Abstract
Latent membrane protein 1 (LMP1) of Epstein-Barr virus induces constitutive signaling in infected cells. LMP1 signaling requires oligomerization of LMP1 via its transmembrane domain, localization to lipid rafts in the membrane, and association of the LMP1 cytoplasmic domain to adaptor proteins, such as the tumor necrosis factor receptor-associated factors (TRAFs). Protein complementation is a novel technique to examine protein-protein interaction through the assembly of functional fluorescent proteins or enzymes from inactive fragments. A previous study in our lab demonstrated the use of bimolecular fluorescence complementation (BiFC) to study the assembly of the LMP1 signaling complexes within the plasma membrane of mammalian cells. In the present study, LMP1 was used as bait in a genome-wide BiFC screen with an enhanced retroviral mutagen to identify new LMP1-binding proteins. Our screen identified a novel LMP1-binding protein, transmembrane protein 134 (Tmem134). Tmem134 is a candidate oncogene that is amplified in breast cancer cell lines. Binding, colocalization, and cofractionation between LMP1 and Tmem134 were confirmed. Finally, Tmem134 affected LMP1-induced NF-κB induction. Together, these data suggest that BiFC is a unique and novel platform to identify proteins recruited to the LMP1-signaling complex.
INTRODUCTION
Epstein-Barr virus (EBV) is a large double-stranded DNA virus that is classified as a gamma-1 herpesvirus of the lymphocryptovirus genus (26, 40). EBV has infected greater than 90% of the world's population and is the etiologic agent of infectious mononucleosis (7, 20). EBV was the first virus to be associated with human cancer (13), and several latent viral proteins are consistently expressed in EBV-associated cancers, including the viral oncogene latent membrane protein 1 (LMP1) (26). EBV efficiently transforms B-lymphocytes in vitro and is associated with a number of epithelial and lymphoid malignancies (26, 39, 49, 50). EBV is associated with nearly all endemic Burkitt's lymphomas (30), T-cell and NK-cell lymphomas, and nasopharyngeal carcinomas (39) and is associated with a high percentage of Hodgkin's lymphomas (18) and gastric carcinomas (26). In the AIDS population, EBV-positive B-cell lymphomas are a significant cause of morbidity and mortality (5, 10). With the exception of Burkitt's lymphoma and gastric carcinoma, all other cancers associated with EBV express LMP1 (26). The correlation between the presence of EBV, expression of LMP1, and development of cancer indicates a role for EBV and LMP1 in oncogenesis and human disease.
LMP1 activates a number of signaling pathways, including nuclear factor kappa B (NF-κB), phosphatidylinositol 3-kinase (PI3K), and mitogen-activated protein kinases (MAPK) (8, 12, 23, 31, 34, 37, 41, 45). Signaling induced by LMP1 is analogous to constitutively activated tumor necrosis factor receptor (TNFR) signaling, and LMP1 serves as a model for TNFR signaling. B-cell transformation by EBV in vitro requires LMP1 (25). Transgenic mice engineered to express LMP1 in either the B cells or epithelial cells develop lymphomas or carcinomas, respectively (19, 27). LMP1 transforms rodent fibroblasts by conferring anchorage-independent growth and loss of contact inhibition (4, 15, 31, 35, 46). Fibroblasts expressing LMP1 form tumors in nude mice and can grow in reduced serum conditions. LMP1 alters the cellular environment by regulating the expression of a number of genes that regulate different aspects of cellular growth and transformation (14–16).
The LMP1 protein has a short cytoplasmic N terminus, a six-pass transmembrane domain (Tm), and a carboxyl-terminal signaling domain. At least three interrelated activities are required for LMP1 signaling: (i) the Tm domain oligomerizes, (ii) the cytoplasmic domains bind TNFR-associated factors (TRAFs), and (iii) LMP1 partitions into lipid rafts. Based on TNFR signaling, it is likely that oligomerization precedes TRAF binding and raft localization (6). However, it is unclear if TRAF binding is required for raft localization, if raft localization is required for TRAF binding, or if they are interdependent. Importantly, the cellular factors required for LMP1 trafficking and recruitment to lipid rafts in the presence or absence of the TRAFs are not known.
Our recently published study demonstrates the utility of using bimolecular fluorescence complementation (BiFC) to study the assembly of the LMP1 signaling complex in the membrane of mammalian cells (44). In BiFC, interacting proteins are expressed as fusion proteins with fragments of yellow fluorescence protein (YFP), amino-YFP (NYFP), or carboxyl-YFP (CYFP). Individual fusion proteins do not possess intrinsic fluorescence, but interaction between proteins leads to assembly of functional YFP which is detected by fluorescence techniques. BiFC was observed with LMP1 and TRAF2 or TRAF3. Mutation of CTAR1 and/or CTAR2 decreased BiFC of LMP1-plus-TRAF combinations. LMP1-plus-LMP1 BiFC was also observed. Both LMP1-plus-TRAF and LMP1-plus-LMP1 BiFC localized to perinuclear and membrane locations, which is consistent with previously described LMP-signaling complexes (21, 42). LMP1-NYFP induced NF-κB reporter activation and transformation of Rat-1 cells, suggesting that LMP1-NYFP can be used as a bait protein in a BiFC screen to identify LMP1-binding proteins that are important for signaling.
In the present study, we validate the use of BiFC as a platform to identify novel LMP1-binding proteins in the membrane of mammalian cells. We identified that transmembrane protein 134 (Tmem134) induces BiFC with LMP1. Tmem134 is a highly conserved, small membrane protein of unknown function and a candidate oncogene that is amplified in breast cancer cells. Tmem134-LMP1 binding was confirmed by coimmunoprecipitation, and Tmem134 and LMP1 colocalized in transfected cells. Tmem134 affected LMP1-induced NF-κB activity. Together, these data demonstrate the utility of using BiFC to screen for novel LMP1-binding proteins.
MATERIALS AND METHODS
Plasmids.
The inducible retrovirus vectors pRetroX-Tet-On Advanced, pRetroX-Tight-Hyg, pRetroX-Tight-Pur, and pRetroX-Tight-Hyg-Luc were purchased from Clontech. The enhanced retroviral mutagen (ERM) vectors VC1, VC2, and VC3, which encode a CYFP tag, were generously provided by Z. Ding and Z. Songyang (11, 28). Babe HA-LMP1 (N-terminal HA tag), M3-LMP1 (N-terminal triple myc tag), LMP1-NYFP (C-terminal NYFP tag), Babe LMP1-NYFP (LMP1-NYFP stable retrovirus vector), and CYFP-TRAF2 (N-terminal CYFP tag) were previously described (15, 33, 44). HA-LMP1 and LMP1-NYFP were cloned into the inducible retrovirus vector pRetroX-Tight-Hyg to produce HA-LMP1-TH and LMP1-NYFP-TH, respectively. CYFP-TRAF2 was cloned into RetroX-Tight-Pur as a BiFC-positive control to produce CYFP-TRAF2 TP. Rat transmembrane protein 134, which was isolated in the BiFC screen, was subcloned with CYFP-HA and HA tags from cDNA into pcDNA3 (Invitrogen).
TriFECTa Dicer-Substrate small interfering RNA (siRNA) for rat Tmem134 was purchased from Integrated DNA Technologies. After validation by knockdown of LMP1-Tmem134 BiFC, two Tmem134 siRNA sequences were cloned into pSilencer 5.1-U6 retro (Ambion). si-302 and si-308 target the sequences CCATGTGATCTTCATCTACTGTGCT and GGCACATGTCTATCCGTAGCTCCCA, respectively.
Cell culture, transfections, and retrovirus.
Human embryonic kidney cells (HEK-293T), SiHa, C33A, and Rat-1 (rodent fibroblasts) cells were maintained in Dulbecco modified Eagle medium (Mediatech) supplemented with antibiotic/antimycotic mixture and 10% (vol/vol) heat-inactivated fetal bovine serum (FBS). Caski, C666-1, BJAB, Raji, LCL, EF3D, Akata, Ramos, and Loukes cells were maintained in RPMI medium (Mediatech) supplemented with antibiotic/antimycotic mixture and 10% (vol/vol) heat-inactivated FBS. Cells were transfected with Transit-LT1 (Mirus) according to the manufacturer's directions. Retrovirus production was accomplished as previously described (16) by transfection of HEK-293T cells with retrovirus expression vectors with plasmids expressing VSV-G and gag-pol. Twenty-four hours posttransfection, the medium was changed and cells were moved to 33°C. Forty-eight hours posttransfection, clarified supernatants were collected and used to infect Rat-1 or C33A cells. Retrovirus infection was performed in the presence of 8 μg/ml Polybrene. Stably transduced cells were selected with G418 (neo) (1 mg/ml; Mediatech), hygromycin B (hygro) (0.5 mg/ml; Mediatech), or puromycin (puro) (5 μg/ml; Mediatech). Tetracycline-inducible promoters were induced with the less toxic tetracycline analog doxycycline (dox) (Clonetech). Inducible Rat-1 and C33A cells were created by infection with pRetroX-Tet-On Advanced retrovirus and selection with neo to yield Rat-1 Tet-On and C33A Tet-On cells, respectively. Tet-On cells were infected with the indicated pRetroX-Tight-Hyg or pRetroX-Tight-Pur retrovirus derivatives and selected with hygro or puro, respectively. Stable C33A cells were produced by infection with control (Babe) or Babe-LMP1 retrovirus and selection with puro.
Western blotting.
Cells were washed with ice-cold phosphate-buffered saline (PBS) (Mediatech) and lysed with radioimmunoprecipitation assay buffer (RIPA) (10 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 1% deoxycholic acid, protease and phosphatase inhibitors [Pierce]). Cell lysates were clarified by centrifugation and quantitated by the Bio-Rad DC protein assay system (Bio-Rad). Samples were then boiled in SDS sample buffer, and indicated amounts of protein were separated using SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (LiCor) for Western blotting. LMP1 was detected with a mixture of four rat monoclonal antibodies diluted 1:500 each (Cao 7E10, Cao 8G3, LMP1 IG6, and Cao 7G8; Ascenion GmbH). TRAF2 antibody was purchased from Santa Cruz, and HA tag and Actin antibodies were purchased from Cell Signaling. Fusion proteins were detected with myc-tag antibody (Upstate) and YFP antibodies (632381; Clontech). Western blot Guaranteed Tmem134 antibody was purchased from GenScript. Primary antibodies were detected with IRDye-labeled secondary antibodies (Li-Cor) and scanning with a Li-Cor Odyssey imaging system. Bands were quantitated using the Li-Cor imaging software.
Reporter assays.
Reporter assays were performed as previously described (16, 44). Rat-1 Tet-On cells were confirmed by transfection with RetroX-Tight-hygro-luc, which inducibly expresses firefly luciferase. Twenty-four hours posttransfection, cells were incubated in media in the absence or presence of 500 ng/ml dox. Forty hours posttransfection, cells were harvested and firefly luciferase activity was assayed using the dual-luciferase reporter assay system (Promega) according to the manufacturer's directions. Triplicate samples were analyzed for each condition.
Rat-1 Tet-On HA-LMP1-TH cells which inducibly express HA-LMP1 were transfected with siRNA expression plasmids NF-κB-luc (Stratagene) and RL-SV40 (control Renilla luciferase). Twenty-four hours posttransfection, cells were incubated in media in the absence or presence of dox. Forty hours posttransfection, cells were harvested and dual-luciferase assays were performed using the dual-luciferase reporter assay system (Promega) according to the manufacturer's directions. Triplicate samples were analyzed for each condition.
ERM screen.
The ERM screen, which is based on previously described strategies (11, 28, 29), is performed by infection with three retroviruses followed by antibiotic selection. First, Rat-1 cells were infected with pRetroX-Tet-On Advanced followed by selection with neo to make Rat-1 Tet-On cells. Second, Rat-1 Tet-On cells were infected with LMP1-NYFP TH and selected with neo and hygro to create an inducible bait cell line. Third, Rat-1 Tet-On LMP1-NYFP cells were infected with the ERM vector VC1 and selected with neo, hygro, and puro. In parallel, BiFC control cells were made by infection of Rat-1 Tet-On LMP1-NYFP cells with CYFP-TRAF2 TP followed by neo, hygro, and puro selection. Induced and uninduced BiFC control cells were used to establish gating for BiFC-positive cells that excluded BiFC-negative cells. VC1-infected cells were induced with 500 ng/ml doxycyline for 8 h and incubated overnight in the absence of doxycycline. BiFC-positive VC1 cells were sorted by fluorescence-activated cell sorting (FACS) using the FACS Aria (BD Biosciences) in the Rosalind Franklin University Flow Cytometry Core Facility. Single-cell clones were isolated by plating the BiFC sorted cells in limiting dilution in 96-well plates. Single-cell clones were expanded, plated in replicates, and retested for BiFC by the addition of 500 ng/ml dox overnight followed by observation by fluorescence microscopy. VC1-clone 1, which reinduced BiFC, was further characterized.
Clone identification.
Total RNA was purified from induced VC1-clone 1 cells and treated with DNase (Qiagen). First-strand cDNA synthesis with SuperScript III (Invitrogen) was performed with either random primer (RT-1, GCTAATACGACTCACTATAGGGATCCNNNNSACG) or anchored dT primer (RT-1T, GCTAATACGACTCACTATAGGGATCCTTTTTTTTTTTTTTTTV). Both RT-1 and RT-1T have T7′ primer sequences (underlined) at their 5′ ends for subsequent PCR and sequencing steps. The cDNA was PCR amplified with the T7-2′ (GCTAATACGACTCACTATAGGGATC) and CYFP′ (ACTTCAAGATCCGCCACAACATCGAG) primers with AccuPrime Pfx (Invitrogen). Reactions with or without reverse transcriptase were used to control for amplification of contaminating genomic DNA. PCR-amplified bands were gel purified and directly sequenced with the CYFP′ primer. The sequence downstream of the ERM splice junction was used to BLAST search the GenBank database to determine the targeted open reading frame.
Pulldown assays.
Pulldown of LMP1-binding proteins using LMP1 with an N-terminal tandem triple myc tag was performed as previously described (33). HEK293T cells were transfected with vector (pcDNA3), CYFP-HA-Tmem134, or HA-Tmem134 in the presence or absence of M3-LMP1. Forty-eight hours posttransfection, cells were harvested for pulldown. Ten percent of the lysates were set aside for direct load samples, and the rest of the lysates were subject to pulldown with the mammalian c-Myc tag IP/Co-IP kit (Pierce) according to the manufacturer's directions. Direct load and pulldowns were analyzed by Western blotting for myc tag (LMP1) and HA tag (Tmem134) constructs. Endogenous pulldowns were performed similarly with 200 μg of protein from EBV-infected LCL cells. Pulldowns were performed with LMP1 antibody (see “Western blotting” above) and protein G beads (Invitrogen). In parallel, control pulldowns and pulldowns with lysate alone (no LMP1 antibody) and antibody alone (no lysate) were performed. Direct load and pulldowns were analyzed by Western blotting for the presence of Tmem134 (custom; Genscript). To control for nonspecific pulldown, blots were stripped and reprobed with caveolin-1 antibody (Sigma).
Fluorescence and confocal microscopy.
Cells were plated on coverslips and fixed 48 h postselection for retrovirus infection or 24 h post-dox induction with 4% paraformaldehyde in PBS for 10 min at 4°C and washed with PBS. Coverslips were blocked for 10 min at room temperature in blocking solution (0.2% fish skin gelatin, 0.2% Triton X-100, phosphate-buffered saline) and stained in primary antibody for 1 h in a humidified chamber. The following primary antibodies were used at a working dilution of 1:100 in blocking solution: the Golgi marker mouse anti-golgin-97 (Invitrogen, Grand Island, NY), the early endosome marker goat anti-EEA1 (Santa Cruz, Santa Cruz, CA), rat anti-LMP1, a mixture of four rat monoclonal antibodies diluted 1:500 each (Cao 7E10, Cao 8G3, LMP1 IG6, and Cao 7G8; Ascenion, GmbH), and rabbit anti-Tmem134 (custom; Genscript). The lipid raft marker goat flotillin-1 (Abcam, Cambridge, MA) was used at a 1:25 working dilution, and Alexa Fluor 647-conjugated concanavalin A (Invitrogen), an ER marker, was used at a 1:2,000 working dilution in blocking solution. Coverslips were then washed with PBS and stained with secondary antibodies for 30 min in a humidified chamber. Donkey anti-rabbit 488, goat anti-rabbit 488, donkey anti-goat 594, goat anti-mouse 594, goat anti-rat 647, and chicken anti-rat 647 fluorescent Alexa Fluor secondary antibodies were used at a 1:1,000 dilution, and goat anti-rat 568 secondary antibody was used at a 1:2,000 dilution in blocking solution. Coverslips were washed and mounted with ProLong Gold Antifade reagent containing 4′-6-diamidino-2-phenylindole (DAPI) (Invitrogen). An Olympus Fluoview 300 fluorescent confocal microscope at the microscopy core of Rosalind Franklin University of Medicine and Science was used for data collection at 60× magnification under oil immersion, and analysis was performed using Fluoview software (Olympus, Melville, NY). Z-stacked images represent a 0.5-micrometer section of the cell in the x, y, and z planes. Secondary-antibody-alone and single-antibody staining were used to ensure specificity.
Lipid raft fractionation.
Lipid rafts were isolated using the caveolae/rafts isolation kit (Sigma) according to the manufacturer's directions. Cells were washed with ice-cold PBS and lysed with bicarbonate lysis buffer (0.5 M, pH 11.0). Approximately 200 mg wet cell pellet for suspension cells was used for each gradient. Lysates were Dounce homogenized, passed through a syringe, and sonicated before loading onto gradients. Optiprep was added to the lysates in centrifuge tubes to a final concentration of 35% (wt/vol) and overlaid with 30%, 25%, 20%, and 0% Optiprep layers. Gradients were spun at 200,000 × g for 4 h at 4°C and allowed to decelerate without braking. Fractions were collected from the top to the bottom of the gradient. Caveolin-positive fractions were determined by dot blot or Western blot analyses. Protein was concentrated by acetone precipitation and resuspended in RIPA buffer. Protein concentration was determined by bicinchoninic acid (BCA) assay, and 40 to 50 μg of protein per fraction was analyzed by Western blotting for LMP1 and Tmem134.
Flow cytometry staining.
Cells were harvested, washed with ice-cold PBS, and fixed by the addition of ice-cold methanol to 90% (vol/vol) while gently vortexing. Fixed cells were incubated on ice for 30 min or at −20°C for later staining. Cells were pelleted by centrifugation at 260 × g for 10 min at 4°C and washed twice with Flow PBS (PBS plus 0.5% [wt/vol] bovine serum albumin). Cells were incubated in 100 μl of primary antibodies (LMP1 [Ascenion GmbH] at a 1:200 dilution) in Flow PBS for 1 h at room temperature and then washed twice with Flow PBS. Cells were then incubated with Alexa Fluor-647-labeled secondary antibodies at a 1:1,000 dilution for 30 min at room temperature and washed twice with Flow PBS. Parallel staining of secondary antibodies alone was also performed. Stained cells were analyzed using the LSRII (BD Biosciences) in the Rosalind Franklin University Flow Cytometry Core. The main cell population was gated based on forward and side scatter dot plot, and 10,000 events were analyzed per condition for Alexa Fluor-647 fluorescence. Flow cytometry data were analyzed with BD FACSDiva (BD Biosciences) and FlowJo (Tree Star) software.
RESULTS
BiFC is a novel approach that can be applied to membrane proteins in live cells to detect protein-protein interactions. We have previously published the detection of LMP1-TRAF and LMP1-LMP1 binding by BiFC in the membrane of live cells (44). LMP1 BiFC was present in physiological subcellular locations, and LMP1-TRAF binding was sensitive to mutations which disrupt TRAF binding and LMP1 signaling. Importantly, LMP1 BiFC proteins were fully functional in rodent fibroblast transformation assays and activated NF-κB reporters, suggesting that BiFC is a suitable in vivo platform to identify LMP1-binding proteins that are important for signaling. In this study, the identification of a novel LMP1-binding protein, Tmem134, is described.
BiFC ERM screen.
The strategy of the BiFC screen is outlined in Fig. 1A. The overall principle is to express LMP1 as a bait protein, LMP1-NYFP, and express different CYFP-fused test proteins. LMP1-binding proteins that are tagged with CYFP will induce BiFC with LMP1-NYFP. Proteins that induce BiFC will be identified and characterized further. Expression of both bait and test proteins is controlled by inducible promoters to minimize any potentially toxic effects of tagged protein expression. The screen is performed by infection with a series of three retroviruses. First, a dox-inducible (Tet-On) cell line is created. Second, the inducible bait cells (LMP1-NYFP) cells are selected. Third, the bait cells are infected with the enhanced retroviral mutagen (ERM). ERM vectors induce the creation of CYFP-tagged cellular proteins, as described previously (11, 28, 29). The ERM vectors act as exon-trapping vectors and preferentially integrate within regions of active transcription throughout the genome (47). CYFP-fusion proteins are created with cellular proteins, where they integrate by inducing splicing between a splice donor site in the ERM and the splice acceptor of the next downstream cellular exon. YFP-positive cells are isolated by fluorescence-activated cell sorting (FACS), and the interacting proteins are identified by reverse genetics.
Fig 1.
ERM strategy. (A) Overall experimental design. (B) Control and induced Tet-On LMP1-NYFP cells were analyzed by Western blotting for LMP1-NYFP expression by blotting for LMP1. Rat-1 cells infected with Babe LMP1-NYFP, which stably expresses LMP1-NYFP, were also harvested. Positions of LMP1-NYFP and molecular weight markers are indicated.
To make an inducible cell line, Rat-1, rodent fibroblasts, cells were infected with RetroX-Tet-On Advanced, a retrovirus that expresses rtTA (dox-controlled transactivator protein), and stable cells were selected. Rat-1 Tet-On cells were confirmed by transfection with RetroX-Tight-Hyg-Luc and luciferase assay (data not shown). To make an inducible bait cell line for the BiFC screen, the LMP1 BiFC construct LMP1-NYFP was cloned into an inducible retrovirus expression vector, pRetroX-Tight-Hyg. The resulting retrovirus, LMP1-NYFP TH, was used to infect Tet-On cells, and a stable cell line was selected. Inducible LMP1-NYFP expression was confirmed by induction with dox and Western blotting (Fig. 1B). Tet-On LMP1-NYFP stable cells were plated, induced with dox for 24 h, and harvested for Western blotting. In parallel, uninduced Tet-On LMP1-NYFP and Rat-1 cells which constitutively express LMP1-NYFP (Babe LMP1-NYFP) were also harvested. Western blotting for LMP1 detected the expression of LMP1-NYFP in the inducible cells in the presence of dox and in the Babe LMP1-NYFP cells at about 80 kDa, which is the predicted molecular size of LMP1 with the NYFP tag. Expression of LMP1-NYFP in uninduced cells was not detected. This confirms the inducible expression of the LMP1-NYFP in the bait cell line.
To perform the BiFC screen, Tet-On LMP1-NYFP cells were infected with the ERM vector VC1. VC1 is compatible with cellular exons in which the first base of the exon is also the first base of the codon triplet for the protein coding sequence. Tet-On LMP1-NYFP cells infected with VC1 were passaged a low density, and infected cells were selected. In parallel for a BiFC-positive control, CYFP-TRAF2 was cloned into the inducible retrovirus vector pRetroX-Tight-Pur. Tet-On LMP1-NYFP cells were infected with CYFP-TRAF2 TP retrovirus and selected.
To isolate cells which exhibit BiFC, FACS was performed. Bait cells stably infected with VC1 or CYFP-TRAF2 were induced for 8 h with 500 ng/ml dox and then incubated overnight in the absence of dox. The gating for BiFC-positive cells was established with the induced CYFP-TRAF2 cells. Approximately 400,000 VC1 cells were analyzed for BiFC, and 5,220 positive events were isolated. BiFC-positive cells were plated in 96-well plates in limiting dilution to expand single-cell clones. Once clones became 50% confluent, they were expanded into multiple wells for further analysis.
Identification of the Tmem134 clone.
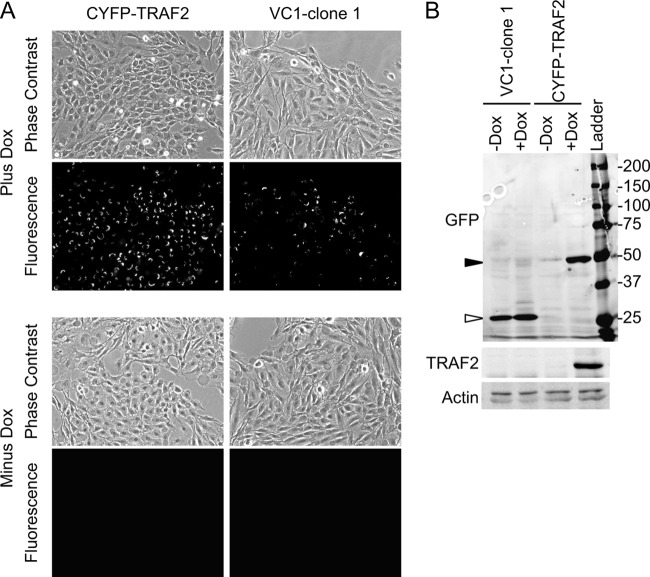
To confirm BiFC, single-cell clones were induced and observed by fluorescence microscopy. Clones were expanded in replicates in 96-well plates, and 1 well for each clone was induced with dox overnight. Induced clones were observed for BiFC by fluorescence microscopy. Uninduced clones were also examined for background fluorescence. VC1-clone 1 was the only clone which exhibited BiFC following induction with dox (Fig. 2A, upper panels). Bright perinuclear fluorescence was observed with VC1-clone 1. Similar fluorescence was observed with induction of CYFP-TRAF2 positive-control cells. Fluorescence was not observed in the uninduced wells (Fig. 2A, lower panels). Subsequent optimization of the BiFC screen has allowed for recovery of >95% clones that induce BiFC upon dox addition (data not shown). Optimized steps include using bait cell line single-cell clones and dox duration and concentration. To confirm new CYFP-fusion protein expression, Western blotting was performed. VC1-clone 1 was expanded, induced overnight with dox, and harvested for Western blotting. In parallel, uninduced VC1-clone 1 and induced and uninduced CYFP-TRAF2 were also harvested. Western blotting with green fluorescent protein (GFP) antibody that reacts with the CYFP-tag showed several bands in various lanes (Fig. 2B). Bands of about 25 kDa were observed in both the induced and uninduced VC1-clone 1 lanes (Fig. 2B, white arrowhead). The 25-kDa band slightly increased in the induced VC1-clone 1 and was not observed in the CYFP-TRAF2 lanes. CYFP-TRAF2 was detected at about 50 kDa in the induced CYFP-TRAF2 lane (Fig. 2B, black arrowhead). Blotting with TRAF2-specific antibody shows reactions with the induced CYFP-TRAF2 but not VC1-clone 1 (Fig. 2B and data not shown, respectively). Blotting for actin confirms equal loading. This indicates that VC1-clone 1 induces a new CYFP protein that is not CYFP-TRAF2. Because the CYFP domain is about 10 kDa, the new VC1-clone 1 has tagged a cellular protein of about 15 kDa. Interestingly, the VC1-clone 1 protein appears to be expressed constitutively, suggesting that its expression is not toxic.
Fig 2.
Isolation of VC1-clone 1. (A) A single-cell clone, VC1-clone 1, was induced and observed for BiFC with fluorescence microscopy. Control cells which inducibly express LMP1-NYFP and CYFP-TRAF2 were also induced. Representative phase-contrast and fluorescence micrographs are shown (upper panels [Plus Dox]). Uninduced cells were also observed (lower panels [Minus Dox]). (B) Control and induced cells were analyzed by Western blotting for CYFP fusion protein expression by blotting with a GFP monoclonal antibody that reacts with the CYFP domain. Position of VC1-clone 1 fusion protein and CYFP-TRAF2 (white and black arrowheads, respectively) and molecular weight markers are indicated. Lysates were also blotted for TRAF2 and actin (loading control).
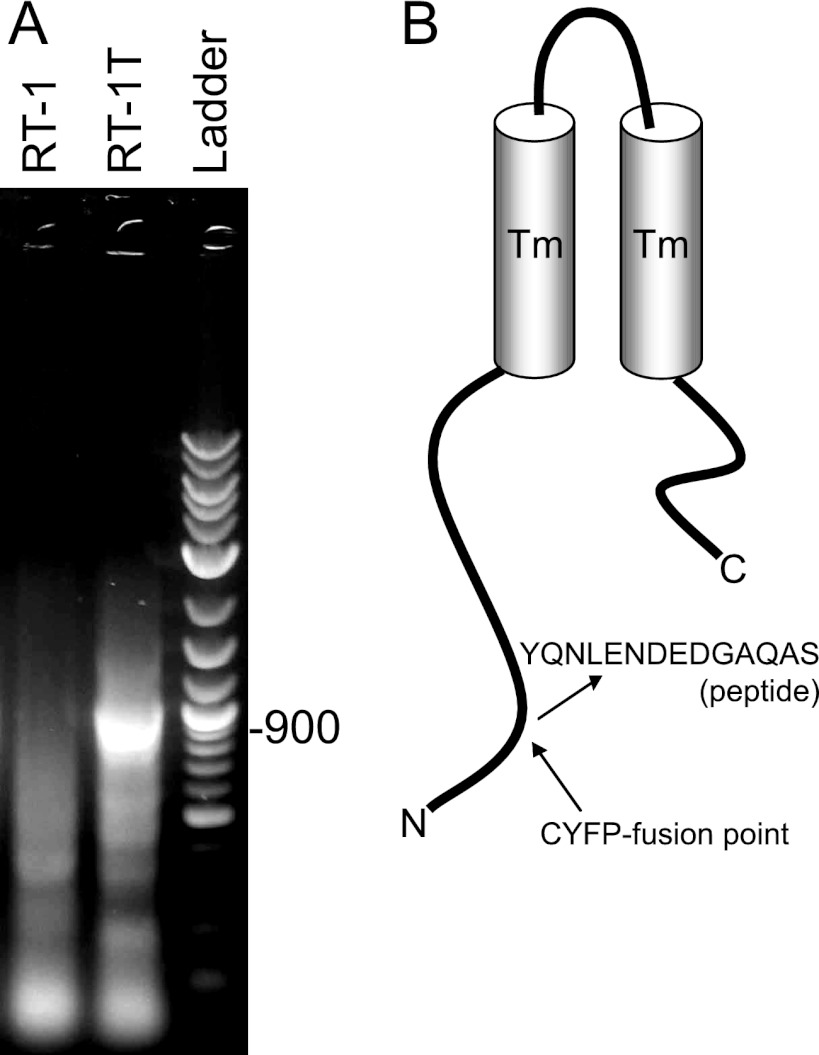
To determine the VC1-clone 1 CYFP-tagged protein, reverse genetics were used as described previously (11, 28, 29). cDNA was synthesized from total RNA with either random primer RT-1′ or anchored dT primer RT-1T′. Both RT-1 and RT-1T have T7′ primer sequences at their 5′ ends for subsequent PCR and sequencing steps. The cDNA was PCR amplified with the T7′ and CYFP′ primers, and PCRs were analyzed by agarose gel electrophoresis (Fig. 3A). The 900-bp band that was observed in the RT-1T′ PCR was gel purified and sequenced with the CYFP′ primer. The presence of CYFP and ERM linker sequence, including the HA tag sequence, confirmed that the sequenced fragment was the CYFP fusion protein mRNA sequence. The sequence downstream of the ERM splice junction was used to search the GenBank database using BLAST. The search determined that the targeted open reading frame was Tmem134 (data not shown).
Fig 3.
Tmem134 clone (VC1-clone 1). (A) PCR amplification of cDNA from reverse transcription reactions with random (RT-1) or anchored oligo(dT) (RT-1T) primers. Ladder, molecular weight of sequenced PCR product in the RT-1T reaction. (B) Structure of Tmem134 and CYFP fusion point. Transmembrane (Tm) domains and peptide sequence used for antibody production are indicated.
Tmem134 is a small protein of unknown function. It is predicted to encode a transmembrane protein with a 122-amino-acid N-terminal cytoplasmic tail, two-pass Tm domain, and 20-amino-acid C-terminal cytoplasmic tail (Fig. 3B). Tmem134 was annotated using high-throughput cDNA sequencing approaches (2, 43, 51). It is highly conserved (>90% identity) through mammalian species which have been sequenced, including human, chimpanzee, mouse, rat, dog, and bovine. Tmem134 also contains a conserved domain (DUF872 [PF05915]) of unknown function that is conserved through zebrafish, Drosophila, and Caenorhabditis elegans (>40% identity, >60% homology) and has homologues into the plant kingdom. The entire DUF872 domain that includes the transmembrane domains is contained within VC1-clone 1. Tmem134 has been identified as a candidate oncogene that is amplified in breast cancer cell lines (24). VC1-clone 1 induces splicing to exon 2 of Tmem134, which includes the putative transmembrane domain. The predicted molecular size of CYFP-Tmem134 is about 25 kDa, which is consistent with the observed band in Fig. 2B. CYFP-Tmem134 was cloned from cDNA, and BiFC with LMP1-NYFP was confirmed in transfected cells (data not shown). These data identify a novel LMP1-binding protein, Tmem134. Custom Tmem134-specific antibody was synthesized from the indicated peptide sequence (Fig. 3B) in the cytoplasmic N terminus.
Tmem134 expression.
Tmem134-LMP1 binding was identified by BiFC in rodent fibroblasts. Tmem134 expression in cell types relevant to EBV infection is unknown. Custom Tmem134 antibody was validated to react with tagged rat and endogenous human Tmem134 (data not shown). To determine Tmem134 expression in human epithelial and B-cell lines, Western blotting was performed. Various human cell lines were harvested, and Western blotting for Tmem134 and actin were performed. Several EBV-positive B-cell lines (Raji, LCL, and EF3D) and EBV-negative B-cell lines (Akata, Ramos, Loukes, and BJAB) were analyzed (Fig. 4A). Tmem134 is expressed at similar levels in the different B-cell lines regardless of the presence of EBV. Various human epithelial cell lines were analyzed in Fig. 4B, including SiHa, C666-1, Caski, 293T, and C33A. Tmem134 is also expressed in the human epithelial cell lines. Actin blotting confirmed relatively equal loading, and blotting for LMP1 confirmed LMP1 expression in the EBV-positive cell lines. This confirms that Tmem134 is expressed in the human cell types that EBV infects.
Fig 4.
Tmem134 protein expression in B-cell and epithelial cell lines. (A) Fifty micrograms of protein for each cell line was analyzed by Western blotting for Tmem134 expression. EBV-positive (EBV+) and EBV-negative (EBV−) B-cell lines were analyzed. (B) A number of human epithelial cell lines were analyzed. Western blotting for actin (loading control) and LMP1 for B-cell lines was also performed.
Confirmation of LMP1-Tmem134 binding by immunoprecipitation.
To confirm binding of LMP1 and Tmem134, coimmunoprecipitations were performed. CYFP-Tmem134 from the ERM screen, which contains an internal HA tag (Fig. 2B), and Tmem134, cloned with the HA tag but lacking CYFP domain (HA-Tmem134), were used in pulldown assays with triple myc-tagged LMP1 (M3-LMP1) as previously described (33). HEK293T cells were transfected with vector (pcDNA3), CYFP-Tmem134, or HA-Tmem134 in the presence or absence of M3-LMP1. Forty-eight hours posttransfection, cells were harvested for pulldown. Direct load and myc pulldowns were analyzed by Western blotting (Fig. 5A). Western blotting for myc-tag (LMP1) and HA-tag (Tmem134 constructs) in the direct load indicated expression of all constructs. The HA-tag antibody reacted with bands of the expected sizes for CYFP-Tmem134 and HA-Tmem134 (Fig. 5A, white and black arrowheads, respectively). There were also other bands of unexpected sizes. There was a higher, more diffuse band in the HA-Tmem134 lane. Many signaling and trafficking proteins are regulated by ubiquitylation, and the higher-molecular-weight band may be due to covalent modification by ubiquitin. There was also a lower-molecular-weight band in the CYFP-Tmem134 lane. This band is likely the result of initiation of translation at one of two in-frame internal methionines in the CYFP-coding sequence that are flanked by Kozak sequences.
Fig 5.
Pulldown of Tmem134 with LMP1. (A) Tmem134 was pulled down by LMP1 in transfected cells. Vector control, HA-Tmem134, and CYFP-HA-Tmem134 (CYFP-T134) were transfected in the presence or absence of myc-tagged LMP1 (LMP1). Forty-eight hours posttransfection, cells were harvested for myc pulldown and 10% of the lysates were saved for direct load. The remaining lysates were pulled down with the mammalian c-Myc tag IP/Co-IP kit (Pierce) according to the manufacturer's directions. Direct load and pulldown were analyzed by Western blotting for LMP1 (myc tag) and Tmem134 (HA tag). Locations of HA-Tmem134 (black arrowhead) and CYFP-HA-Tmem134 (white arrowhead) are indicated. (B) Endogenous Tmem134 was pulled down by LMP1 in EBV-transformed cells. Cells were harvested, and 200 μg of protein were used for pulldown with LMP1 antibody (LMP1 IP). Control pulldowns without LMP1 antibody (lysate alone) and without lysate (LMP1 Ab alone) were processed in parallel. Direct load and pulldowns were analyzed by Western blotting for Tmem134. To control for nonspecific pulldown the blot was stripped and blotted for caveolin. Location of caveolin-specific (black arrowhead) and Tmem134-specific (white arrowhead) bands are indicated.
Western blotting of the myc-pull down demonstrates efficient pulldown of LMP1. In the absence of transfected LMP1, faint bands were observed by blotting the myc pulldowns with the HA antibody. This suggests that a small amount of Tmem134 binds to the myc beads nonspecifically in the absence of LMP1. Strong pulldown of both CYFP-Tmem134 and HA-Tmem134 was observed in pulldowns containing LMP1. Interestingly the lower-molecular-weight band in the CYFP-Tmem134, which presumably encodes all of the Tmem134 protein sequence but less of the CYFP sequence, was also pulled down by LMP1. The fact that Tmem134 was pulled down by LMP1 is indicative of binding between LMP1 and Tmem134.
To determine pulldown of endogenous Tmem134 with LMP1, LMP1 pulldowns were performed from the cell lysates of EBV-transformed cells (LCL). Control pulldowns without antibody or without cell lysates were also performed. Endogenous LMP1 pulldowns from LCLs were analyzed for Tmem134 by Western blotting (Fig. 5B). Tmem134 was observed in the direct load and LMP1 IP lanes (Fig. 5B, white arrowhead). A faint band was observed in the pulldown without antibody (lysate alone), which suggests that Tmem134 weakly binds to the beads during pulldown nonspecifically. Tmem134 bands were quantified, and there was a 9-fold increase in Tmem134 pulldown in the LMP1 IP compared to the lysate-alone IP. No band was observed in the lane without lysate (LMP1 Ab alone). To control for nonspecific pulldown of all raft-associated proteins, the blots was stripped and blotted for raft protein caveolin-1. Caveolin-1 was observed in the direct load lane (Fig. 5B, black arrowhead) but not in the LMP1 IP or control pulldown lanes. Pulldown of Tmem134 with LMP1 antibody suggests binding between Tmem134 and LMP1 when the proteins are expressed at physiological levels within EBV-infected cells. Failure to pull down caveolin-1 suggests that the LMP1-Tmem134 pulldown does not pull down all lipid raft-associated proteins nonspecifically. This suggests that the pulldown of Tmem134 with LMP1 is specific.
Tmem134-LMP1 subcellular localization.
Since the function of Tmem134 is unknown, the discrete locations in the cell where Tmem134 is located in the presence or absence of LMP1 may yield insight into Tmem134 function and Tmem134 function in LMP1 signaling. Custom Tmem134 antibody was validated for use in immunofluorescence microscopy (data not shown). This enabled the staining of endogenous rather than transfected Tmem134. In order to examine LMP1 at physiologically relevant expression levels, LMP1 expression was determined in a variety of cell lines by flow cytometry and Western blotting (Fig. 6). EBV-infected cell lines express physiological levels of LMP1 compared to C33A infected with LMP1 retrovirus (Babe-LMP1) (black histograms, Fig. 6A and B, respectively, and 6C). C33A LMP1-inducible cells express slightly elevated levels of LMP1 compared to the EBV-infected cells even at the lowest dox concentrations (20 ng/ml) which are able to induce LMP1 expression (Fig. 6). To determine the subcellular localization of Tmem134 in the presence or absence of LMP1, confocal microscopy was performed (Fig. 7). Control and LMP1 cells were fixed and stained by immunofluorescence. Four color channels were acquired by confocal microscopy. The large micrographs contain stains for Tmem134 (green), LMP1 (red), and subcellular markers (blue). The small inset micrographs contain the DAPI counterstain to locate the corresponding nuclei. Representative confocal x, y, and z planes in which overlapping signals were observed are presented (Fig. 7).
Fig 6.

Relative LMP1 expression in cell lines. BJAB (EBV-negative), EF3D and LCL (EBV-infected), and various C33A stable cells were compared for LMP1 expression by flow cytometry (A and B) and Western blotting (C). C33A Babe and C33A Babe-LMP1 were stably transduced with control and LMP1-expressing retroviruses, respectively. C33A-Tet-On HA-LMP1 cells inducibly express LMP1 in the presence of dox. Inducible cells were treated in the absence (no dox) or in the presence of dox (20, 50, or 100 ng/ml) overnight. For flow cytometry, cells were fixed and stained with LMP1 antibody and anti-rat AF647 antibody. Histograms from secondary-antibody-alone-stained cells for both pairs were similar to control cells stained with LMP1 antibody (data not shown). EBV-positive and EBV-negative cells are shown in panel A, and C33A stable cells are shown in panel B. In parallel, cells were harvested and 60 μg of protein per cell type was analyzed by Western blotting for LMP1 and actin (loading control) in panel C.
Fig 7.
Tmem134 and LMP1 subcellular localization. Immunofluorescence staining of control and LMP1 cells were performed. C33A cells were stably infected with control (A, C, and E) or LMP1 (B, D, and F) retrovirus. Control (G) and induced (20 ng/ml dox) C33A Tet-On LMP1-inducible cells (H) were also examined. Control and LMP1 cells were fixed and stained for immunofluorescence with Tmem134 (green), LMP1 (red), and subcellular markers (blue). Cells were also stained with DAPI (smaller insets). Subcellular markers include endoplasmic reticulum (concanavalin A) (A and B), Golgi apparatus (golgin) (C and D), early endosome (EEA1) (E and F), and lipid raft (flotillin-1) (G and H). Confocal images were acquired, and representative x, y, and z planes are depicted. Overlap of Tmem134 and LMP1 (red and green, respectively) is shown as yellow (arrow with open arrowhead), Tmem134 and subcellular markers (green and blue, respectively) is shown as cyan (white arrow), and LMP1 and subcellular markers (red with blue, respectively) is shown as purple (white arrowhead). Triple colocalization of Tmem134, LMP1, and subcellular markers (green, red, and blue, respectively) is shown as white (open arrowhead).
Tmem134 could function in one or more levels in LMP1 maturation, signaling, or turnover. To examine a role in protein synthesis and maturation, cells were stained for subcellular markers for colocalization with the endoplasmic reticulum (ER) (concanavalin A) (Fig. 7A and B) and Golgi apparatus (golgin) (Fig. 7C and D). To examine a role in protein turnover or secretion in exosomes, cells were stained for colocalization with the early endosome (EEA1) (Fig. 7E and F). Finally, to examine a role in signaling, cells were stained for colocalization with lipid rafts (flotillin-1) (Fig. 7G and H). Staining for ER, Golgi apparatus, and early endosome was performed with stable cells infected with control (Fig. 7A, C, And E) or LMP1 retrovirus (Fig. 7B, D, And F), which expresses physiological levels of LMP1 compared to EBV-infected cells (Fig. 6). The most extensive overlap was observed with Tmem134 and ER (Fig. 7A, white arrow and cyan color). Punctate Tmem134 staining, which was not ER associated, was also observed toward the cell periphery. Small areas of overlap were also observed with Tmem134 and Golgi apparatus (Fig. 7C, white arrow and cyan color) and early endosome (Fig. 7E, white arrow and cyan color). In the LMP1 stable cells (Fig. 7B, D, And F) there was overlap with Tmem134 and LMP1 (arrow with open arrowhead, yellow color) and subcellular markers and LMP1 (white arrowhead, purple color). Triple colocalization (open arrowhead, white color) was observed for Tmem134 and LMP1 with ER (Fig. 7B) and Golgi apparatus (Fig. 7D). In the LMP1 stable cells, triple colocalization was not observed between LMP1, Tmem134, and EEA1 (Fig. 7F). This was surprising because we did observe pairwise colocalization with each of the three proteins as described above in multiple fields. It is unclear as to whether the lack of triple colocalization is a technical limitation or indicates an important functional clue; i.e., does LMP1 which moves into the endosome no longer bind Tmem134? Although the presented micrographs, which were chosen to highlight regions of colocalization, suggest altered Tmem134 staining in the presence of LMP1, Tmem134 stains in control and LMP1 cells in similar confocal planes do not appear differently (data not shown).
Control and dox-induced C33A Tet-On LMP1-inducible cells were stained for Tmem134, LMP1, and flotillin-1 (lipid raft marker) (Fig. 7G and H) and examined by confocal microscopy for colocalization. Although cells were induced with the minimum dox concentration required to observe LMP1 expression (20 ng/ml), LMP1 is expressed at slightly higher levels than in EBV-infected cells (Fig. 6C). There was colocalization between Tmem134 and flotillin-1 in control and induced cells (Fig. 7G, white arrows, and Fig. 7H, cyan color). As expected, there was colocalization between LMP1 and flotillin-1 (Fig. 7H, white arrowhead, purple color) and LMP1 and Tmem134 (arrow with open arrowhead, yellow color). There was also triple colocalization (open arrowhead, white color). Together, these data suggest that Tmem134 is most strongly associated with the ER compartment but is present in other membranous compartments throughout the cell.
Tmem134 and LMP1 lipid raft fractionation.
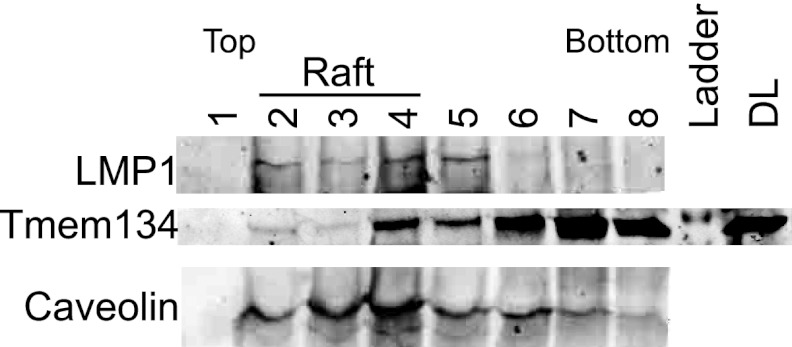
Tmem134 and LMP1 colocalized in lipid rafts in C33A-inducible cells. To determine if Tmem134 is present in lipid rafts with LMP1 in infected cells, lipid raft gradient fractionation was performed. To examine endogenous LMP1 and Tmem134, an EBV-infected lymphocyte cell line was used for lipid raft fractionation. Raft fractions were analyzed by Western blotting for caveolin-1 (raft marker), LMP1, and Tmem134 (Fig. 8). Caveolin-1 was enriched in fractions 2 to 4, which confirmed successful raft fractionation. LMP1 was present in the raft fractions as well as fraction 5. The presence of LMP1 in fraction 5 is consistent with a previously described lipid raft “barge” fraction (48). Tmem134 was present across the gradient fractions in both raft and nonraft fractions, which is consistent with lipid raft, ER, Golgi apparatus, and early endosome colocalization (Fig. 7). This confirms that Tmem134 is present with LMP1 in raft fractions in EBV-infected cells and also indicates that Tmem134 is may serve both raft and nonraft functions in cells.
Fig 8.
Tmem134 and LMP1 cofractionation in caveolin-positive lipid raft fractions. An EBV-positive lymphocyte cell line was harvested, and lipid raft fractionation was performed. Fractionated protein was concentrated by acetone precipitation and quantitated, and 50 μg of protein per fraction was analyzed by Western blotting. Lipid raft fractions were analyzed for LMP1, Tmem134, and caveolin-1 (lipid raft marker). Top, bottom, and lipid raft gradient fractions are indicated.
Tmem134 affects LMP1 signaling.
Since endogenous Tmem134 expression was difficult to detect in Rat-1 cells (data not shown), Tmem134-specific siRNAs were screened against CYFP-Tmem134 expression in transfected cells. The two most potent Tmem134 siRNA sequences, si-302 and si-308, were cloned into siRNA expression vectors. Plasmids expressing negative-control siRNA (si-Neg), si-302, or si-308 were transfected with CYFP-Tmem134 expression plasmid into 293T cells. Forty-eight hours posttransfection, cells were harvested and analyzed for CYFP-Tmem134 expression by Western blotting (Fig. 9A). si-302 inhibits CYFP-Tmem134 expression by greater than 95%, while si-308 decreases expression by about 50%. Blotting for actin indicates equal loading between samples.
Fig 9.
Regulation of LMP1-dependent NF-κB reporter activity by Tmem134. (A) Tmem134 siRNA against Rat Tmem134 was validated by transfection of 293T cells with CYFP-Tmem134 and two Tmem134 siRNA expression plasmids (si-302 and si-308) or negative-control siRNA (si-Neg) and Western blotting for fusion protein expression with custom Tmem134 antibody (T134) and loading control actin. Rat-1 Tet-LMP1 cells were transfected with pRL-SV40 (control Renilla luciferase plasmid), NF-κB reporter plasmid (NF-κB-luc), and indicated siRNA expression plasmids. (B) At 24 h posttransfection, cells were changed to media lacking (no dox) or containing 100 ng/ml dox (+dox), and at 48 h posttransfection, dual-luciferase assays were performed. The mean of triplicate samples for each condition is plotted relative to uninduced reporter activity for each siRNA group. (D) NF-κB dual-luciferase assays were also performed in 293T cells with CYFP-Tmem134. 293T cells were transfected with LMP1, LMP1-signaling mutant (Mut), or vector (Vec), CYFP-zip (zip, control plasmid), CYFP-T134, or CYFP-dnTRAF3 (dnT3), and RL-SV40 and NF-κB-luc. Dual luciferase assays were performed 48 h posttransfection, and the mean luciferase activity of triplicate samples for each condition is plotted. (C and E) LMP1 expression was determined in Rat-1 Tet-LMP1 (C) and 293T (E) cells using Western blotting for LMP1 and actin. Western blots are aligned under the corresponding bars of the reporter assays. LMP1 expression relative to actin is given in panel E. Both assays were repeated at least four times, and statistical significance (P < 0.05) using Student's t test is indicated with an asterisk.
Rat-1 Tet-On HA-LMP1 cells, which inducibly express HA-LMP1, were transfected with siRNA expression plasmids NF-κB-luc and RL-SV40. Twenty-four hours posttransfection, cells were induced to express LMP1 with 100 ng/ml dox, and 48 h posttransfection, dual-luciferase assays were performed. Uninduced cells were also analyzed for each condition. Luciferase activity was determined relative to that in uninduced cells transfected with each siRNA (Fig. 9B). Cells transfected with control siRNA expression vector (siNeg) activated NF-κB activity about 5-fold in the induced cells. Tmem134 siRNA inhibited NF-κB activity relative to Tmem134 knockdown. si-302, which inhibits >95% Tmem134 expression, had lower NF-κB activation, approximately 3-fold induced. si-308, which had an intermediate knockdown of Tmem134, had decreased NF-κB induction to about 4-fold. Altered NF-κB activity in Tmem134 knockdown cells suggests a role for Tmem134 in LMP1 function. The triplicate samples from the reporter assay were pooled, quantitated for protein concentration, and analyzed by Western blotting for LMP1 and actin expression (Fig. 9C). Actin expression and LMP1 induction appeared equal in different samples.
Since Tmem134 expression was low in Rat-1 cells compared to human cell lines, 293T cells were also examined for Tmem134 effects. To determine if CYFP-Tmem134 from the BiFC screen that lacks Tmem134 exon 1 affects LMP1 NF-κB activity, dual-luciferase assays were performed (Fig. 9D). 293T cells were transfected with M3-LMP1, M3-LMP1(A5-Y384G) (Mut), or pcDNA3 vector (Vec) plasmids and NF-κB-luc, RL-SV40 (control luciferase), and CYFP-tagged effector plasmids. M3-LMP1(A5-Y384G) contains mutations in CTAR1 and CTAR2 signaling domains. CYFP-tagged effector plasmids include CYFP-zip (BiFC vector control plasmid) (zip), CYFP-Tmem134 (T134), and CYFP-TRAF3 (dnT3). The TRAF3 in CYFP-TRAF3 contains the TRAF3 LMP1-binding domain and has previously been shown to act as a dominant negative TRAF in signaling assays (32). CYFP-zip with LMP1 activated NF-κB activity, while LMP1 with CYFP-Tmem134 increased NF-κB activation to about twice that of the cells transfected with zip. Cotransfection of dnT3 had little effect on LMP1-induced NF-κB activity. The LMP1 signaling mutant (Mut) and vector cells (Vec) had little NF-κB activity. The triplicate samples from the reporter assay were pooled, quantitated for protein concentration, and analyzed by Western blotting for LMP1 and actin expression (Fig. 9E). Actin expression appeared relatively equal between samples, and LMP1 expression was determined relative to actin expression. LMP1 expression was greater in combination with the zip construct but relatively equal in the Tmem134 and dnT3 lanes and similar to the expression of the LMP1 mutant. This suggests that the increased NF-κB activity in the presence of CYFP-Tmem134 is not due to greater LMP1 expression. In contrast, greater NF-κB activity was observed with slightly lower LMP1 expression in Tmem134 than in either dnTRAF3 or vector (zip) samples.
DISCUSSION
In the present study, we describe the development of a screen for LMP1-binding proteins based on BiFC. LMP1-NYFP was used as a bait protein, and the CYFP domain was supplied by an enhanced retroviral mutagen that acts as an exon trap. A clone which was sorted based on YFP fluorescence was recovered by FACS, and the CYFP-fusion protein was determined to be a new LMP1-binding protein, Tmem134. Tmem134-LMP1 binding was confirmed by coimmunoprecipitation and colocalization. Tmem134 affected LMP1-induced NF-κB activity and fractionated into lipid raft fractions with LMP1 in EBV-infected cells.
The current screen was performed with the ERM vector VC1. VC1 is compatible with exons in which the first base of the exon is also the first base of the codon triplet within the coding sequence. Known LMP1-binding proteins like TRAF2 and TRAF3 are expected to be isolated by the screen. However, the first bases of the target exons for TRAF2 and TRAF3 are bases three and two of the codon triplet, respectively, which means that these genes would be isolated only in screens with VC3 and VC2, respectively. To obtain full genome coverage the screens must be performed with all three ERM vectors.
It was surprising that only one BiFC-positive clone was recovered out of nearly 5,000 sorted events. Based on subsequent experiments, several factors likely contributed to recovering only a single positive clone. First, overexpression of LMP1 can lead to cytotoxicity. In the current screen, BiFC was induced with high dox concentrations, and we have subsequently determined that at least 10- to 20-fold-lower dox concentrations are sufficient to induce BiFC. Many BiFC-positive cells may have failed to be recovered due to the decreased viability. Second, analyzing the sorted clones by fluorescence microscopy may have incorrectly judged some clones to be false positives. Although in theory any fluorescent technique can detect BiFC, in practice these techniques vary greatly in their sensitivity. Flow cytometry is a much more sensitive method to detect BiFC than fluorescence microscopy. Some BiFC-positive clones may have been discarded following screening by fluorescence microscopy.
The fact that we identified a membrane protein and a possible oncogene in our BiFC ERM screen highlights the utility of our approach of identifying LMP1-binding proteins within the membrane of mammalian cells. LMP1-binding proteins were initially identified using Y2H screens with the cytoplasmic domain of LMP1 (36). Although Y2H screens are powerful tools for identifying and characterizing protein-protein interactions, Y2H requires interacting proteins to be transported to the nucleus to induce transcription of reporter genes, which generally precludes the inclusion of transmembrane domains. LMP1 signaling occurs in the cholesterol-rich lipid raft domains of the membrane (3, 9, 17, 22, 38, 48). The contribution of the membrane domain of LMP1 to recruitment of downstream effector proteins can generally not be determined by Y2H. In contrast, BiFC does not require nuclear localization and can be performed with LMP1 in the membrane of mammalian cells. Determining LMP1-binding proteins with a BiFC screen and full-length membrane-bound LMP1 will give a more complete picture of the cellular proteins required for LMP1 signaling.
Tmem134 is a highly conserved small transmembrane protein of unknown function. It is overexpressed in several breast cancer cell lines, which suggests that it could contribute to carcinogenesis (24). It also has a highly conserved domain, suggestive of a critical cellular function. The specific role of Tmem134 in LMP1-mediated signaling is currently unknown. Tmem134 could function in the proper assembly of the LMP1-signaling complex or trafficking of LMP1 to lipid rafts or could be important for LMP1 turnover. Recently an integrated approach identified several proteins involved in vesicular trafficking as critical drivers of cancer development (1); such proteins represent novel pharmacological targets to inhibit cancer growth. Determining how Tmem134 affects LMP1 signaling in EBV-infected cells expressed at physiological levels will be critical to determine Tmem134 function. Future studies with Tmem134 siRNA or Tmem134 mutants will yield insight as to Tmem134 function and its role in LMP1 signaling in infected cells. Furthermore, Tmem134 and LMP1 staining was only partially colocalized by immunofluorescence microscopy. This suggests that Tmem134 may be important for only some but not all aspects of LMP1 function. In addition, Tmem134 undoubtedly performs critical functions in concert with other cellular proteins independent of LMP1. It will also be important to determine how Tmem134 affects the signaling of other TNFR proteins, CD40-LMP1 chimeras, and other membrane proteins.
Considering that few clones were recovered by FACS, possibly from toxicity related to LMP1 overexpression, it is tempting to speculate that Tmem134 might inhibit an LMP1 function that would have enabled it to be isolated in our screen. In this case, the BiFC screen might be enriched in proteins which are able to act as dominant negative proteins to block LMP1 function. The current study provides critical proof-of-principle identification of a novel LMP1-binding protein, Tmem134. We anticipate in the future that the BiFC approach will identify other novel proteins that will more fully delineate the mechanisms of LMP1 signaling. Such identification highlights new potential points of therapeutic intervention. These points of intervention have the potential to arrest the growth of tumors that require LMP1 signaling and block the establishment of EBV latency.
ACKNOWLEDGMENTS
This work was supported by the H.M. Bligh Cancer Research Laboratories, grant 08-35 of the American Cancer Society, Illinois Division, Inc., and The American Cancer Society Illinois Division Research Scholar Grant, RSG-12-229-01-MPC, to D.N.E.
We thank Bob Dickinson, Rita Levine, Neelam Sharma-Walia, and Virginie Bottero of the RFUMS Flow Cytometry Core Facility for assistance with FACS analysis.
Footnotes
Published ahead of print 1 August 2012
REFERENCES
- 1. Akavia UD, et al. 2010. An integrated approach to uncover drivers of cancer. Cell 143:1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersson B, Wentland MA, Ricafrente JY, Liu W, Gibbs RA. 1996. A “double adaptor” method for improved shotgun library construction. Anal. Biochem. 236:107–113 [DOI] [PubMed] [Google Scholar]
- 3. Ardila-Osorio H, et al. 2005. TRAF interactions with raft-like buoyant complexes, better than TRAF rates of degradation, differentiate signaling by CD40 and EBV latent membrane protein 1. Int. J. Cancer 113:267–275 [DOI] [PubMed] [Google Scholar]
- 4. Baichwal VR, Sugden B. 1988. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461–467 [PubMed] [Google Scholar]
- 5. Carbone A. 2002. AIDS-related non-Hodgkin's lymphomas: from pathology and molecular pathogenesis to treatment. Hum. Pathol. 33:392–404 [DOI] [PubMed] [Google Scholar]
- 6. Chan FK, et al. 2000. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288:2351–2354 [DOI] [PubMed] [Google Scholar]
- 7. Crawford DH, et al. 2006. A cohort study among university students: identification of risk factors for Epstein-Barr virus seroconversion and infectious mononucleosis. Clin. Infect. Dis. 43:276–282 [DOI] [PubMed] [Google Scholar]
- 8. Dawson CW, Eliopoulos AG, Blake SM, Barker R, Young LS. 2000. Identification of functional differences between prototype Epstein-Barr virus-encoded LMP1 and a nasopharyngeal carcinoma-derived LMP1 in human epithelial cells. Virology 272:204–217 [DOI] [PubMed] [Google Scholar]
- 9. Devergne O, et al. 1996. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol. Cell. Biol. 16:7098–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diamond C, Taylor TH, Aboumrad T, Anton-Culver H. 2006. Changes in acquired immunodeficiency syndrome-related non-Hodgkin lymphoma in the era of highly active antiretroviral therapy: incidence, presentation, treatment, and survival. Cancer 106:128–135 [DOI] [PubMed] [Google Scholar]
- 11. Ding Z, et al. 2006. A retrovirus-based protein complementation assay screen reveals functional AKT1-binding partners. Proc. Natl. Acad. Sci. U. S. A. 103:15014–15019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eliopoulos AG, Young LS. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 16:1731–1742 [DOI] [PubMed] [Google Scholar]
- 13. Epstein MA, Achong BG, Barr YM. 1964. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet i:702–703 [DOI] [PubMed] [Google Scholar]
- 14. Everly DN, Jr, Mainou BA, Raab-Traub N. 2008. The ID proteins contribute to the growth of rodent fibroblasts during LMP1-mediated transformation. Virology 376:258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Everly DN, Jr, Mainou BA, Raab-Traub N. 2004. Induction of Id1 and Id3 by latent membrane protein 1 of Epstein-Barr virus and regulation of p27/Kip and cyclin-dependent kinase 2 in rodent fibroblast transformation. J. Virol. 78:13470–13478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Everly DN, Jr, Mainou BA, Raab-Traub N. 2009. Transcriptional downregulation of p27KIP1 through regulation of E2F function during LMP1-mediated transformation. J. Virol. 83:12671–12679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Franken M, et al. 1996. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J. Virol. 70:7819–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gandhi MK, Tellam JT, Khanna R. 2004. Epstein-Barr virus-associated Hodgkin's lymphoma. Br. J. Haematol. 125:267–281 [DOI] [PubMed] [Google Scholar]
- 19. Hannigan A, et al. 2011. Lymphocyte deficiency limits Epstein-Barr virus latent membrane protein 1 induced chronic inflammation and carcinogenic pathology in vivo. Mol. Cancer 10:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henle G, Henle W, Diehl V. 1968. Relation of Burkitt's tumor-associated herpes-ytpe [sic] virus to infectious mononucleosis. Proc. Natl. Acad. Sci. U. S. A. 59:94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hennessy K, Fennewald S, Hummel M, Cole T, Kieff E. 1984. A membrane protein encoded by Epstein-Barr virus in latent growth-transforming infection. Proc. Natl. Acad. Sci. U. S. A. 81:7207–7211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Higuchi M, Izumi KM, Kieff E. 2001. Epstein-Barr virus latent-infection membrane proteins are palmitoylated and raft-associated: protein 1 binds to the cytoskeleton through TNF receptor cytoplasmic factors. Proc. Natl. Acad. Sci. U. S. A. 98:4675–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Izumi KM, Kieff ED. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc. Natl. Acad. Sci. U. S. A. 94:12592–12597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kao J, et al. 2009. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One 4:e6146 doi:10.1371/journal.pone.0006146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaye KM, Izumi KM, Kieff E. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. U. S. A. 90:9150–9154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kieff ED, Rickinson AB. 2007. Epstein-Barr virus and its replication, p 2603–2654 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol II Lippincott Williams and Wilkins, Philadelphia. PA [Google Scholar]
- 27. Kulwichit W, et al. 1998. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 95:11963–11968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu D, Songyang Z. 2008. ERM-mediated genetic screens in mammalian cells. Methods Enzymol. 446:409–419 [DOI] [PubMed] [Google Scholar]
- 29. Liu D, Yang X, Yang D, Songyang Z. 2000. Genetic screens in mammalian cells by enhanced retroviral mutagens. Oncogene 19:5964–5972 [DOI] [PubMed] [Google Scholar]
- 30. Magrath I. 1990. The pathogenesis of Burkitt's lymphoma. Adv. Cancer Res. 55:133–270 [DOI] [PubMed] [Google Scholar]
- 31. Mainou BA, Everly DN, Jr, Raab-Traub N. 2005. Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene 24:6917–6924 [DOI] [PubMed] [Google Scholar]
- 32. Mainou BA, Everly DN, Jr, Raab-Traub N. 2007. Unique signaling properties of CTAR1 in LMP1-mediated transformation. J. Virol. 81:9680–9692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mainou BA, Raab-Traub N. 2006. LMP1 strain variants: biological and molecular properties. J. Virol. 80:6458–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller WE, Mosialos G, Kieff E, Raab-Traub N. 1997. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J. Virol. 71:586–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moorthy RK, Thorley-Lawson DA. 1993. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of rat-1 fibroblasts. J. Virol. 67:1638–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mosialos G, et al. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80:389–399 [DOI] [PubMed] [Google Scholar]
- 37. Paine E, Scheinman RI, Baldwin AS, Jr, Raab-Traub N. 1995. Expression of LMP1 in epithelial cells leads to the activation of a select subset of NF-κ B/Rel family proteins. J. Virol. 69:4572–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pioche-Durieu C, et al. 2005. In nasopharyngeal carcinoma cells, Epstein-Barr virus LMP1 interacts with galectin 9 in membrane raft elements resistant to simvastatin. J. Virol. 79:13326–13337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raab-Traub N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 12:431–441 [DOI] [PubMed] [Google Scholar]
- 40. Rickinson AB, Kieff E. 2007. Epstein-Barr Virus, p 2655–2700 In Knipe DM, Howley PM. (ed), Fields virology, fifth ed, vol II Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 41. Roberts ML, Cooper NR. 1998. Activation of a ras-MAPK-dependent pathway by Epstein-Barr virus latent membrane protein 1 is essential for cellular transformation. Virology 240:93–99 [DOI] [PubMed] [Google Scholar]
- 42. Schultheiss U, et al. 2001. TRAF6 is a critical mediator of signal transduction by the viral oncogene latent membrane protein 1. EMBO J. 20:5678–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strausberg RL, et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. U. S. A. 99:16899–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Talaty P, Emery A, Everly DN., Jr 2011. Characterization of the latent membrane protein 1 signaling complex of Epstein-Barr virus in the membrane of mammalian cells with bimolecular fluorescence complementation. Virol. J. 8:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thornburg NJ, et al. 2006. LMP1 signaling and activation of NF-kappaB in LMP1 transgenic mice. Oncogene 25:288–297 [DOI] [PubMed] [Google Scholar]
- 46. Wang D, Liebowitz D, Kieff E. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831–840 [DOI] [PubMed] [Google Scholar]
- 47. Wu X, Li Y, Crise B, Burgess SM. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749–1751 [DOI] [PubMed] [Google Scholar]
- 48. Yasui T, Luftig M, Soni V, Kieff E. 2004. Latent infection membrane protein transmembrane FWLY is critical for intermolecular interaction, raft localization, and signaling. Proc. Natl. Acad. Sci. U. S. A. 101:278–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Young L, et al. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 321:1080–1085 [DOI] [PubMed] [Google Scholar]
- 50. Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757–768 [DOI] [PubMed] [Google Scholar]
- 51. Yu W, et al. 1997. Large-scale concatenation cDNA sequencing. Genome Res. 7:353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]