Abstract
Parvoviruses cause a variety of mild to severe symptoms or asymptomatic infections in humans and animals. During a viral metagenomic analysis of feces from children with acute diarrhea in Burkina Faso, we identified in decreasing prevalence nucleic acids from anelloviruses, dependoviruses, sapoviruses, enteroviruses, bocaviruses, noroviruses, adenoviruses, parechoviruses, rotaviruses, cosavirus, astroviruses, and hepatitis B virus. Sequences from a highly divergent parvovirus, provisionally called bufavirus, were also detected whose NS1 and VP1 proteins showed <39% and <31% identities to those of previously known parvoviruses. Four percent of the fecal samples were PCR positive for this new parvovirus, including a related bufavirus species showing only 72% identity in VP1. The high degree of genetic divergence of these related genomes from those of other parvoviruses indicates the presence of a proposed new Parvoviridae genus containing at least two species. Studies of the tropism and pathogenicity of these novel parvoviruses will be facilitated by the availability of their genome sequences.
INTRODUCTION
Metagenomic analyses of human fecal samples have shown frequent coinfections with known enteric pathogens (26, 70) and the presence of numerous novel viral species, particularly members of the Picornaviridae (20, 31, 33, 37, 40, 51), Astroviridae (26, 27, 41) Parvoviridae (39, 45), and Circoviridae (49, 58) families of small enteric viruses. Metagenomics studies of animal feces have similarly shown frequent coinfections and yielded highly divergent viral species (10, 16, 25, 30, 43, 44, 50, 52, 53, 57, 60, 61, 64).
Parvoviruses are small nonenveloped icosahedral viruses with linear single-stranded DNA genomes of 4.5 to 5.5 kb, infect a variety of mammals, and are associated with a wide spectrum of acute and chronic diseases (14, 32, 55). The family Parvoviridae is divided into two subfamilies based on their host range, Parvovirinae and Densovirinae, which infect vertebrate and nonvertebrate animals, respectively (46). Under the International Committee on Taxonomy of Viruses (ICTV), the mammal- and bird-infecting family Parvovirinae is currently divided into five genera, Bocavirus, Erythrovirus, Dependovirus, Amdovirus, and Parvovirus (46). Other, still unclassified, parvoviruses have also been reported (46). At least four different groups of parvoviruses have been reported to infect human (14). Adeno-associated viruses (AAV) in the Dependovirus genus, initially isolated in 1967 (9), were the first parvoviruses identified in humans and are not known to cause any symptoms. AAV are dependent on coinfection with a helper adenovirus or herpesvirus for efficient replication. The second identified human parvovirus, B19V, was identified in 1975 (22) and sequenced in 1984 (23). B19V consists of three genotypes and is the prototype of the Erythrovirus genus, which includes related viruses infecting nonhuman primates and chipmunks (46). B19V infection in utero can cause fetal hydrops and developmental abnormalities (4, 15), while infection of children typically leads to the minor rash erythema infectiosum (5, 6, 59). In children and adults with sickle cell anemia, B19V can cause arrest of erythropoiesis and chronic anemia in AIDS patients (28). In 2005, a human bocavirus was characterized in respiratory secretions of Swedish children (2) and has been associated with lower respiratory tract infections worldwide, often in combination with other viral infections (1, 21, 35, 38, 62). Related parvoviruses named human bocaviruses 2 to 4 have also been detected in the feces of children with acute flaccid paralysis (39, 45) and diarrhea (7). Members of the Bocavirus genus also infect bats (GenBank accession no. JQ814850), cows (17), dogs (42, 63), pigs (11, 19, 24, 48, 64), chimpanzees (67), and gorillas (43). Human parvovirus 4 (PARV4) was initially described in 2007 in the plasma of a febrile injection drug user (36). Because of the high prevalence of anti-PARV4 antibodies in hemophiliacs using plasma pool-derived coagulation factors and in hepatitis C virus (HCV)- and HIV-positive injection drug users, the transmission of PARV4 in developed countries is thought to be mainly parenteral (29, 54, 66, 72, 73). The detection of PARV4 DNA in the blood of sub-Saharan African children indicates that other routes of PARV4 transmission may exist (56, 68). Related viruses have been detected in chimpanzees (67), baboons (67), bats (16), sheep (69), pigs (47), and cows (47), and a Parvoviridae genus named Partetravirus has been proposed (69). PARV4 has been reported in patients with different symptoms, but the full extent of its pathogenicity remains uncertain (8, 18, 36, 65).
Here we report on a viral metagenomic analysis of feces of children from Burkina Faso with acute diarrhea (12) allowing the characterization of their enteric viruses, including the genetic characterization of novel parvoviruses from a previously unrecognized genus (12, 13).
MATERIALS AND METHODS
Biological samples.
A prior study analyzing feces from children <5 years of age with acute diarrhea reported that 34% of them were positive for rotavirus by an immunochromatographic assay (SD Bioline Rota/Adeno; Standard Diagnostics, Inc., Kyonggi-Do, South Korea) (12, 13). Samples were collected between November 2008 and February 2010 from the capital city of Ouagadougou in Burkina Faso (12). Rotavirus antigen-negative samples were used in this study approved by the University of California at San Francisco committee on human research.
Viral metagenomics.
Viral particles were first enriched by filtration and nuclease treatment to digest non-particle-protected nucleic acids (3). Nonspecific RNA and DNA amplification was then performed by random reverse transcription (RT)-PCR using primers with randomized 3′ ends (70). Forty-nine different primers with distinct 5′ regions were used to molecularly label 49 random RT-PCRs. Amplicons were then pooled, and 454 libraries were generated and pyrosequenced using the 454 Titanium FLX+ sequencer (70). Each of 49 molecular tags was used on two fecal samples, for a total of 98 diarrhea samples. Because fecal samples were analyzed in pairs, the viruses identified are reported per sample pair rather than for individual samples. The 454 singlet reads and assembled contigs of >100 bp were compared to the GenBank protein databases using BLASTx.
Bufavirus PCR.
Primers BF.F1 (5′-TCAACAATCACTCAGGCAAATGG-3′) and BR.R1 (5′-AGTTTGCCTGGATGTTCTTTGA-3′) were used for the first round of PCR, and primers BF.F2 (5′-CTAACACTGGTACTTGCTATGGAC-3′) and BF.R2 (5′-TTCTCTGGTGATGATTCTTTTGTC-3′) were used for the nested PCR round, resulting in an expected amplicon of ∼440 bp. The PCR protocol for the first and second rounds was as follows: denaturation at 95°C for 5 min and 35 cycles of 95°C for 30s, 48°C for 30s, and 72°C for 1 min.
RESULTS
Viral particles in fecal samples from children with acute diarrhea were first enriched by filtration. Non-particle-protected nucleic acids were digested with nucleases to reduce the nonviral background, and the remaining nucleic acids were extracted, amplified by random RT-PCR, tagged, and then pyrosequenced, generating over half a million reads (see Materials and Methods). Using a BLASTx E score cutoff of 10−5, we identified 24,626 reads to known human viruses whose translated sequences were >90% identical to those of viral sequences already in GenBank. The distributions of these viral hits among the 49 pairs of samples analyzed are shown in Table 1. Because diarrhea samples were pooled in pairs prior to random RT-PCR, viruses detected could only be assigned to pairs of samples. The most common human viral sequences amplified were those of members of the Anelloviridae family (9,093 reads), sapoviruses (6,586 reads), dependovirus (aka AAV) (2,798 reads), bocaviruses (2,773 reads), enteroviruses (2,349 reads), noroviruses (682 reads), human astroviruses (194 reads), parechovirus (55 reads), rotaviruses (41 reads), hepatitis B virus (HBV; 30 reads), adenoviruses (23 reads), cosaviruses (5 reads), and human astrovirus HMO-A/VA2 (2 reads) (Table 1). The most prevalent enteric viruses detected were Anelloviridae (found in 34 sample pairs), dependoviruses (12 sample pairs), sapoviruses (10 sample pairs), enteroviruses (9 sample pairs), bocaviruses (8 sample pairs), noroviruses (7 sample pairs), adenoviruses (5 sample pairs), parechoviruses (3 sample pairs), rotaviruses (2 sample pairs), cosavirus (2 sample pairs), astroviruses (1 sample pair), human astrovirus HMO-A/VA2 (1 sample pair), and HBV (1 sample pair) (Table 1). From 0 to 6 eukaryotic viruses (average, 1.9) were detected in fecal sample pairs. When only pathogens known to cause diarrhea were included (counting sapovirus, enterovirus, norovirus, human astrovirus, parechovirus, rotavirus, and adenovirus but excluding Anelloviridae, dependoviruses, bocaviruses, cosaviruses, human astrovirus HMO-A/VA2, and HBV) from 0 to 3 pathogens (average, 0.77) were detected per sample pair at this depth of sequencing (Table 1).
Table 1.
Distribution of sequence reads to different viral species and total of human viruses in 49 sample pairs
| Sample paira | No. of human viruses detected | No. of known human pathogens | Anelloviridae (n = 34) | Dependovirus (n = 12) | Sapovirus (n = 10) | Enterovirus (n = 9) | Bocavirus (n = 8) | Norovirus (n = 7) | Adenovirus (n = 5) | Parechovirus (n = 3) | Rotavirus (n = 2) | Astrovirus (n = 2) | Cosavirus (n = 2) | HBV (n = 1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27 | 6 | 3 | 220 | 1,025 | 11 | 1 | 1 | 19 | ||||||
| 37 | 4 | 1 | 794 | 1 | 1 | 30 | ||||||||
| 47 | 4 | 2 | 6 | 1 | 3 | 38 | ||||||||
| 96 | 4 | 2 | 2 | 1 | 3 | 1 | ||||||||
| 20 | 3 | 1 | 210 | 1 | 15 | |||||||||
| 43 | 3 | 2 | 128 | 1,639 | 194b | |||||||||
| 28 | 3 | 1 | 32 | 1 | 38 | |||||||||
| 30 | 3 | 2 | 31 | 7 | 4 | |||||||||
| 22 | 3 | 1 | 27 | 1 | 663 | |||||||||
| 7 | 3 | 2 | 17 | 1 | 35 | |||||||||
| 13 | 3 | 0 | 16 | 1,309 | 2c | |||||||||
| 49 | 3 | 1 | 1 | 11 | 6 | |||||||||
| 26 | 3 | 3 | 1 | 1 | 1 | |||||||||
| 41 | 3 | 2 | 24 | 16 | 1 | |||||||||
| 44 | 2 | 1 | 2,379 | 1 | ||||||||||
| 11 | 2 | 1 | 1,446 | 1 | ||||||||||
| 4 | 2 | 1 | 1,193 | 19 | ||||||||||
| 38 | 2 | 0 | 134 | 2 | ||||||||||
| 15 | 2 | 1 | 112 | 1 | ||||||||||
| 10 | 2 | 0 | 93 | 1 | ||||||||||
| 3 | 2 | 1 | 21 | 1 | ||||||||||
| 33 | 2 | 1 | 17 | 1 | ||||||||||
| 19 | 2 | 1 | 13 | 13 | ||||||||||
| 42 | 2 | 2 | 13 | 647 | ||||||||||
| 17 | 2 | 0 | 6 | 95 | ||||||||||
| 35 | 2 | 0 | 5 | 1 | ||||||||||
| 34 | 2 | 1 | 3 | 1 | ||||||||||
| 32 | 2 | 2 | 11 | 6 | ||||||||||
| 8 | 2 | 1 | 43 | 1,227 | ||||||||||
| 23 | 2 | 0 | 2 | 1,375 | ||||||||||
| 25 | 2 | 0 | 2 | 9 | ||||||||||
| 31 | 1 | 0 | 1,799 | |||||||||||
| 18 | 1 | 0 | 182 | |||||||||||
| 29 | 1 | 0 | 98 | |||||||||||
| 21 | 1 | 0 | 35 | |||||||||||
| 12 | 1 | 0 | 24 | |||||||||||
| 1 | 1 | 0 | 16 | |||||||||||
| 2 | 1 | 0 | 12 | |||||||||||
| 5 | 1 | 0 | 4 | |||||||||||
| 6 | 1 | 0 | 4 | |||||||||||
| 9 | 1 | 1 | 6,488 | |||||||||||
| 86 | 1 | 0 | 36 | |||||||||||
| 45 | 1 | 1 | 7 | |||||||||||
| 40 | 1 | 0 | 452 | |||||||||||
| 14 | 0 | 0 | ||||||||||||
| 16 | 0 | 0 | ||||||||||||
| 24 | 0 | 0 | ||||||||||||
| 39 | 0 | 0 | ||||||||||||
| 48 | 0 | 0 | ||||||||||||
| Total no. of reads | 95 | 38 | 9,093 | 2,798 | 6,586 | 2,349 | 2,773 | 682 | 23 | 55 | 41 | 196 | 16 | 30 |
The sample pair underlined contained the initial bufavirus sequence reads. The samples shaded in gray were positive for bufavirus by nested PCR.
HAstV.
HMO-A/VA2.
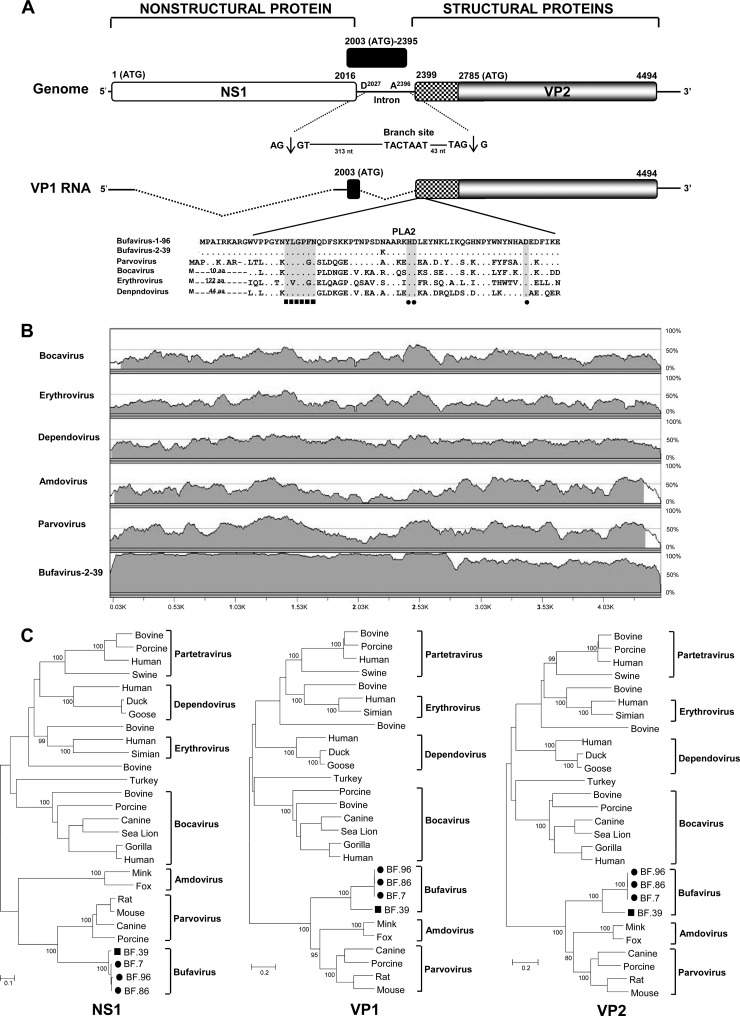
Ten sequences out of a total of 1,057 contigs plus singlets from one specimen pair (Table 1, pair 96) showed significant similarities (E values, 1 × 10−2 to 4 × 10−28) to parvovirus proteins. Gaps between pyrosequences were filled by PCR, and 5′ and 3′ regions were amplified by rapid amplification of cDNA ends. The nearly complete genome (4,921 bp) of bufavirus 1-BF96 (for Burkina Faso parvovirus species 1 strain 96), including a partial 5′ untranslated region (UTR) (18 bp), the complete NS1 sequence (671 aa), the complete VP1 (707 aa) and VP2 (569 aa) sequences, and a partial 3′ UTR (400 bp), was acquired (GenBank accession no. JQ918261). The genome contained two major open reading frames (ORFs), with the left and right ORFs encoding NS1 and VP1, respectively (Fig. 1A). A middle ORF (130 aa) was also found that did not show any similarity to other parvovirus genomes by BLAST (Fig. 1A). Sequence alignment showed a high degree of sequence divergence, with <50% overall nucleotide identity to other parvoviruses (Fig. 1B). The putative bufavirus NS1 start codon was located in a strong Kozak sequence, CACCATGG. The ATP- or GTP-binding Walker loop motif (GXXXXGK[T/S]) was found in NS1 (402GPASTGKS409) (71). In addition, NS1 also contained two conserved replication initiator motifs, GLHIHVLVC and IANYFLIKKP (where conserved amino acids are in boldface type) (34). A sequence identity matrix was then generated (Table 2). NS1 showed less than 39% identity with other parvoviral NS1 sequences, including its closest relatives in the Parvovirus genus.
Fig 1.
Bufavirus genome and phylogeny. (A) Organization of the bufavirus genome. The PLA2 similarity region, including the calcium-binding region and catalytic residues, is shown. Theoretical splicing for expression of VP1 is shown. nt, nucleotides. (B) Pairwise sliding window of percent nucleotide identity of bufavirus aligned with representatives of other parvovirus genera and bufavirus 2. (C) Phylogenetic analyses of the NS1, VP1, and VP2 proteins of bufavirus and related parvoviruses. Genetic distances were calculated by Kimura's two-parameter method (PHYLIP), and a phylogenetic tree with 100 bootstrap resampling of the alignment data sets was generated using the neighbor-joining method. Each scale indicates the number of amino acid substitutions per position. Proposed and official Parvoviridae genera are labeled. The names and accession numbers of the taxa used are listed in Table S1 in the supplemental material.
Table 2.
Pairwise percent amino acid sequence identities among NS1, VP1, and VP2 regions of bufavirus 1-BF96 and the prototypes of different parvovirus genera
| Protein and virus | % Amino acid sequence identity |
|||||
|---|---|---|---|---|---|---|
| Bufavirus | Bocavirus | Erythrovirus | Dependovirus | Amdovirus | Parvovirus | |
| NS1 | ||||||
| Bufavirus | 100 | |||||
| Bocavirus | 14 | 100 | ||||
| Erythrovirus | 13 | 15 | 100 | |||
| Dependovirus | 18 | 15 | 20 | 100 | ||
| Amdovirus | 18 | 15 | 11 | 14 | 100 | |
| Parvovirus | 38 | 16 | 13 | 18 | 18 | 100 |
| VP1 | ||||||
| Bufavirus | 100 | |||||
| Bocavirus | 17 | 100 | ||||
| Erythrovirus | 10 | 13 | 100 | |||
| Dependovirus | 15 | 24 | 19 | 100 | ||
| Amdovirus | 26 | 10 | 10 | 10 | 100 | |
| Parvovirus | 31 | 19 | 10 | 17 | 28 | 100 |
| VP2 | ||||||
| Bufavirus | 100 | |||||
| Bocavirus | 14 | 100 | ||||
| Erythrovirus | 10 | 15 | 100 | |||
| Dependovirus | 14 | 21 | 20 | 100 | ||
| Amdovirus | 32 | 13 | 10 | 13 | 100 | |
| Parvovirus | 29 | 14 | 12 | 14 | 32 | 100 |
Two potential splice sites were detected, a potential donor site (AG↓GT) at nucleotide 2027 and an acceptor site (AG↓G) at nucleotide 2396. The putative VP1 sequence started at the first ATG of the middle ORF at nucleotide 2003 upstream of a splice donor site at nucleotide 2027 (Fig. 1A). The phospholipase A2 (PLA2) motif (Fig. 1A), with its highly conserved calcium-binding site (YLGPF), was found in the main ORF of VP1. The phospholipase catalytic residues (HD and D) were present at amino acid positions 40 to 41 and 62 (Fig. 1A). Bufavirus VP1 showed <31% amino acid identity to other genera of the family Parvovirinae, including its closest relative in the Parvovirus genus (Table 2). The N terminus of the bufavirus VP2 protein contained a glycine-rich sequence (GGGGGGGGSGVG) also present in other parvoviral VP2 proteins. The VP2 protein was most closely related to those of amdoviruses (Table 2). Amdoviruses lack the PLA2 motif found in the N terminus of VP1 in bufavirus and other parvoviruses. Three tandem repeats of the sequence TAGTTGATAAGT were seen in the 400-base-long 3′ UTR. Two other repeats, TAGTTTATAAGT and TAGTTTATAAAT (where mutated bases are in boldface italics), were also found in the 3′ UTR region. Such repeated sequences were not reported in other parvoviruses.
The three major proteins of bufavirus were aligned with those of other parvoviruses and phylogenetically analyzed (Fig. 1C). Bufavirus clustered with the members of the Parvovirus genus in NS1 but was basal to both the Parvovirus and Amdovirus genera in its capsid region. The VP2 protein identity among the five ICTV-approved genera of the subfamily Parvovirinae ranged from 10% to 28%. Bufavirus showed a range of 10% to 31% identity to the VP2 proteins of these five genera and may therefore qualify as the reference genome for a novel genus.
To investigate the prevalence of this novel parvovirus, nested PCR primers targeting the NS1 region were designed. Bufavirus DNA was detected in an additional 3 out of 98 fecal samples from children with acute gastroenteritis in Burkina Faso, with a total frequency of virus detection of 4% (4/98). PCR amplicons were directly sequenced to confirm their identification. The complete protein-coding regions of these three related viral genomes were then determined. The coding regions of two strains, BF7 (GenBank accession no. JX027295) and BF86 (GenBank accession no. JX027296), showed a high level of nucleotide identity (99%) to prototype bufavirus 1-BF96, but the coding regions of the bufavirus BF39 genome (GenBank accession no. JX027297) showed an overall nucleotide identity of only 87%. The VP2 proteins of the three closely related strains BF96, BF7, and BF86 showed only 72% amino acid identity to that of strain BF39, indicating that it might represent a second species in the proposed genus. The NS1 of BF39 was >95% identical to those of the other bufaviruses. We therefore provisionally named this genome bufavirus 2-BF39 to reflect the presence of a second tentative species highly divergent in its VP2 protein sequence. The high degree of identity between the two species in their NS genes relative to their more divergent capsid genes may indicate that the VP region was acquired by recombination from a still uncharacterized viral genome or that the structural region diverged at a much higher rate than the nonstructural region. An alignment of NS1 and VP1 from both bufavirus species with the most closely related parvoviruses is shown in Fig. S1 in the supplemental material.
In order to investigate the wider distribution of bufavirus, we tested fecal specimens collected from children with diarrhea in Chile (n = 100) and with nonpolio acute flaccid paralysis in Tunisia (n = 63) by nPCR. One bufavirus 1 was amplified from the feces of one Tunisian child, indicating that these viruses are not geographically restricted.
DISCUSSION
A metagenomic analysis showed the presence of a diverse enteric virome in diarrheic children <5 years old in Burkina Faso. A prior study of this population showed 34% of the diarrhea cases to involve rotaviruses (12). The detection of two samples containing rotavirus sequences despite the exclusion of rotavirus-positive samples likely reflects viral loads below the assay's antigen detection level. The most common infections detected were of anelloviruses and dependoviruses, neither of which are recognized pathogens, likely reflecting common and/or chronic commensal intestinal infections in these children. The hierarchy of the next most common viruses detected was sapoviruses > enteroviruses > bocaviruses > noroviruses > adenoviruses > parechoviruses > astroviruses > cosaviruses, most of which have been associated with diarrhea, while a few of the more recently characterized viruses are of unknown pathogenicity (bocaviruses, astrovirus HMO-A/VA2, cosaviruses). The detection of blood-borne HBV DNA sequences may be the result of bleeding into the intestinal tract. The metagenomic detection of bufavirus sequences in only one sample while three more samples were positive by nested PCR reflects the lower sensitivity of pyrosequencing at the depth used here. The rate of infection with other viruses measured here by metagenomics is therefore likely also an underestimate of their actual prevalence in feces.
A potential new genus in the Parvoviridae family was genetically characterized, and a PCR survey showed a prevalence of 4% among the rotavirus antigen-negative cases of childhood diarrhea. The genetic characterization of a second Bufavirus species with a highly divergent capsid gene indicates that wider geographic sampling for related viruses will likely reveal other related species. The genetic diversity within this proposed genus may also indicate a range of phenotypes upon their hosts.
Wider geographic sampling of human and animal fecal samples will provide a better description of the genetic diversity of this proposed Parvoviridae genus. Serological assays will help determine whether bufaviruses infect humans or are simply passing through the gut from a dietary source. Case-control studies comparing the presence and viral loads of bufavirus DNA in feces or the prevalence of anti-bufavirus IgM will also help determine whether members of this viral clade are associated with diarrhea or other symptoms.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge National Heart, Lung, and Blood Institute R01HL083254 and support from the Blood Systems Research Institute to E.D.
Footnotes
Published ahead of print 1 August 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Allander T. 2008. Human bocavirus. J. Clin. Virol. 41:29–33 [DOI] [PubMed] [Google Scholar]
- 2. Allander T, et al. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 102:12891–12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. U. S. A. 98:11609–11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anand A, Gray ES, Brown T, Clewley JP, Cohen BJ. 1987. Human parvovirus infection in pregnancy and hydrops fetalis. N. Engl. J. Med. 316:183–186 [DOI] [PubMed] [Google Scholar]
- 5. Anderson MJ, Lewis E, Kidd IM, Hall SM, Cohen BJ. 1984. An outbreak of erythema infectiosum associated with human parvovirus infection. J. Hyg. 93:85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson MJ, et al. 1985. Experimental parvoviral infection in humans. J. Infect. Dis. 152:257–265 [DOI] [PubMed] [Google Scholar]
- 7. Arthur JL, Higgins GD, Davidson GP, Givney RC, Ratcliff RM. 2009. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog. 5:e1000391 doi:10.1371/journal.ppat.1000391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benjamin LA, et al. 2011. Human parvovirus 4 as potential cause of encephalitis in children, India. Emerg. Infect. Dis. 17:1484–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blacklow NR, Hoggan MD, Rowe WP. 1967. Isolation of adenovirus-associated viruses from man. Proc. Natl. Acad. Sci. U. S. A. 58:1410–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blinkova O, et al. 2010. Novel circular DNA viruses in stool samples of wild-living chimpanzees. J. Gen. Virol. 91:74–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blomström AL, et al. 2010. Studies of porcine circovirus type 2, porcine boca-like virus and torque teno virus indicate the presence of multiple viral infections in postweaning multisystemic wasting syndrome pigs. Virus Res. 152:59–64 [DOI] [PubMed] [Google Scholar]
- 12. Bonkoungou IJ, et al. 2010. Epidemiology of rotavirus infection among young children with acute diarrhoea in Burkina Faso. BMC Pediatr. 10:94 doi:10.1186/1471-2431-10-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bonkoungou IJ, et al. 2011. Genotype diversity of group A rotavirus strains in children with acute diarrhea in urban Burkina Faso, 2008-2010. J. Med. Virol. 83:1485–1490 [DOI] [PubMed] [Google Scholar]
- 14. Brown KE. 2010. The expanding range of parvoviruses which infect humans. Rev. Med. Virol. 20:231–244 [DOI] [PubMed] [Google Scholar]
- 15. Brown T, Anand A, Ritchie LD, Clewley JP, Reid TM. 1984. Intrauterine parvovirus infection associated with hydrops fetalis. Lancet ii:1033–1034 [DOI] [PubMed] [Google Scholar]
- 16. Canuti M, et al. 2011. Two novel parvoviruses in frugivorous New and Old World bats. PLoS One 6:e29140 doi:10.1371/journal.pone.0029140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen KC, et al. 1986. Complete nucleotide sequence and genome organization of bovine parvovirus. J. Virol. 60:1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen MY, Yang SJ, Hung CC. 2011. Placental transmission of human parvovirus 4 in newborns with hydrops, Taiwan. Emerg. Infect. Dis. 17:1954–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheung AK, et al. 2010. Identification and molecular cloning of a novel porcine parvovirus. Arch. Virol. 155:801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chiu CY, et al. 2008. Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proc. Natl. Acad. Sci. U. S. A. 105:14124–14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chow BD, Esper FP. 2009. The human bocaviruses: a review and discussion of their role in infection. Clin. Lab Med. 29:695–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cossart YE, Field AM, Cant B, Widdows D. 1975. Parvovirus-like particles in human sera. Lancet i:72–73 [DOI] [PubMed] [Google Scholar]
- 23. Cotmore SF, Tattersall P. 1984. Characterization and molecular cloning of a human parvovirus genome. Science 226:1161–1165 [DOI] [PubMed] [Google Scholar]
- 24. Cságola A, et al. 2012. Detection, prevalence and analysis of emerging porcine parvovirus infections. Arch. Virol. 157:1003–1010 [DOI] [PubMed] [Google Scholar]
- 25. Donaldson EF, et al. 2010. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 84:13004–13018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Finkbeiner SR, Kirkwood CD, Wang D. 2008. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol. J. 5:117 doi:10.1186/1743-422X-5-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Finkbeiner SR, et al. 2009. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J. Virol. 83:10836–10839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frickhofen N, et al. 1990. Persistent B19 parvovirus infection in patients infected with human immunodeficiency virus type 1 (HIV-1): a treatable cause of anemia in AIDS. Ann. Intern. Med. 113:926–933 [DOI] [PubMed] [Google Scholar]
- 29. Fryer JF, Lucas SB, Padley D, Baylis SA. 2007. Parvoviruses PARV4/5 in hepatitis C virus-infected patient. Emerg. Infect. Dis. 13:175–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ge X, et al. 2011. Genetic diversity of novel circular ssDNA viruses in bats in China. J. Gen. Virol. 92(Pt 11):2646–2653 [DOI] [PubMed] [Google Scholar]
- 31. Greninger AL, et al. 2009. The complete genome of klassevirus—a novel picornavirus in pediatric stool. Virol. J. 6:82 doi:10.1186/1743-422X-6-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoelzer K, Parrish CR. 2010. The emergence of parvoviruses of carnivores. Vet. Res. 41:39 doi:10.1051/vetres/2010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holtz LR, et al. 2009. Klassevirus 1, a previously undescribed member of the family Picornaviridae, is globally widespread. Virol. J. 6:86 doi:10.1186/1743-422X-6-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ilyina TV, Koonin EV. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jartti T, et al. 2012. Human bocavirus-the first 5 years. Rev. Med. Virol. 22:46–64 [DOI] [PubMed] [Google Scholar]
- 36. Jones MS, et al. 2005. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 79:8230–8236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones MS, Lukashov VV, Ganac RD, Schnurr DP. 2007. Discovery of a novel human picornavirus in a stool sample from a pediatric patient presenting with fever of unknown origin. J. Clin. Microbiol. 45:2144–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kahn J. 2008. Human bocavirus: clinical significance and implications. Curr. Opin. Pediatr. 20:62–66 [DOI] [PubMed] [Google Scholar]
- 39. Kapoor A, et al. 2009. A Newly Identified Bocavirus Species in Human Stool. J. Infect. Dis. 199:196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kapoor A, et al. 2008. A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc. Natl. Acad. Sci. U. S. A. 105:20482–20487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kapoor A, et al. 2009. Multiple novel astrovirus species in human stool. J. Gen. Virol. 90:2965–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kapoor A, et al. 2012. Characterization of novel canine bocaviruses and their association with respiratory disease. J. Gen. Virol. 93:341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kapoor A, et al. 2010. Identification and characterization of a new bocavirus species in gorillas. PLoS One 5:e11948 doi:10.1371/journal.pone.0011948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kapoor A, et al. 2011. Characterization of a canine homolog of human Aichivirus. J. Virol. 85:11520–11525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kapoor A, et al. 2010. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 201:1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. King AMQ, Lefkowitz EJ, Adams MJ, Carstens EB. 2012. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, New York, NY [Google Scholar]
- 47. Lau SK, et al. 2008. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J. Gen. Virol. 89:1840–1848 [DOI] [PubMed] [Google Scholar]
- 48. Li B, et al. 2012. Complete genome sequence of a novel species of Porcine Bocavirus, PBoV5. J. Virol. 86:1286–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li L, et al. 2010. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J. Virol. 84:1674–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li L, et al. 2010. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 84:6955–6965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li L, et al. 2009. A Novel Picornavirus Associated With Gastroenteritis. J. Virol. 83:12002–12006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li L, et al. 2011. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 92:2534–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li L, et al. 2011. The fecal viral flora of California sea lions. J. Virol. 85:9909–9917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Manning A, Willey SJ, Bell JE, Simmonds P. 2007. Comparison of tissue distribution, persistence, and molecular epidemiology of parvovirus B19 and novel human parvoviruses PARV4 and human bocavirus. J. Infect. Dis. 195:1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Manteufel J, Truyen U. 2008. Animal bocaviruses: a brief review. Intervirology 51:328–334 [DOI] [PubMed] [Google Scholar]
- 56. Panning M, et al. 2010. Novel human parvovirus 4 genotype 3 in infants, Ghana. Emerg. Infect. Dis. 16:1143–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Phan TGAK, et al. 2011. The Fecal Viral Flora of Wild Rodents. PLoS Pathog. 7:e1002218 doi:10.1371/journal.ppat.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Phan TG, et al. 2012. A third gyrovirus species in human faeces. J. Gen. Virol. 93(Pt 6):1356–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Plummer FA, et al. 1985. An erythema infectiosum-like illness caused by human parvovirus infection. N. Engl. J. Med. 313:74–79 [DOI] [PubMed] [Google Scholar]
- 60. Reuter G, et al. 2012. Astrovirus in wild boars (Sus scrofa) in Hungary. Arch. Virol. 157:1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reuter G, Pankovics P, Delwart E, Boros Á. 2012. Identification of a novel astrovirus in domestic sheep in Hungary. Arch. Virol. 157:323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schildgen O, et al. 2008. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin. Microbiol. Rev. 21:291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schwartz D, Green B, Carmichael LE, Parrish CR. 2002. The canine minute virus (minute virus of canines) is a distinct parvovirus that is most similar to bovine parvovirus. Virology 302:219–223 [DOI] [PubMed] [Google Scholar]
- 64. Shan T, et al. 2011. The fecal virome of pigs on a high-density farm. J. Virol. 85:11697–11708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sharp CP, Lail A, Donfield S, Gomperts ED, Simmonds P. 2012. Virologic and clinical features of primary infection with human parvovirus 4 in subjects with hemophilia: frequent transmission by virally inactivated clotting factor concentrates. Transfusion 52:1482–1489 [DOI] [PubMed] [Google Scholar]
- 66. Sharp CP, et al. 2009. High frequencies of exposure to the novel human parvovirus PARV4 in hemophiliacs and injection drug users, as detected by a serological assay for PARV4 antibodies. J. Infect. Dis. 200:1119–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sharp CP, et al. 2010. Widespread infection with homologues of human parvoviruses B19, PARV4, and human bocavirus of chimpanzees and gorillas in the wild. J. Virol. 84:10289–10296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Simmonds P, et al. 2008. A third genotype of the human parvovirus PARV4 in sub-Saharan Africa. J. Gen. Virol. 89:2299–2302 [DOI] [PubMed] [Google Scholar]
- 69. Tse H, et al. 2011. Discovery and genomic characterization of a novel ovine partetravirus and a new genotype of bovine partetravirus. PLoS One 6:e25619 doi:10.1371/journal.pone.0025619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Victoria JG, et al. 2009. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol. 83:4642–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Walker JE, Saraste M, Runswick MJ, Gay NJ. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yang SJ, Hung CC, Chang SY, Lee KL, Chen MY. 2011. Immunoglobulin G and M antibodies to human parvovirus 4 (PARV4) are frequently detected in patients with HIV-1 infection. J. Clin. Virol. 51:64–67 [DOI] [PubMed] [Google Scholar]
- 73. Yu X, et al. 2012. High prevalence of human parvovirus 4 infection in HBV and HCV infected individuals in shanghai. PLoS One 7:e29474 doi:10.1371/journal.pone.0029474 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.