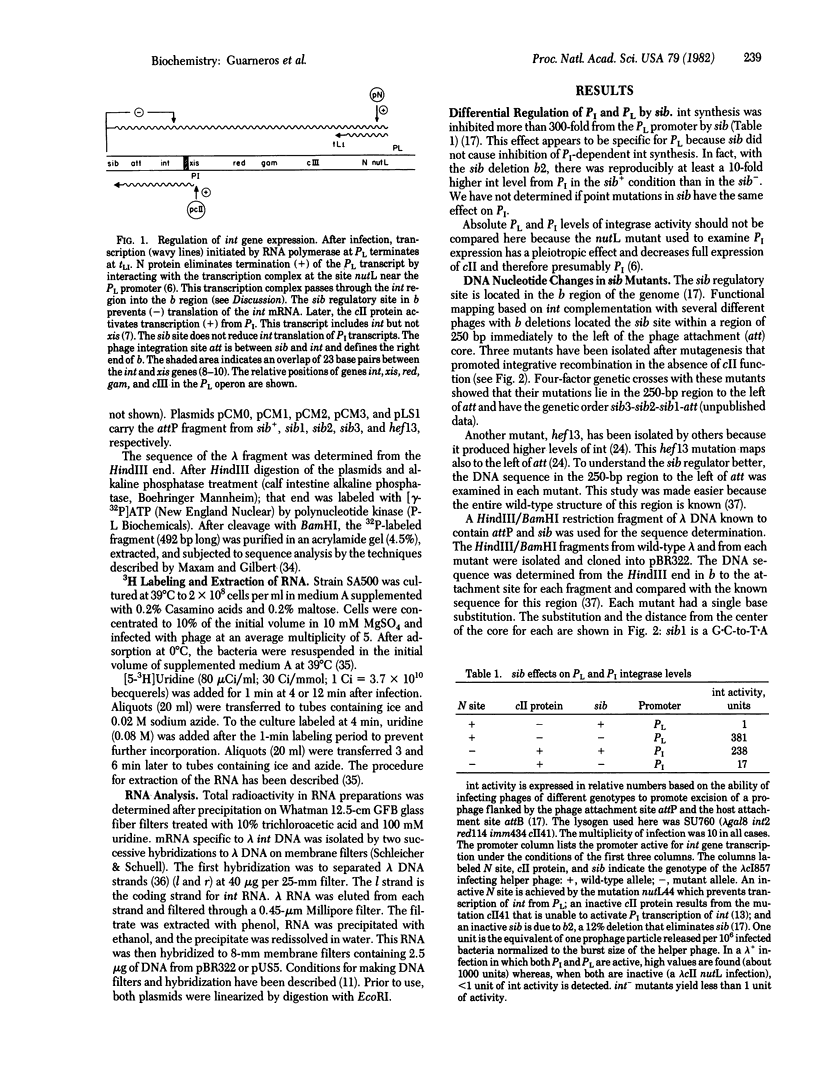
Abstract
The bacteriophage lambda int gene product, integrase, recombines the phage DNA with the host DNA at specific sites on each to accomplish lysogeny. The int gene is transcribed from two promoters, PL and PI, each regulated positively by lambda proteins. The expression of integrase is also controlled from a site, sib, in the b region of the phage genome. This is a unique regulatory site because it is located distal to the structural gene in relation to the promoters. The expression of int from the PL promoter is inhibited when sib is present. This effect appears to be specific for PL because sib does not cause inhibition of PI-dependent int synthesis. lambda mutants that contain alterations in the site have been isolated. Sequence analyses of the mutations reveal single base changes, spanning 37 base pairs (bp) in the b region, some 240 bp beyond the int gene. Another mutant, hef13, which has a phenotype similar to that of sib, introduces a nucleotide change within the same 37-bp region. The sib and hef mutations cluster within a region of dyad symmetry. Regulation of int synthesis by sib occurs after transcription of the int gene. There is no difference in the rate of PL-promoted int mRNA synthesis in either sib+ or sib- phage infections, yet int mRNA is less stable in the sib+ infection. Because RNase III host mutants are defective in sib regulation, processing of the PL mRNA at sib by this endoribonuclease may cause int mRNA decay and decrease int synthesis.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J., Mascarenhas D., Fischer R., Benedik M., Campbell A., Echols H. DNA sequence of regulatory region for integration gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1980 May;77(5):2477–2481. doi: 10.1073/pnas.77.5.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhya S., Cleary P., Campbell A. A deletion analysis of prophage lambda and adjacent genetic regions. Proc Natl Acad Sci U S A. 1968 Nov;61(3):956–962. doi: 10.1073/pnas.61.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhya S., Gottesman M., De Crombrugghe B. Release of polarity in Escherichia coli by gene N of phage lambda: termination and antitermination of transcription. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2534–2538. doi: 10.1073/pnas.71.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M., Noff D., Oppenheim A. B. Isolation, characterization and deletion mapping of amber mutations in the cll gene of phage lambda. Virology. 1975 Jan;63(1):147–159. doi: 10.1016/0042-6822(75)90380-3. [DOI] [PubMed] [Google Scholar]
- Belfort M. The cII-independent expression of the phage lambda int gene in RNase III-defective E. coli. Gene. 1980 Oct;11(1-2):149–155. doi: 10.1016/0378-1119(80)90094-3. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bovre K., Szybalski W. Patterns of convergent and overlapping transcription within the b2 region of coliphage lambda. Virology. 1969 Aug;38(4):614–626. doi: 10.1016/0042-6822(69)90181-0. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Chung S., Echols H. Positive regulation of integrative recombination by the cII and cIII genes of bacteriophase lambda. Virology. 1977 Jun 15;79(2):312–319. doi: 10.1016/0042-6822(77)90358-0. [DOI] [PubMed] [Google Scholar]
- Court D., de Crombrugghe B., Adhya S., Gottesman M. Bacteriophage lambda hin function. II. Enhanced stability of lambda messenger RNA. J Mol Biol. 1980 Apr 25;138(4):731–743. doi: 10.1016/0022-2836(80)90062-5. [DOI] [PubMed] [Google Scholar]
- Davies R. W. DNA sequence of the int-xis-Pi region of the bacteriophage lambda; overlap of the int and xis genes. Nucleic Acids Res. 1980 Apr 25;8(8):1765–1782. doi: 10.1093/nar/8.8.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Effect of RNAase III, cleavage on translation of bacteriophage T7 messenger RNAs. J Mol Biol. 1975 Dec 15;99(3):487–499. doi: 10.1016/s0022-2836(75)80140-9. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Nucleotide sequence from the genetic left end of bacteriophage T7 DNA to the beginning of gene 4. J Mol Biol. 1981 Jun 5;148(4):303–330. doi: 10.1016/0022-2836(81)90178-9. [DOI] [PubMed] [Google Scholar]
- Epp C., Pearson M. L., Enquist L. Downstream regulation of int gene expression by the b2 region in phage lambda. Gene. 1981 May;13(4):327–337. doi: 10.1016/0378-1119(81)90012-3. [DOI] [PubMed] [Google Scholar]
- Feiss M., Adyha S., Court D. L. Isolation of plaque-forming, galactose-transducing strains of phage lambda. Genetics. 1972 Jun;71(2):189–206. doi: 10.1093/genetics/71.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin N. C. Altered reading of genetic signals fused to the N operon of bacteriophage lambda: genetic evidence for modification of polymerase by the protein product of the N gene. J Mol Biol. 1974 Oct 15;89(1):33–48. doi: 10.1016/0022-2836(74)90161-2. [DOI] [PubMed] [Google Scholar]
- Gingery R., Echols H. Mutants of bacteriophage lambda unable to integrate into the host chromosome. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1507–1514. doi: 10.1073/pnas.58.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarneros G., Galindo J. M. The regulation of integrative recombination by the b2 region and the cII gene of bacteriophage lambda. Virology. 1979 May;95(1):119–126. doi: 10.1016/0042-6822(79)90406-9. [DOI] [PubMed] [Google Scholar]
- Herskowitz I., Hagen D. The lysis-lysogeny decision of phage lambda: explicit programming and responsiveness. Annu Rev Genet. 1980;14:399–445. doi: 10.1146/annurev.ge.14.120180.002151. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Foeller C., Bidwell K., Landy A. Site-specific recombination functions of bacteriophage lambda: DNA sequence of regulatory regions and overlapping structural genes for Int and Xis. Proc Natl Acad Sci U S A. 1980 May;77(5):2482–2486. doi: 10.1073/pnas.77.5.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. L., Ross W., Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980 May 8;285(5760):85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KELLENBERGER G., ZICHICHI M. L., WEIGLE J. A mutation affecting the DNA content of bacteriophage lambda and its lysogenizing properties. J Mol Biol. 1961 Aug;3:399–408. doi: 10.1016/s0022-2836(61)80053-3. [DOI] [PubMed] [Google Scholar]
- Katzir N., Oppenheim A., Belfort M., Oppenheim A. B. Activation of the lambda int gene by the cii and ciii gene products. Virology. 1976 Oct 15;74(2):324–331. doi: 10.1016/0042-6822(76)90339-1. [DOI] [PubMed] [Google Scholar]
- Kravchenko V. V., Vasilenko S. K., Grachev M. A. A rightward promoter to the left of the att site of lambda phage DNA: possible participant in site-specific recombination. Gene. 1979 Nov;7(3-4):181–195. doi: 10.1016/0378-1119(79)90045-3. [DOI] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly K. F., Signer E. R., Radding C. M. Nonessential functions of bacteriophage lambda. Virology. 1969 Feb;37(2):177–188. doi: 10.1016/0042-6822(69)90197-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. I., Abraham J., Benedik M., Campbell A., Court D., Echols H., Fischer R., Galindo J. M., Guarneros G., Hernandez T. Regulation of the integration-excision reaction by bacteriophage lambda. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):439–445. doi: 10.1101/sqb.1981.045.01.058. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Integration and excision of bacteriophage lambda. Curr Top Microbiol Immunol. 1977;78:171–199. doi: 10.1007/978-3-642-66800-5_6. [DOI] [PubMed] [Google Scholar]
- Oppenheim A., Oppenheim A. B. Regulation of the int gene of bacteriophage lambda: activation by the cII and cIII gene products and the role of the Pi and Pl promoters. Mol Gen Genet. 1978 Sep 20;165(1):39–46. doi: 10.1007/BF00270374. [DOI] [PubMed] [Google Scholar]
- Roehrdanz R. L., Dove W. F. A factor in the b2 region affecting site-specific recombinations in lambda. Virology. 1977 Jun 1;79(1):40–49. doi: 10.1016/0042-6822(77)90332-4. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Ross W., Landy A., Kikuchi Y., Nash H. Interaction of int protein with specific sites on lambda att DNA. Cell. 1979 Oct;18(2):297–307. doi: 10.1016/0092-8674(79)90049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUSSMAN R., JACOB F. [On a thermosensitive repression system in the Escherichia coli lambda bacteriophage]. C R Hebd Seances Acad Sci. 1962 Feb 19;254:1517–1519. [PubMed] [Google Scholar]
- Salstrom J. S., Szybalski W. Coliphage lambdanutL-: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J Mol Biol. 1978 Sep 5;124(1):195–221. doi: 10.1016/0022-2836(78)90156-0. [DOI] [PubMed] [Google Scholar]
- Salstrom J. S., Szybalski W. Transcription termination sites in the major leftward operon of coliphage lambda. Virology. 1978 Jul 15;88(2):252–262. doi: 10.1016/0042-6822(78)90282-9. [DOI] [PubMed] [Google Scholar]
- Schindler D., Echols H. Retroregulation of the int gene of bacteriophage lambda: control of translation completion. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4475–4479. doi: 10.1073/pnas.78.7.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeissner U., Court D., McKenney K., Rosenberg M. Positively activated transcription of lambda integrase gene initiates with UTP in vivo. Nature. 1981 Jul 9;292(5819):173–175. doi: 10.1038/292173a0. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A., Adhya S. L. The galactose operon of E. coli K-12. II. A deletion analysis of operon structure and polarity. Genetics. 1969 Jun;62(2):249–264. doi: 10.1093/genetics/62.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Campbell A. Int-constitutive mutants of bacteriophage lambda. Proc Natl Acad Sci U S A. 1974 Jan;71(1):237–241. doi: 10.1073/pnas.71.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman M. J., Hallick L. M., Echols H., Signer E. R. Properties of recombination-deficient mutants of bacteriophage lambda. J Mol Biol. 1970 Sep 28;52(3):501–520. doi: 10.1016/0022-2836(70)90416-x. [DOI] [PubMed] [Google Scholar]
- Signer E. R. On the control of lysogeny in phage lambda. Virology. 1970 Mar;40(3):624–633. doi: 10.1016/0042-6822(70)90207-2. [DOI] [PubMed] [Google Scholar]
- Simatake H., Rosenberg M. Purified lambda regulatory protein cII positively activates promoters for lysogenic development. Nature. 1981 Jul 9;292(5819):128–132. doi: 10.1038/292128a0. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Weisberg R. A., Gallant J. A. Dual function of the lambda prophage repressor. J Mol Biol. 1967 May 14;25(3):537–544. doi: 10.1016/0022-2836(67)90204-5. [DOI] [PubMed] [Google Scholar]
- Wilder D. A., Lozeron H. A. Differential modes of processing and decay for the major N-dependent RNA transcript of coliphage lambda. Virology. 1979 Dec;99(2):241–256. doi: 10.1016/0042-6822(79)90004-7. [DOI] [PubMed] [Google Scholar]


