalities and hypertrophic cardiomyopathy. Eur. J. Hum. Genet. 19: 394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solignac M., Genermont J., Monnerot M., Mounolou J., 1987. Drosophila mitochondrial genetics: evolution of heteroplasmy through germ line cell divisions. Genetics 117: 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman B. M., 2007. Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators. Novartis Found. Symp. 287: 60–63. [PubMed] [Google Scholar]
- Stouthamer R., Breeuwer J. A. J., Hurst G. D. D., 1999. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53: 71–102. [DOI] [PubMed] [Google Scholar]
- Suissa S., Wang Z., Poole J., Wittkopp S., Feder J., et al. , 2009. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 5: e1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira L., Ferreira A., Ashburner M., 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiefenböck S. K., Baltzer C., Egli N. A., Frei C., 2010. The Drosophila PGC-1 homologue Spargel coordinates mitochondrial activity to insulin signalling. EMBO J. 29: 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivonen J. M., O’Dell K. M., Petit N., Irvine S. C., Knight G. K., et al. , 2001. technical knockout, a Drosophila model of mitochondrial deafness. Genetics 159: 241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivonen J. M., Manjiry S., Touraille S., Alziari S., O’Dell K. M. C., et al. , 2003. Gene dosage and selective expression modify phenotype in a Drosophila model of human mitochondrial disease. Mitochondrion 3: 83–96. [DOI] [PubMed] [Google Scholar]
- Toivonen J. M., Walker G. A., Martinez-Diaz P., Bjedov I., Driege Y., et al. , 2007. No influence of Indy on lifespan in Drosophila after correction for genetic and cytoplasmic background effects. PLoS Genet. 3: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend J. P., Rand D. M., 2004. Mitochondrial genome size variation in New World and Old World populations of Drosophila melanogaster. Heredity 93: 98–103. [DOI] [PubMed] [Google Scholar]
- Turelli M., 1994. Evolution of incompatibility-inducing microbes and their hosts. Evolution 48: 1500–1513. [DOI] [PubMed] [Google Scholar]
- Tyynismaa H., Suomalainen A., 2009. Mouse models of mitochondrial DNA defects and their relevance for human disease. EMBO Rep. 10: 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H., Sembongi H., Bokori-Brown M., Granycome C., Ashley N., et al. , 2004. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 13: 3219–3227. [DOI] [PubMed] [Google Scholar]
- Wallace D. C., 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39: 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett C. S., Burton R. S., 2004. Evolution of interacting proteins in the mitochondrial electron transport system in a marine copepod. Mol. Biol. Evol. 21: 443–453. [DOI] [PubMed] [Google Scholar]
- Wodarz A., Hinz U., Engelbert M., Knust E., 1995. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell 82: 67–76. [DOI] [PubMed] [Google Scholar]
- Yang J., Tower J., 2009. Expression of hsp22 and hsp70 transgenes is partially predictive of Drosophila survival under normal and stress conditions. J. Gerontol. A Biol. Sci. Med. Sci. 64: 828–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylikallio E., Suomalainen A., 2012. Mechanisms of mitochondrial diseases. Ann. Med. 44: 41–59. [DOI] [PubMed] [Google Scholar]
- Ylikallio E., Tyynismaa H., Tsutsui H., Ide T., Suomalainen A., 2010. High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 19: 2695–2705. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Q., Roote J., Brogna S., Davis A. W., Barbash D. A., et al. , 1999. stress sensitive B encodes an adenine nucleotide translocase in Drosophila melanogaster. Genetics 153: 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.