Abstract
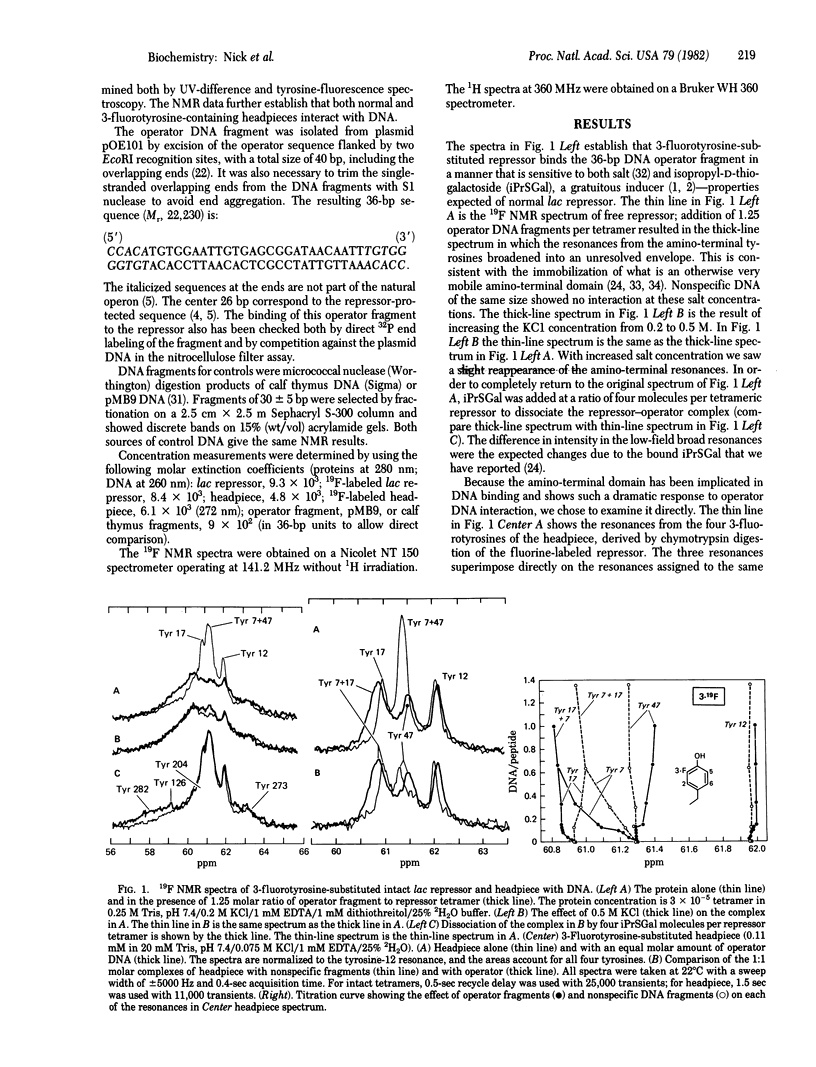
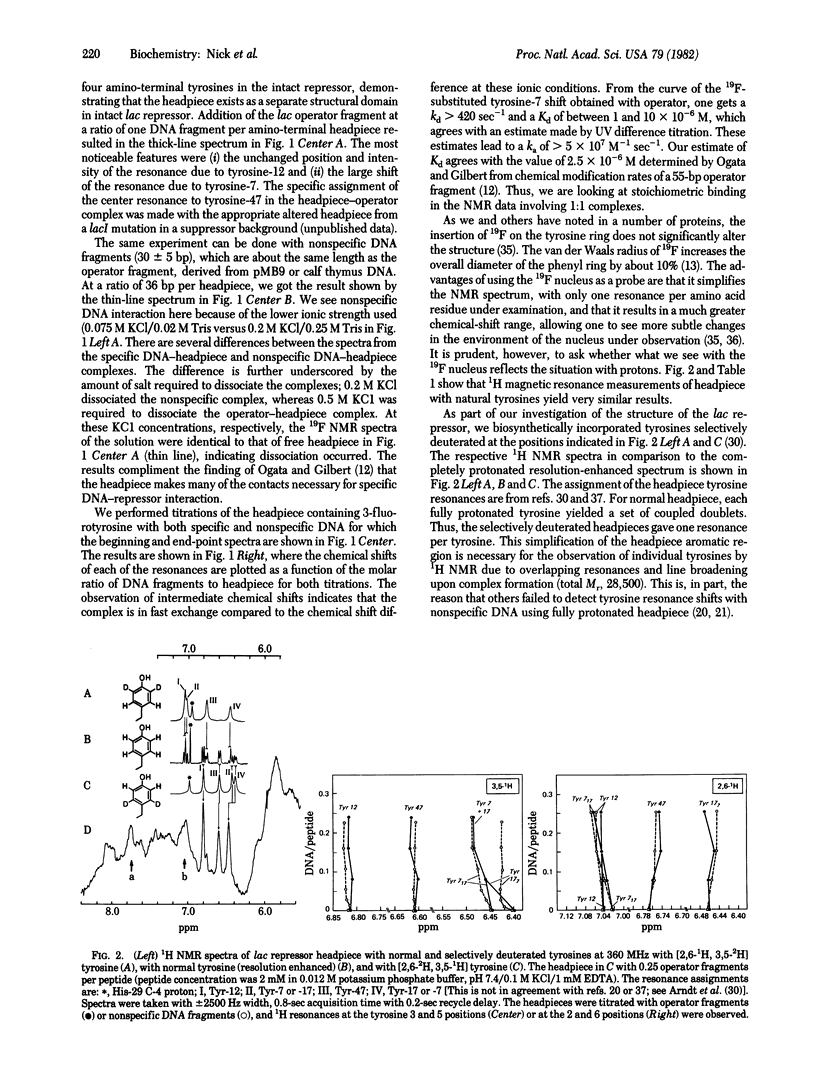
We show here the changes in the NMR spectra of the Escherichia coli lac repressor when bound to isolated lac operator DNA. The observations focus on the aromatic residues--four tyrosines and a single histidine--in the amino-terminal DNA binding domain of the lac repressor. There is a good correlation between chemical shift changes seen by 19F NMR when compared with 1 H NMR of otherwise identical repressor--DNA complexes. The results suggest that the tyrosines do not intercalate in the DNA. The NMR spectral changes with similarly sized DNA fragments, not containing the lac operator DNA sequence, are different. Thus, the amino-terminal domain of the lac repressor is independently capable of discriminating between lac operator and nonspecific DNA sequences. There can be two amino-terminal fragments per operator in the specific complex.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt K. T., Boschelli F., Lu P., Miller J. H. lac Repressor: a proton magnetic resonance look at the deoxyribonucleic acid binding fragment. Biochemistry. 1981 Oct 13;20(21):6109–6118. doi: 10.1021/bi00524a030. [DOI] [PubMed] [Google Scholar]
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois S., Pfahl M. Repressors. Adv Protein Chem. 1976;30:1–99. doi: 10.1016/s0065-3233(08)60478-7. [DOI] [PubMed] [Google Scholar]
- Buck F., Rüterjans H., Beyreuther K. 1H NMR study of the lactose repressor from Escherichia coli. FEBS Lett. 1978 Dec 15;96(2):335–338. doi: 10.1016/0014-5793(78)80430-x. [DOI] [PubMed] [Google Scholar]
- Buck F., Rüterjans H., Kaptein R., Beyreuther K. Photochemically induced dynamic nuclear polarization investigation of complex formation of the NH2-terminal DNA-binding domain of lac repressor with poly[d(AT)]. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5145–5148. doi: 10.1073/pnas.77.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran R., Jardetzky T. S., Jardetzky O. The delta helix--a possible left-handed stable polypeptide structure in the N-terminal segment of the lac repressor. FEBS Lett. 1979 May 1;101(1):11–14. [PubMed] [Google Scholar]
- Clement R., Daune M. P. Binding of lactose repressor to poly d(A-T) : OD AND CD melting of the complex. Nucleic Acids Res. 1975 Mar;2(3):303–318. doi: 10.1093/nar/2.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J. E., Anderson R. A., Ratcliffe R. G., Armitage I. M. Structure of gene 5 protein-oligodeoxynucleotide complexes as determined by 1H, 19F, and 31P nuclear magnetic resonance. Biochemistry. 1976 Dec 14;15(25):5419–5430. doi: 10.1021/bi00670a001. [DOI] [PubMed] [Google Scholar]
- Cuatrecasas P., Fuchs S., Anfinsen C. B. Catalytic properties and specificity of the extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1967 Apr 10;242(7):1541–1547. [PubMed] [Google Scholar]
- Dunaway M., Manly S. P., Matthews K. S. Model for lactose repressor protein and its interaction with ligands. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7181–7185. doi: 10.1073/pnas.77.12.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G. Iodination of Escherichia coli lac repressor. Effect of tyrosine modification on repressor activity. Biochemistry. 1975 Jun 3;14(11):2512–2520. doi: 10.1021/bi00682a034. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J. Sequence of the lacI gene. Nature. 1978 Aug 24;274(5673):765–769. doi: 10.1038/274765a0. [DOI] [PubMed] [Google Scholar]
- Geisler N., Weber K. Escherichia coli lactose repressor: isolation of two different homogeneous headpieces and the existence of a hinge region between residues 50 and 60 in the repressor molecule. FEBS Lett. 1978 Mar 15;87(2):215–218. doi: 10.1016/0014-5793(78)80335-4. [DOI] [PubMed] [Google Scholar]
- Giessner-Prettre C., Pullman B. Intermolecular nuclear shielding due to the aromatic amino acids of proteins and to porphyrins. J Theor Biol. 1971 May;31(2):287–294. doi: 10.1016/0022-5193(71)90188-3. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Müller-Hill B. The lac operator is DNA. Proc Natl Acad Sci U S A. 1967 Dec;58(6):2415–2421. doi: 10.1073/pnas.58.6.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeddel D. V., Yansura D. G., Caruthers M. H. How lac repressor recognizes lac operator. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3578–3582. doi: 10.1073/pnas.75.8.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursky G. V., Tumanyan V. G., Zasedatelev A. S., Zhuze A. L., Grokhovsky S. L., Gottikh B. P. A model for the binding of lac repressor to the lac operator. Mol Biol Rep. 1976 Apr;2(5):427–434. doi: 10.1007/BF00366265. [DOI] [PubMed] [Google Scholar]
- Helene C., Maurizot J. C. Interactions of oligopeptides with nucleic acids. CRC Crit Rev Biochem. 1981;10(3):213–258. doi: 10.3109/10409238109113600. [DOI] [PubMed] [Google Scholar]
- Hogan M., Wemmer D., Bray R. P., Wade-Jardetzky N., Jardetzky O. On the interaction of the lac repressor headpiece with nucleic acids. FEBS Lett. 1981 Feb 23;124(2):202–203. doi: 10.1016/0014-5793(81)80136-6. [DOI] [PubMed] [Google Scholar]
- Hsieh W. T., Matthews K. S. Tetranitromethane modification of the tyrosine residues of the lactose repressor. J Biol Chem. 1981 May 25;256(10):4856–4862. [PubMed] [Google Scholar]
- JACOB F., MONOD J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961 Jun;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Jarema M. A., Lu P., Miller J. H. Genetic assignment of resonances in the NMR spectrum of a protein: lac repressor. Proc Natl Acad Sci U S A. 1981 May;78(5):2707–2711. doi: 10.1073/pnas.78.5.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe A., Riggs A. D., Bourgeois S. Lac repressor-operator interaction. V. Characterization of super- and pseudo-wild-type repressors. J Mol Biol. 1972 Feb 28;64(1):181–199. doi: 10.1016/0022-2836(72)90328-2. [DOI] [PubMed] [Google Scholar]
- Jones C. E., Olson O. M. Sequence-specific DNA-protein interaction: the lac repressor. J Theor Biol. 1977 Jan 21;64(2):323–332. doi: 10.1016/0022-5193(77)90360-5. [DOI] [PubMed] [Google Scholar]
- Jovin T. M., Geisler N., Weber K. Amino-terminal fragments of Escherichia coli lac repressor bind to DNA. Nature. 1977 Oct 20;269(5630):668–672. doi: 10.1038/269668a0. [DOI] [PubMed] [Google Scholar]
- Lu P., Jarema M., Mosser K., Daniel W. E. lac repressor: 3-fluorotyrosine substitution for nuclear magnetic resonance studies. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3471–3475. doi: 10.1073/pnas.73.10.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizot J. C., Charlier M., Hélène C. Lac repressor binding to poly (d(A-T)). Conformational changes. Biochem Biophys Res Commun. 1974 Oct 8;60(3):951–957. doi: 10.1016/0006-291x(74)90406-9. [DOI] [PubMed] [Google Scholar]
- Maurizot J. C., Charlier M. Lac repressor binding to single-stranded polyadenylic acid. FEBS Lett. 1977 Nov 1;83(1):107–110. doi: 10.1016/0014-5793(77)80652-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Hofer M., Schmeissner U., Sommer H., Schmitz A., Lu P. Genetic studies of the lac repressor. IX. Generation of altered proteins by the suppression of nonsence mutations. J Mol Biol. 1979 Jun 25;131(2):191–222. doi: 10.1016/0022-2836(79)90073-1. [DOI] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. DNA-binding site of lac repressor probed by dimethylsulfate methylation of lac operator. J Mol Biol. 1979 Aug 25;132(4):709–728. doi: 10.1016/0022-2836(79)90384-x. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Gabbay E. J. Nitroaniline diamine.poly(dA-dT) complexes: 1H and 19F NMR parameters for full intercalation of aromatic rings into DNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1351–1355. doi: 10.1073/pnas.78.3.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M. T., Jr, deHaseth P. L., Lohman T. M. Interpretation of monovalent and divalent cation effects on the lac repressor-operator interaction. Biochemistry. 1977 Nov 1;16(22):4791–4796. doi: 10.1021/bi00641a005. [DOI] [PubMed] [Google Scholar]
- Ribeiro A. A., Wemmer D., Bray R. P., Wade-Jardetzky N. G., Jardetzky O. High-resolution nuclear magnetic resonance studies of the Lac repressor. 1. Assignments of tyrosine resonances in the N-terminal headpiece. Biochemistry. 1981 Feb 17;20(4):818–823. doi: 10.1021/bi00507a025. [DOI] [PubMed] [Google Scholar]
- Sadler J. R., Betz J. L., Tecklenburg M., Goeddel D. V., Yansura D. G., Caruthers M. H. Cloning of chemically synthesized lactose operators. II. EcoRI-linkered operators. Gene. 1978 May;3(3):211–232. doi: 10.1016/0378-1119(78)90033-1. [DOI] [PubMed] [Google Scholar]
- Sadler J. R., Tecklenburg M., Betz J. L. Plasmids containing many tandem copies of a synthetic lactose operator. Gene. 1980 Feb;8(3):279–300. doi: 10.1016/0378-1119(80)90005-0. [DOI] [PubMed] [Google Scholar]
- Sykes B. D., Hull W. E. Fluorine nuclear magnetic resonance studies of proteins. Methods Enzymol. 1978;49:270–295. doi: 10.1016/s0076-6879(78)49015-9. [DOI] [PubMed] [Google Scholar]
- Wade-Jardetzky N., Bray R. P., Conover W. W., Jardetzky O., Geisler N., Weber K. Differential mobility of the N-terminal headpiece in the lac-repressor protein. J Mol Biol. 1979 Feb 25;128(2):259–264. doi: 10.1016/0022-2836(79)90129-3. [DOI] [PubMed] [Google Scholar]
- Wu R., Bahl C. P., Narang S. A. Lactose operator--repressor interaction. Curr Top Cell Regul. 1978;13:137–178. [PubMed] [Google Scholar]


