Abstract
Epstein-Barr virus (EBV) is an oncogenic human herpesvirus that dramatically reorganizes host gene expression to immortalize primary B cells. In this study, we analyzed EBV-regulated host gene expression changes following primary B-cell infection, both during initial proliferation and through transformation into lymphoblastoid cell lines (LCLs). While most EBV-regulated mRNAs were changed during the transition from resting, uninfected B cells through initial B-cell proliferation, a substantial number of mRNAs changed uniquely from early proliferation through LCL outgrowth. We identified constitutively and dynamically EBV-regulated biological processes, protein classes, and targets of specific transcription factors. Early after infection, genes associated with proliferation, stress responses, and the p53 pathway were highly enriched. However, the transition from early to long-term outgrowth was characterized by genes involved in the inhibition of apoptosis, the actin cytoskeleton, and NF-κB activity. It was previously thought that the major viral protein responsible for NF-κB activation, latent membrane protein 1 (LMP1), is expressed within 2 days after infection. Our data indicate that while this is true, LCL-level LMP1 expression and NF-κB activity are not evident until 3 weeks after primary B-cell infection. Furthermore, heterologous NF-κB activation during the first week after infection increased the transformation efficiency, while early NF-κB inhibition had no effect on transformation. Rather, inhibition of NF-κB was not toxic to EBV-infected cells until LMP1 levels and NF-κB activity were high. These data collectively highlight the dynamic nature of EBV-regulated host gene expression and support the notion that early EBV-infected proliferating B cells have a fundamentally distinct growth and survival phenotype from that of LCLs.
INTRODUCTION
Epstein-Barr virus (EBV) infection of primary resting human B cells transforms them into lymphoblastoid cell lines (LCLs). EBV-mediated growth transformation depends on the expression of a set of viral genes collectively called the latency III gene expression program. Initially after B-cell infection, EBV genomes enter the nucleus, circularize, and are chromatinized (24). The earliest latent genes are expressed from the viral W promoter (Wp) and encode the EBNA-LP and EBNA2 proteins (1, 59). EBNA2 acts as a potent transcriptional trans-activator and is complemented by EBNA-LP to induce viral genes, through the upstream C promoter (Cp), as well as cellular genes that control entry into the cell cycle (47, 58, 59). Cp-driven mRNAs accumulate through the first several cell divisions following infection, leading to increased production of the EBNA3 family of proteins (EBNA3A, -3B, and -3C), EBNA1, and the viral BHRF1 microRNAs (miRNAs) (1, 61). Cp-driven gene expression attenuates Wp usage initially (59), and ultimately, Wp is silenced through DNA methylation (53). The viral latent membrane proteins (LMPs) 1, 2A, and 2B also accumulate following EBNA2-mediated activation of their divergent promoters (19, 44, 54, 56). Additionally, an increasing body of literature indicates that a set of lytic cycle genes are expressed transiently early after B-cell infection, although no virion DNA synthesis occurs, no late gene products are made, and no virions are produced (22). Despite our understanding of the kinetic order of viral gene expression during early B-cell infection, little is understood regarding the cellular genes that are regulated during these early times after infection as B cells begin to proliferate.
Previous studies of early events following B-cell infection have identified a relatively small set of B-cell activation-, cell cycle-, and apoptosis-associated genes (8, 15, 18, 43, 47). In these studies, cyclin D2 and cyclin E1 were found to be induced concomitant with phosphorylation of Rb family members and E2F1 gene expression, at approximately 3 days postinfection (47). These changes are consistent with the initiation of EBV-driven B-cell proliferation. Furthermore, we recently identified the DNA damage response (DDR) as a potent restrictor of EBV-mediated B-cell immortalization (35). The DDR is activated during a period of hyperproliferation, 4 to 8 days following primary B-cell infection, in which cells divide once per 8 to 12 h. After the first 4 or 5 cell divisions, EBV-infected B cells attenuate this proliferation rate such that they divide once per ∼24 h, similar to LCLs (35). Genes associated with an activated DDR and proliferation follow a similar pattern of high expression early after infection and then attenuated expression in LCLs. A more comprehensive analysis of EBV-regulated host gene expression during the transition from early hyperproliferating cells to LCLs is lacking.
Several studies have been performed to analyze EBV-regulated gene expression upon de novo infection of Burkitt's lymphoma (BL) cells as well as to compare EBV-positive to EBV-negative cell lines and tumors (7). These studies collectively indicate that the core gene regulatory activities in an EBV-immortalized LCL include (i) EBNA2-driven, RBP-Jκ/CBF1/CSL-dependent activation of cell cycle regulatory and B-cell activation genes (31, 50, 63); and (ii) LMP1-mediated, constitutive, antiapoptotic NF-κB, AP1 (c-Fos/c-Jun), and ATF2 activities (6, 11). Other viral latency genes can also impact the LCL transcriptome, including the genes encoding EBNA1 (3), the EBNA3 proteins (57, 62), and viral miRNAs (46). In particular, the EBNA3 proteins impinge on host gene expression through recruitment of histone deacetylases and other chromatin modifying proteins that epigenetically modify the promoter regions of cyclin-dependent kinase inhibitor genes such as the p16(INK4a) gene (32, 45) and of apoptotic genes such as the Bim gene (4).
EBNA2 targets cellular genes primarily through its association with RBP-Jκ, in essence mimicking downstream Notch signaling (14, 17). In fact, canonical Notch targets such as Hes1 and Hey1 are also EBNA2 targets (31, 50). Furthermore, several studies have identified direct EBNA2 targets, including the transcription factors c-Myc, Ets1, and Runx3, as well as indirect EBNA2 targets, such as cyclin D2 and E2F1 (21, 48, 49, 63). A recent study of genome-wide EBNA2 targets indicates that in addition to RBP-Jκ sites, EBNA2 is also directed to cellular genes by early B-cell factor (EBF), RUNX, ETS, NF-κB, and PU.1 motifs (64). Thus, the complexity and integrative nature of EBNA2-mediated gene regulation are likely more sophisticated than was previously appreciated.
EBV latency III conversion of BL41 cells is known to alter the expression of several hundred host genes, and most of these changes are also regulated by heterologous LMP1 expression in BL41 cells (6). These data suggest an important role for the NF-κB signaling pathway as a primary mediator of EBV latency III-regulated host gene expression. In fact, the importance of these changes has been well validated by genetic and pharmacological approaches, indicating a requirement for NF-κB in the survival of LCLs (6, 23). The role of NF-κB signaling during the earliest stages of B-cell immortalization has not been studied extensively.
In this report, we comprehensively describe the dynamic changes in host gene expression during primary B-cell infection by EBV. We have identified gene ontology (GO) groups that are constitutively altered following B-cell infection and through immortalization and also those that are uniquely changed from early to late times after infection. Surprisingly, the major genes regulated from approximately 1 to 2 weeks after B-cell infection are the NF-κB-regulated genes. We further characterized and substantiated this finding with complementary experiments assessing LMP1-induced NF-κB activity and its role at different times during B-cell outgrowth.
MATERIALS AND METHODS
Cell lines and culture conditions.
Human peripheral blood mononuclear cells (PBMCs) were obtained by Ficoll purification (Histopaque-1077 column; Sigma) of buffy coats from healthy donors (Carolina Red Cross) and were kept in RPMI 1640 medium (Invitrogen) supplemented with 15% fetal bovine serum (Gemini Bio Products), 100 U/ml penicillin, and 2 mM l-glutamine (Invitrogen). B cells were separated using human B-lymphocyte enrichment set DM (IMag; BD) as recommended by the manufacturer. PBMCs were infected with limiting amounts of B95-8 virus (0.4 to 12 μl EBV B95-8 Z-HT supernatant/1 × 106 PBMCs) in the presence of cyclosporine (0.5 μg/ml) for 1 h at 37°C. The cells were washed once in phosphate-buffered saline (PBS) and then resuspended in fresh medium. For each infection condition, 105 cells were plated in a 96-well plate (10 wells in total) to grow as LCLs. Microarray samples were generated from monoclonal LCLs produced from the smallest amount of virus used. For the purification of EBV-positive and proliferating B cells, 1.5 × 108 PBMCs were stained with 4 μM 6-carboxyfluorescein succinimidyl ester (CFSE) for 3 min and then washed three times in washing buffer (PBS plus 5% fetal bovine serum). Stained PBMCs (106/ml) were infected with 25 μl/1 × 106 PBMCs of B95-8 virus as already described. The cells were washed once in PBS and then resuspended in fresh medium. At 6 days postinfection, CFSE-labeled PBMCs were washed in washing buffer and stained with allophycocyanin (APC)-labeled anti-CD19 (BD Biosciences) (2 μl/3 × 105 PBMCs) for 30 min on ice. After 3 washes in washing buffer, CD19+ CFSElow cells were sorted on a FACSVantage flow cytometer. Sorted B cells were analyzed for the presence of the EBV genome by fluorescence in situ hybridization and for EBNA-LP by immunofluorescence assay (35).
Antibodies.
Primary antibodies to RelA, RelB, IκBα, and NF-κB2/p52 were purchased from Cell Signaling. Phospho-specific antibodies to Ser176/180 or Ser177/181 of IκB kinase α/β (IKKα/β), Ser32 of IκBα, and Ser536 of p65 were also purchased from Cell Signaling. Primary antibodies to TRAF1, α-tubulin, and MAGOH were purchased from Santa Cruz Biotechnology. Horseradish peroxidase (HRP)-conjugated anti-rabbit and anti-mouse secondary antibodies were purchased from Cell Signaling. Mouse anti-human CD19 antibody was purified from the supernatant of the 33-6-6 hybridoma cell line (a kind gift of T. Tedder, Duke University Medical Center) and conjugated to either phycoerythrin (PE) or APC for use as a surface B-cell marker in flow cytometry. An agonistic anti-CD40 antibody was purified from the supernatant of the G28-5 hybridoma cell line (a kind gift of E. Kieff, Harvard Medical School).
Compounds.
IKKβ was inhibited using the Calbiochem IKK-2 inhibitor IV. Cell proliferation was measured by staining PBMCs with CFSE (Sigma) or CellTrace violet (Invitrogen).
Microarray analysis and gene enrichment.
Total mRNAs from four healthy donors were prepared under three conditions: (i) uninfected purified CD19+ B cells; (ii) infected PBMCs, sorted at 6 days postinfection for population doubling 1 to population doubling 4 (PD1-4; Prolif cells); and (iii) monoclonal LCLs derived by limiting virus dilution on PBMCs. cDNA preparation, labeling, and fragmentation were performed using a GeneChip wild-type cDNA synthesis and amplification kit (Affymetrix) and Exon array labeling kits (Affymetrix). Twelve samples (four for each condition) were hybridized to HuEx 1.0ST Exon arrays (Affymetrix), and the chips were scanned at the Duke Microarray Facility. The resultant CEL files were RMA normalized, and the data were analyzed with GenePattern (36) and GSEA v2 (52) to analyze c3 transcription factor sets. Differential expression of genes was evaluated by applying 2-way analysis of variance (ANOVA) or significance analysis of microarrays (SAM) (55). Gene ontology categories were assessed using the PANTHER algorithm (34). K-means clustering of genes (2-way ANOVA P value of <0.01; 2-fold change) with 8 nodes was performed using GenePattern (36).
B-cell proliferation assays.
Human PBMCs were stained with CellTrace violet and then infected in bulk by incubation with supernatant from B95-8 Z-HT cells. IKK-2 inhibitor IV (1 μM) or the agonistic CD40 antibody G28-5 (1 μg/ml) was added to the cells at the various time points described. The inhibitor or agonistic antibody was washed out of the medium every 7 days and added back or not, depending on the time. After 21 days, the PBMCs were washed with PBS, stained with CD19-PE antibody and 7-aminoactinomycin D (7-AAD), and analyzed by flow cytometry in the presence of 10,000 Sepharose AccuCount blank particles (Spherotech). Live proliferated B cells (CD19+ CellTrace violetlow 7-AADlow) were counted and normalized to the bead count. The experiment was replicated for eight healthy human PBMC donors.
Transformation assays.
Infection of human PBMCs by EBV B95-8 was performed in the presence of 0.1% dimethyl sulfoxide (DMSO), 1 μM IKK-2 inhibitor IV, or 1 μg/ml of agonistic CD40 antibody (G28-5), added at different times postinfection. B95-8 Z-HT supernatant was titrated from 1 ml/107 PBMCs to 30 μl/107 PBMCs. A total of 2.5 × 106 infected PBMCs were seeded in 24 wells of a 96-well plate for each infection point. The percentage of wells positive for B-cell outgrowth (LCLs) at 5 weeks postinfection was plotted relative to the amount of virus used per well. The efficiency of transformation was determined as published previously (16), with the amount of B95-8 virus necessary to yield 62.5% positive wells considered 1 transforming unit (TU) per well.
Western blotting.
Samples were lysed in a 1% Triton X-100-containing buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 1 mM dithiothreitol [DTT], 20 mM NaF, 10 mM sodium pyrophosphate, and Complete protease inhibitors without EDTA) and normalized to total protein content by the Bradford assay (Bio-Rad). All samples were run in Novex 4 to 12% Bis-Tris gels and blotted using standard procedures. The antibodies used are described above.
RNA extraction and quantitative RT-PCR (qRT-PCR).
RNA was extracted from purified primary B cells by use of a Qiagen RNeasy kit. Reverse transcription (RT) to generate cDNA was performed with a High Capacity cDNA kit (Applied Biosystems). Real-time PCR was performed using either SYBR green (Quanta Biosciences) or specific TaqMan probes in an Applied Biosystems Step One Plus instrument. A20 and TRAF1 were detected using the exon-spanning TaqMan probes Hs00560402 and Hs00194639, respectively (Applied Biosystems). ICAM1, c-FLIP, and LMP1 were detected using exon-spanning probes purchased from Integrated DNA Technologies, along with SYBR green.
Microarray data accession number.
Exon array data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE20200.
RESULTS
EBV dynamically regulates mRNA transcript levels during primary B-cell outgrowth following infection.
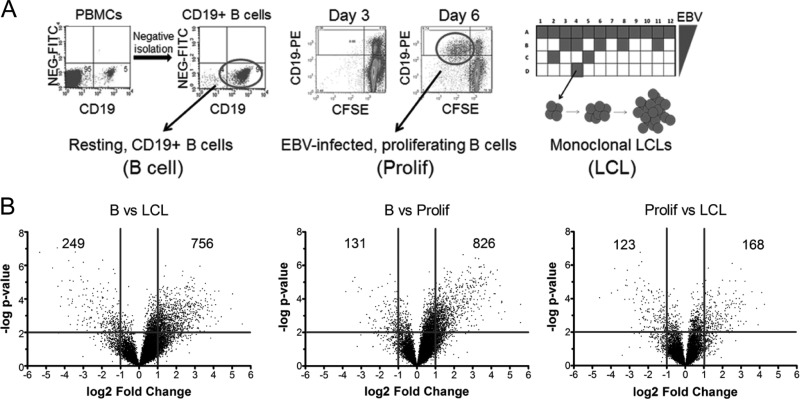
The global host gene expression profile in primary B cells at early times after EBV infection is poorly characterized. Therefore, we performed gene expression analysis using Affymetrix Human Exon 1.0 ST (HuEx) microarrays and mRNAs from 4 biological replicates of (i) purified adult human CD19+ peripheral blood B cells, (ii) EBV-infected and sorted proliferating B blasts at 6 days postinfection (Prolif cells), and (iii) monoclonal LCLs (Fig. 1A). More than 90% of the sorted proliferating B cells were infected with EBV, as determined by fluorescence in situ hybridization of the EBV genome and immunofluorescence assay of EBNA-LP (35). Hybridization data were RMA normalized, and significant gene expression changes were visualized using volcano plots of the transitions from B cells to Prolif cells, Prolif cells to LCLs, and B cells to LCLs (Fig. 1B). EBV more strongly increased than decreased mRNA levels upon initial B-cell proliferation, as 826 genes were significantly induced (>2-fold; P < 0.01), while only 131 genes were significantly repressed (>2-fold). However, mRNA levels changed more evenly during the transition from Prolif cells to LCLs, where 168 mRNAs were increased >2-fold and 123 mRNAs were reduced >2-fold. Overall, from resting B cells to LCLs, 756 mRNAs were increased >2-fold, while 249 mRNAs were reduced >2-fold.
Fig 1.
EBV regulates host gene expression throughout B-cell outgrowth. (A) Schematic of samples used for HuEX microarray analysis. (Left) CD19+ B cells were purified from PBMCs (Carolina Red Cross) by negative isolation. Dot plots show CD19-PE staining on the x axis and the nonspecific fluorescein isothiocyanate (FITC) channel on the y axis for PBMCs (left) or negatively isolated cells (right). (Middle) Dot plots of CD19-PE (y axis) versus CFSE (x axis) indicate B cells proliferating after EBV infection. The left panel shows cells at 3 days postinfection, when few cells are dividing (i.e., CD19+ CFSElow), while by 6 days postinfection, proliferating B cells are readily visible (right). The circle indicates the population of cells that was sorted for each donor. (Right) Schematic diagram of a plate in which EBV infection of PBMCs leads to monoclonal LCL outgrowth. (B) Volcano plots showing gene expression changes between time points. The y axis depicts the negative log of the P value, and the x axis depicts the log2 value for the observed fold change. Vertical lines indicate a 2-fold span (P value of 0.01).
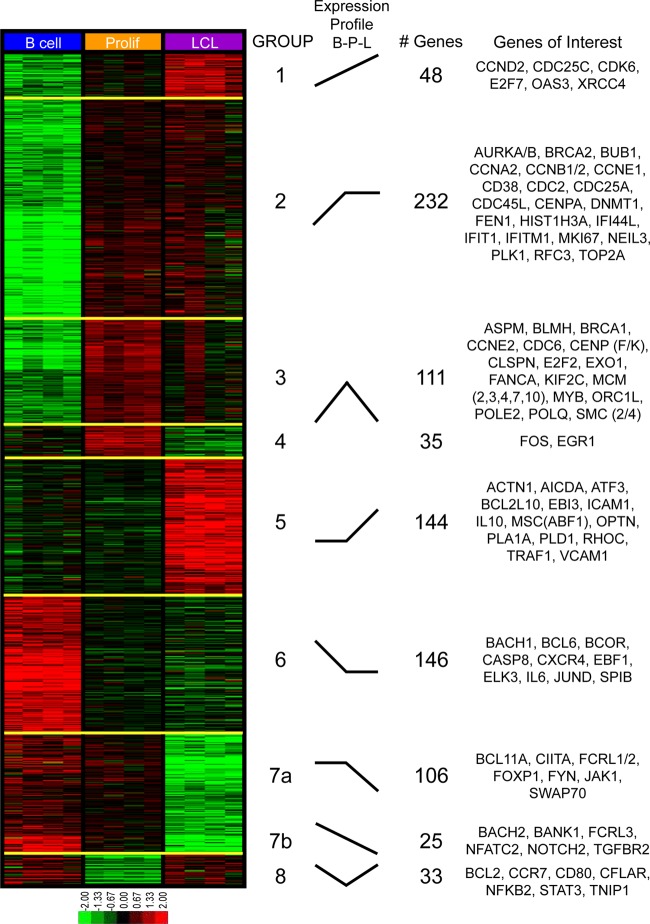
Genes exhibiting significant (2-way ANOVA P value of <0.01) and robust (2-fold) expression changes across any of the three temporal states generated a universe of 874 transcripts that we analyzed further to assess dynamic gene expression changes and gene ontology. We performed K-means clustering on these mRNAs, using 8 groups to represent all possible changes between the three temporal states. This analysis clearly revealed that early proliferating cells are distinct from both resting B cells and LCLs and that transcriptomic changes induced by EBV are diverse and temporally complex (Fig. 2). Groups 1 and 2 contain genes induced in the transition from B cells to LCLs, most of which were induced primarily during the B-cell-to-Prolif-cell transition. Groups 3 and 4 contain genes that were highly induced in the transition from B cells to Prolif cells and then repressed to different extents through LCL outgrowth. Group 5 contains genes that were statistically unchanged in the transition from B cells to Prolif cells but were induced in the transition from Prolif cells to LCLs. Group 6 genes were repressed early and remained low during LCL outgrowth, while groups 7a and 7b were repressed at the late transition from Prolif cells to LCLs. Finally, group 8 represents genes that were repressed early after infection and strongly induced during the transition from Prolif cells to LCLs.
Fig 2.
K-means clustering reveals distinct patterns of EBV-induced transcriptomic changes throughout outgrowth. Four biological replicates of negatively isolated resting B cells, sorted early proliferating CD19+ CFSElow cells (Prolif), and monoclonal LCLs were isolated as described in the legend to Fig. 1A and then subjected to microarray analysis. Genes exhibiting significant (two-way ANOVA P value of <0.01) and robust (>2-fold) gene expression changes were subjected to K-means clustering analysis. The gene expression level is represented by a two-tone color scale with log2 values (shown at the bottom). Eight groups (groups 1 to 8) categorize the EBV-induced expression changes during B-cell outgrowth, and their relative expression changes are depicted schematically under “expression profile.” Each cluster contains a distinct number of genes, and representative genes from each cluster are listed to the right.
Functional annotation of EBV-regulated temporal gene expression profiles.
We next characterized the GO categories of the EBV-regulated genes during early versus late stages of B-cell growth transformation, using PANTHER analysis of the K-means-clustered groups (34). PANTHER allows for GO analysis of the following components: (i) pathways, (ii) biological processes, (iii) molecular functions, (iv) cellular components, and (v) protein classes. Importantly, PANTHER determines whether the observed number of genes from a given GO group is enriched or depleted relative to that predicted based on a reference list, which in our case was the universe of genes expressed above background at any of the three sample points.
PANTHER analysis revealed expected and also novel changes in GO classes during transformation (Table 1). K-means groups 1 and 2, describing the early induction of B-cell proliferation, were characterized by robust increases in genes associated with the cell cycle, mitosis, p53 pathway signaling, and responses to stress. Similarly, the genes that were induced initially and repressed later in transformation (groups 3 and 4) included those associated with the cell cycle, microtubule function, and DNA helicase and DNA binding activities. These findings are consistent with those reported previously by our group indicating an early phase of cellular hyperproliferation characterized by an activated DNA damage response, which was attenuated from the early to late stages of transformation (35).
Table 1.
Summary of gene ontology analysis results
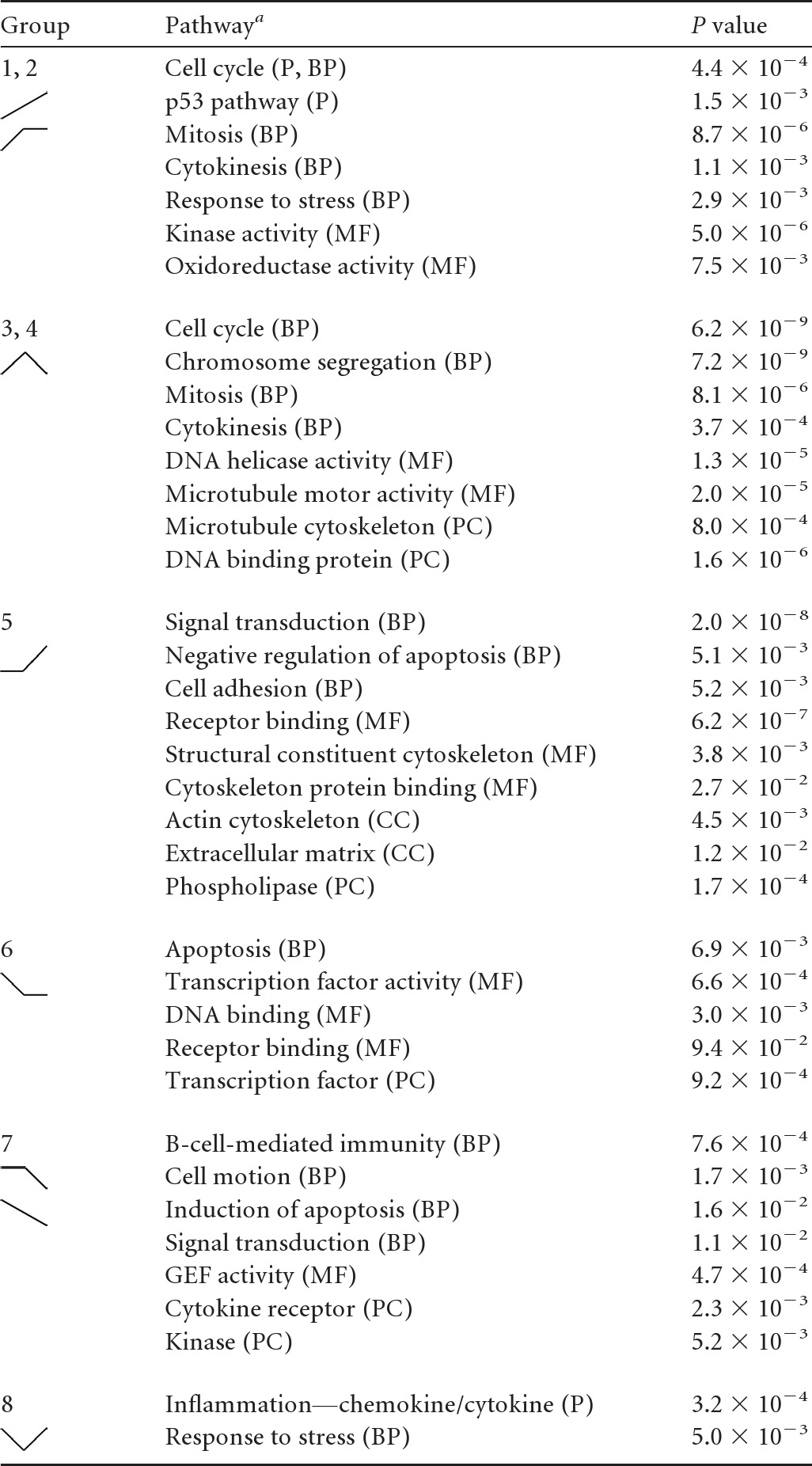
P, pathways; BP, biological processes; MF, molecular functions; CC, cellular component; PC, protein class.
Unexpected changes were observed in group 5, where GO groups associated with signal transduction, cell adhesion, negative regulation of apoptosis, the actin cytoskeleton, and cytoskeleton protein binding were enriched in genes upregulated uniquely in the transition from early proliferating cells to LCLs. These changes are consistent with known activities associated with LMP1 and NF-κB signaling (6). In fact, a simple analysis of the genes included in this group revealed many known NF-κB targets, including TRAF1, EBI3, and ICAM1, among others (Fig. 2).
K-means groups 6, 7, and 8 comprise genes that were repressed upon EBV infection and are characterized by a diverse set of GO classes. K-means group 6 genes were repressed upon early proliferation and were enriched for genes encoding DNA binding proteins, specifically transcription factors, including both repressors such as BCL6, BCOR, and BACH1 and activators such as JUND, EBF1, and ELK3. Furthermore, chemokine receptor genes were enriched in this group, including the most EBV-downregulated gene in our analysis, the CXCR4 gene. K-means group 7 was characterized by genes that were repressed uniquely late during the transition from early proliferating B cells to LCLs. This group was enriched for genes associated with B-cell immunity, cell motion, the induction of apoptosis, and cytokine receptors. The genes in these groups included those encoding TGFBR2, NOTCH2, and FCRL1, -2, and -3. K-means group 8 included genes that were initially repressed and then induced from proliferating cells to LCLs. Interestingly, these genes were highly enriched for those involved in inflammation due to chemokines and cytokines. Furthermore, among the genes present in this group were those for NFKB2 and STAT3, which regulate chemokine and cytokine expression and signaling.
Transcriptional regulation during EBV immortalization.
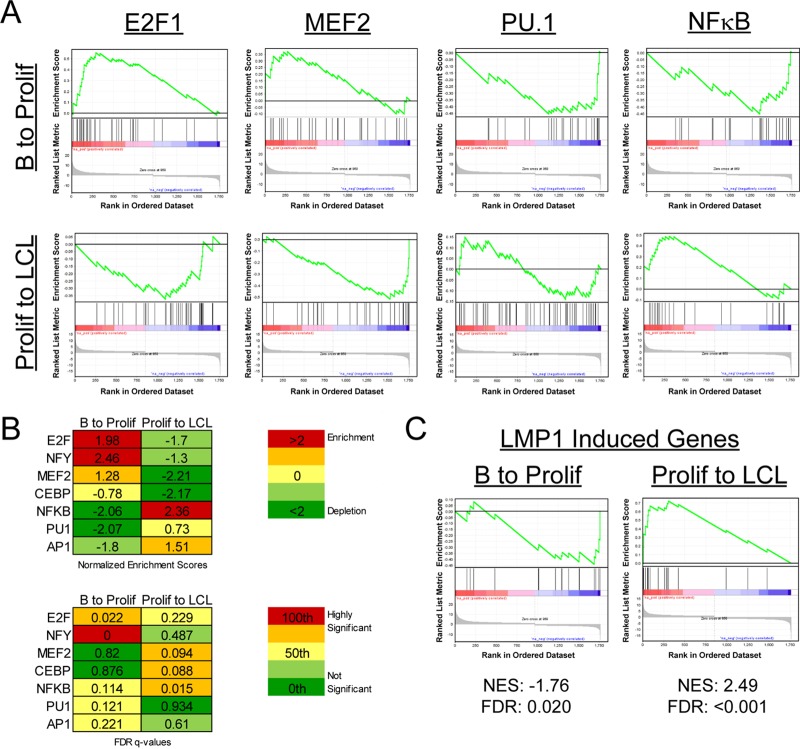
We next sought to define the transcriptional regulatory elements that controlled the dynamic mRNA expression changes that we observed from early EBV-induced proliferation through immortalization. Therefore, we used gene set enrichment analysis (GSEA) to define the transcription factor binding sites that were enriched or depleted in the upstream regions of genes changing through EBV-driven B-cell outgrowth. Enrichment or depletion is shown by a normalized enrichment score (NES), which is calculated by the degree to which a gene set is overrepresented at the extremes of a ranked list of genes and then normalized to the gene set size (52). NES are also reported with false discovery rate (FDR) values that represent the possibility that a given NES is a false-positive finding (52). Considering first the transition from resting B cells to early proliferating cells (B to Prolif), we observed a strong enrichment in S-phase-inducing transcription factors such as E2F1 (Fig. 3A and B). Upregulated genes containing binding sites for NF-Y were also significantly enriched in the B-to-Prolif transition set. Concurrently, a significant depletion of genes containing consensus upstream NF-κB, PU.1, and AP1 elements was observed in the B-to-Prolif data set (Fig. 3A and B).
Fig 3.
Gene set enrichment analysis reveals differential transcription factor usage between early and late times postinfection. (A) GSEA of transcription factor usage (MSigDB c3) was performed on our set of genes that changed significantly (>2-fold) in the transitions from resting B cells to early proliferating cells (B to Prolif) and from proliferating B cells to monoclonal LCLs (Prolif to LCL). Enrichment plots for E2F1, MEF2, PU.1, and the GGGNNTTTCC consensus site for NF-κB are shown. (B) (Top) NES for each transcription factor for B to Prolif as well as Prolif to LCL. (Bottom) False discovery rate Q values (FDR Q values) for the plots. (C) A set of LMP1-induced genes was constructed based on previously published data (6), and GSEA was performed on the previously mentioned gene sets. LMP1-induced genes were repressed early after infection (B to Prolif) and enriched later (Prolif to LCL). NES and FDR Q values are shown beneath the plots.
Genes changing in the transition from Prolif cells to LCLs were regulated differently from those from the B-to-Prolif transition. For example, genes containing targets of the lytic gene Zta-inducing transcription factors CEBP and MEF2 were strongly and significantly depleted from the Prolif-to-LCL transition (Fig. 3A and B). Consistent with attenuation of the B-cell proliferation rate (35), E2F1 targets were strongly downregulated from the Prolif-to-LCL data set. Finally, consistent with our GO analysis of K-means clustering data, NF-κB targets were robustly and significantly upregulated in this later phase of transformation. Furthermore, GSEA analysis indicated that a known set of LMP1-induced genes (6) was also initially depleted and then significantly enriched from Prolif cells to LCLs (Fig. 3C).
NF-κB targets are not induced until high levels of LMP1 mRNA and protein are detected in infected lymphoblasts.
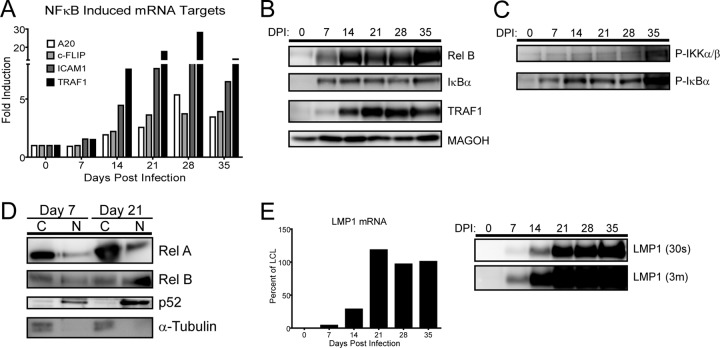
The induction of NF-κB- and LMP1-regulated genes at late times during EBV-driven B-cell outgrowth was unexpected. Therefore, we validated our microarray results by using quantitative RT-PCR, Western blotting, and activation analysis of the NF-κB pathway during a time course experiment following primary B-cell infection with EBV. First, we confirmed that mRNA induction of the NF-κB targets A20, c-FLIP, ICAM1, and TRAF1 occurred after the first week following EBV infection (Fig. 4A). Western analysis of RelB, IκBα, and TRAF1 corroborated the delayed NF-κB mRNA target induction (Fig. 4B).
Fig 4.
NF-κB targets and activity, as well as LMP1, are not induced until late after EBV infection. (A) qRT-PCR analysis of NF-κB-targeted genes relative to resting B-cell levels. RNA was isolated from EBV-infected B cells every 7 days for 35 days postinfection. Data are representative of four experiments. (B) Western blot of NF-κB-targeted proteins. Protein was isolated from EBV-infected B cells every 7 days for 35 days postinfection. MAGOH is a ubiquitously expressed splicing factor that was used as a loading control because it does not significantly change at the mRNA or protein level during EBV infection of primary B cells. (C) Western blots depict the activity of the NF-κB pathway through phospho-specific Western blotting of IκBα and IKKα/β following primary B-cell infection. (D) Western blots depicting the activity of both canonical and noncanonical NF-κB pathways by showing nuclear (N) versus cytoplasmic (C) distributions of RelA, RelB, and p52. α-Tubulin is the loading and fractionation control. (E) LMP1 does not accumulate to LCL levels until 21 days postinfection. (Left) qRT-PCR analysis of the LMP1 mRNA transcript, normalized to the day 35 level as 100% of the LCL level. Data are representative of four experiments. (Right) Western blot showing LMP1 protein levels (S12 antibody, 30-s exposure). An overexposed blot (3m = 3 min) is provided to show that while the LMP1 protein was detected as early as 7 days postinfection, it was detectable only as a fraction of LCL LMP1 protein levels.
NF-κB target gene activation in LCLs is dependent on LMP1 signaling and proceeds through activation of the IκB kinase complex, phosphorylation and ubiquitin-dependent degradation of IκB family members, and NF-κB nuclear translocation (6, 30). To corroborate the gene expression changes we observed at late times following infection, we analyzed the activating phosphorylation of IKKα/β, degradation-inducing phosphorylation of IκBα, and nuclear translocation of p65/RelA, RelB, and processed NF-κB2/p52. Consistent with our gene expression data, each of these activities increased significantly from day 7 postinfection to day 21 postinfection (Fig. 4C and D). Finally, we observed that the steady-state LMP1 mRNA and protein levels were induced to near LCL levels only after approximately 14 to 21 days postinfection (Fig. 4E). These data suggest that the delay in NF-κB activation was likely due to the delay in LMP1 protein expression following primary B-cell infection.
Rescuing NF-κB activity specifically favors growth of early proliferating cells.
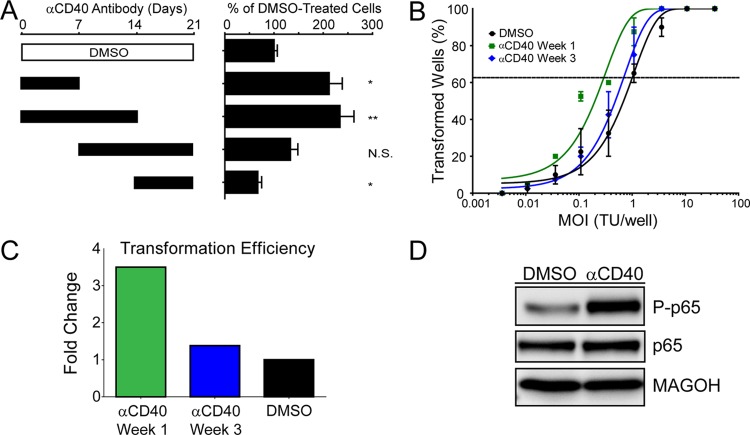
The lag in NF-κB activity in early EBV-infected proliferating B cells prompted us to ask whether heterologous activation of NF-κB would rescue EBV-induced proliferation or transformation efficiency. It is anecdotally known that EBV-infected B cells grow out at a higher frequency in the presence of CD40 ligand (CD40L)-expressing feeder cells. However, we wondered whether this result was strictly dependent on the time frame within which the EBV-infected blasts must be exposed to this heterologous NF-κB activating signal. We specifically asked whether EBV-induced B-cell outgrowth would be affected by CD40 activation by using the agonistic antibody G28-5 during 1- or 2-week intervals following EBV infection of primary B cells. As shown in Fig. 5A, when the G28-5 CD40 antibody was present only during the first week after EBV infection, the number of proliferating B cells at 21 days was double that if the antibody was not present. A similar result was obtained if the antibody was present during the first 2 weeks. However, if the antibody was present only during the third week or during the second and third weeks following B-cell infection, then no increase in EBV-driven B-cell proliferation was observed (Fig. 5A). Since the proliferation assay was performed at 21 days postinfection, close to the time point of the late G28-5 additions, we also performed transformation assays (5-week outgrowth with a limiting dilution of virus). Indeed, the efficiency of B-cell transformation was increased approximately 4-fold if G28-5 was present during the first week after infection, whereas no increase was observed if G28-5 was present during the third week after infection (Fig. 5B and C). Importantly, the G28-5 antibody activated the NF-κB pathway in LCLs, as indicated by the IKKβ-dependent phosphorylation of the NF-κB transcription factor subunit p65 at serine 536 (Fig. 5D) (33, 39).
Fig 5.
Rescuing NF-κB activity specifically favors growth of early proliferating cells. (A) NF-κB was activated via CD40 signaling with 1 μg/ml of the agonistic antibody G28-5. The antibody was present in the culture for 1- or 2-week increments, as denoted by the solid black bars, and was washed out at either 7, 14, or 21 days postinfection. After 21 days, samples were analyzed by fluorescence-activated cell sorting (FACS), and events were normalized to a known number of Sepharose beads present in the sample. Total CD19+ CFSElow (proliferating) B cells present after 21 days were normalized to a DMSO vehicle control. Error bars denote standard errors of the means (SEM) for eight biological replicates. Significance compared to the DMSO vehicle control was calculated by Student's t test. N.S., not significant; *, P < 0.01; **, P < 0.001. (B) Quantification of EBV-induced B-cell outgrowth following PBMC infection in the presence of DMSO vehicle control (black) or the agonistic anti-CD40 antibody G28-5, present in the culture from days 0 to 7 (αCD40 Week 1; green) or days 14 to 21 (αCD40 Week 3; blue). Error bars are SEM for two biological replicates. (C) Transformation efficiencies for the αCD40 Week 1 and αCD40 Week 3 groups are shown as fold changes over the DMSO control level. (D) LCLs were treated with DMSO vehicle control or 1 μg/ml G28-5 antibody. Activity of the NF-κB pathway is shown by the presence of the phosphorylated form of p65 (P-p65).
Early EBV-infected proliferating cells are less NF-κB dependent than later proliferating cells and LCLs.
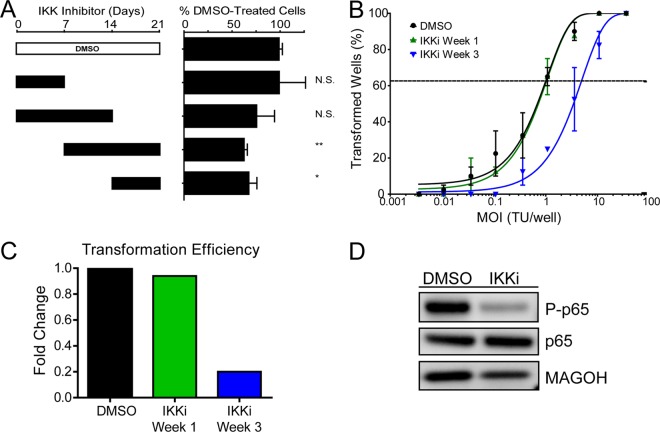
The dependence of EBV-transformed cells on NF-κB for survival is firmly established (6, 23). However, the point after EBV infection when B cells become sensitive to NF-κB inhibition has not been established. Given our gene expression data, we hypothesized that early proliferating B cells during the first week following infection would be significantly less sensitive to NF-κB inhibition than infected cells at 3 weeks postinfection, when LMP1 is expressed at high levels, along with many NF-κB targets. Therefore, we performed experiments similar to those described above for CD40 agonism, but using a specific inhibitor of the IKKβ subunit of the IKK complex. EBV-induced B-cell proliferation at 21 days postinfection was unchanged upon IKK inhibition during week 1 after infection only (Fig. 6A). In contrast, cells inhibited at weeks 2 and 3 or only during week 3 showed significantly impaired outgrowth. Consistently, EBV transformation was reduced 5-fold if IKKβ was inhibited during week 3 only, whereas inhibition during week 1 had no impact on transformation (Fig. 6B and C). Inhibiting the IKKβ complex in LCLs reduces NF-κB activity, as shown by a decrease in the phosphorylated form of the NF-κB transcription factor p65 (Fig. 6D). Therefore, the temporal delay in expression of LMP1 and NF-κB target genes results in a period during which EBV-infected proliferating B cells are insensitive to IKK inhibition.
Fig 6.
Early proliferating cells are less NF-κB dependent than later proliferating cells and LCLs. (A) NF-κB activity was inhibited via a small-molecule inhibitor of IKKβ (IKKi) (1 μM). The inhibitor was present in the culture for 1- or 2-week increments, as denoted by the solid black bars, and was washed out at either 7, 14, or 21 days postinfection. After 21 days, samples were analyzed by FACS, and events were normalized to a known number of Sepharose beads present in the sample. Total CD19+ CFSElow (proliferating) B cells present after 21 days were normalized to a DMSO vehicle control. Error bars denote SEM for eight biological replicates. Significance compared to the DMSO vehicle control was calculated by Student's t test. N.S., not significant; *, P < 0.05; **, P < 0.01. (B) Quantification of EBV-induced B-cell outgrowth following PBMC infection in the presence of DMSO vehicle control (black) or the IKKβ small-molecule inhibitor IKKi, present in the culture from days 0 to 7 (IKKi Week 1; green) or days 14 to 21 (IKKi Week 3; blue). Error bars are SEM for two biological replicates. (C) Transformation efficiencies for the IKKi Week 1 and IKKi Week 3 groups are shown as fold changes over the DMSO control level. (D) LCLs were treated with DMSO vehicle control or with 1 μM IKKi. Activity of the NF-κB pathway is shown by the presence of the phosphorylated form of p65 (P-p65).
DISCUSSION
In this report, we have comprehensively analyzed the changes in cellular mRNA transcript abundance between primary human resting CD19+ B cells, EBV-infected early hyperproliferating B blasts, and EBV-immortalized LCLs. Early studies of EBV-regulated gene expression changes identified the induction of E2F family members and the phosphorylation of pocket proteins enabling E2F-mediated transcription and S-phase induction within the first few days postinfection (8, 18, 43, 47). The induction of B-cell activation markers at early times after infection has also been reported (15, 42). Here we extended these studies to transcriptome-wide analysis and found substantially more dynamic regulation of host gene expression from 6 days to 35 days postinfection than was previously appreciated. Genes activated early after infection were enriched for those with E2F1 and NF-Y binding sites in their promoters and depleted for those with PU.1, AP1, and NF-κB sites. At late times after infection, genes with MEF2 and CEBP sites were depleted, while NF-κB targets were strongly enriched. Surprisingly, the LMP1-mediated NF-κB signaling pathway was not induced robustly until approximately 2 weeks postinfection. Consistently, early EBV-infected blasts were not sensitive to NF-κB inhibition, and heterologous NF-κB activation through CD40 agonism increased the transformation efficiency.
Gene ontology categories associated with the dynamic gene expression changes confirmed and extended previous observations. As expected, proliferation- and DNA damage-responsive genes were highly induced early after infection and then attenuated during B-cell outgrowth (35). E2F1 and NF-Y targets were robustly induced early, consistent with induction of S phase and with reports that NF-Y contributes to upregulation of the EBV latency C promoter, respectively (5). At later times, the observed repression of MEF2 and CEBP targets is consistent with suppression of the lytic Zta gene promoter and tight control of viral latency during B-cell outgrowth (28, 60). Finally, and most surprisingly, was the delay in LMP1-regulated gene expression that occurred during the transition from early proliferating cells to LCLs. This late transition to LCLs was characterized by an enrichment in genes associated with the IκB kinase cascade, actin cytoskeleton dynamics, inhibition of apoptosis, and a strong upregulation of NF-κB targets.
Several hypotheses may account for the delay in NF-κB activity. First, in our GSEA analysis of transcription factors regulated in the transitions from B cells to Prolif cells and from Prolif cells to LCLs, we identified PU.1/SPI1 targets as being depleted early and then not changing significantly at later times after infection. PU.1 is a critical transcription factor targeted by EBNA2 on the LMP1 promoter (19). Therefore, it is conceivable that genome-wide PU.1 activity is a proxy for LMP1 promoter activity, indicating that the promoter may not be activated sufficiently early after infection to drive LMP1 expression. At later times, other cofactors may be increased in expression or activity, cooperating with PU.1 to drive LMP1 expression.
An alternative to this hypothesis relates to the high-level c-Myc activity found in early hyperproliferating cells (35), which may directly attenuate NF-κB activity, and consequently LMP1 levels, through inhibition of an LMP1/NF-κB feed-forward loop (10, 20). This would be consistent with work indicating that high-level c-Myc expression inhibits NF-κB activity and with the notion that Myc-driven tumors such as Burkitt's lymphoma are poorly immunogenic, due to low-level NF-κB activity, compared to diffuse large B cell lymphomas (DLBCLs) or LCLs (9, 12, 25). Therefore, a set of c-Myc targets may be responsible in part for low LMP1 levels.
We recently identified the miRNA expression profiles of resting and early proliferating EBV-infected B cells and LCLs (13). The set of miRNAs distinctive for the early hyperproliferating cells included the c-Myc-regulated miRNA clusters miR-17∼92, miR-106a∼363, and miR-106b∼25. Concurrently, in collaboration with Bryan Cullen's group, we also recently identified the set of mRNAs bound by viral and cellular miRNAs in LCLs (46). In this study, the c-Myc-regulated miR-17 seed family was found to be a suppressor of LMP1 mRNA and protein levels. Therefore, in LCLs, the equilibrium of this interaction may be such that LMP1 is expressed at a high level, while it is possible that during early outgrowth, when miR-17 levels are highest, the effects on LMP1 are more profound. Therefore, we propose that LMP1 may be suppressed early after infection by the EBNA2-driven, c-Myc-regulated miR-17 miRNA seed family.
It is also important that we performed all of our infections with the B95-8 strain of EBV. While this could have an effect on our overall expression profiling due to the loss of the BART miRNAs, it was recently shown that B95-8 complemented with the deleted BART miRNAs induces normal B-cell proliferation and transformation (41). While it has been reported that some of the deleted BART miRNAs might target LMP1 (29, 37), these interactions would have a repressive effect on LMP1 expression, and in their absence, one might expect premature LMP1 expression. Since we characterized a long delay in LMP1 expression and activity following primary B-cell infection, it is unlikely that these BART miRNA-LMP1 interactions play a significant role in this phenotype.
One final and complementary hypothesis for the delay in LMP1 levels at early times after B-cell infection is that LMP1 mRNA stability may increase during outgrowth. Previous studies indicate that the LMP1 gene is the most poorly transcribed gene of the viral latent genes in LCLs, but its mRNA is the most abundant transcript (40). Therefore, the LMP1 mRNA is likely stabilized in LCLs, and the trans-acting factors required for this stability increase through B-cell outgrowth. Perhaps these putative factors are also suppressed by Myc-regulated miRNAs or directly by Myc.
Irrespective of the mechanism by which LMP1 expression is attenuated until 2 weeks postinfection, our data raise the question of how these proliferating B cells are able to survive in the absence of high-level NF-κB activity. There are two plausible explanations for this that will be intriguing to characterize in the future. First, Altmann and Hammerschmidt found that the EBV Bcl-2 homologs BHRF1 and BALF1 are both required for primary B-cell transformation (2). These genes are both expressed transiently upon primary B-cell infection and are required to suppress apoptosis. While it is unclear how these genes are regulated throughout B-cell outgrowth, their expression at early times postinfection may be required for survival in the absence of LMP1-mediated NF-κB signaling.
A second and complementary hypothesis is that EBNA2-dependent mRNA targets may be sufficient for survival during early outgrowth (26, 27). Since EBV induces a robust DDR during this early period, it is conceivable that EBNA2-mediated survival is initially more important than LMP1 effects. However, for long-term outgrowth, the trade-offs associated with the growth-suppressive effects of the DDR are too great, and LMP1 is then critically important for survival.
The interplay between c-Myc and NF-κB is important in the balance between proliferation, survival, and immunogenicity of EBV-infected cells. As described above, c-Myc and NF-κB gene expression programs are incompatible in B-cell lymphomas (9). One consequence of the c-Myc-driven gene expression program associated with low-level NF-κB activation is greatly decreased immunogenicity (38, 51). This is best observed in Burkitt's lymphoma cells, which display poor immunogenicity due to decreased expression of major histocompatibility complex (MHC) class I molecules, antigen processing machinery, and proinflammatory cytokines and chemokines (9). Thus, during primary B-cell infection, EBV may attenuate NF-κB activity (dependent or independent of c-Myc) in order to evade the immune system, while generating many progeny latently infected cells through hyperproliferation. In acute infection, this immune evasion could occur through suppression of proinflammatory cytokine production, while in the context of naïve B-cell infection following reactivation, this could be due to depressed cytotoxic T-lymphocyte (CTL) recognition of early proliferating cells. Studies of CTL responses to early EBV-infected B cells compared to LCLs and of the levels of proinflammatory cytokines produced in these cells will be informative. Such future studies would support an important and novel mechanism by which EBV establishes and maintains latency in the host.
ACKNOWLEDGMENTS
We thank the Duke Cancer Institute Flow Cytometry Core Facility for help on this project (specifically Mike Cook, Nancy Martin, and Lynn Martinek). We thank Elliott Kieff (Harvard Medical School) for the G28-5 (anti-CD40) hybridoma and Thomas Tedder (Duke University) for the 33-6-6 (anti-CD19) hybridoma. Also, we thank Neel Mukherjee as well as members of the Luftig laboratory for their helpful comments and discussions.
This work was supported by pilot grants to M.A.L. from Golfers Against Cancer, the American Cancer Society, and the Duke CFAR, an NIH-funded program (5P30 AI064518), as well as by NIH grant 1R01-CA140337. A.M.P. was supported by NIH grant 5T32CA009111, and J.P.T. was supported by a SMART fellowship from the Department of Defense.
Footnotes
Published ahead of print 1 August 2012
REFERENCES
- 1. Alfieri C, Birkenbach M, Kieff E. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595–608 [DOI] [PubMed] [Google Scholar]
- 2. Altmann M, Hammerschmidt W. 2005. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. 3:e404 doi:10.1371/journal.pbio.0030404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altmann M, et al. 2006. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc. Natl. Acad. Sci. U. S. A. 103:14188–14193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderton E, et al. 2008. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt's lymphoma. Oncogene 27:421–433 [DOI] [PubMed] [Google Scholar]
- 5. Borestrom C, Zetterberg H, Liff K, Rymo L. 2003. Functional interaction of nuclear factor y and sp1 is required for activation of the Epstein-Barr virus C promoter. J. Virol. 77:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cahir-McFarland ED, et al. 2004. Role of NF-kappa B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J. Virol. 78:4108–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calderwood MA, Johannsen E. 2010. Chapter 6: molecular profiles of EBV latently infected cells, p 101–116 In Robertson ES. (ed), Epstein-Barr virus: latency and transformation, vol 1 Caister Academic Press, Norwich, Norfolk, United Kingdom [Google Scholar]
- 8. Cannell EJ, Farrell PJ, Sinclair AJ. 1996. Epstein-Barr virus exploits the normal cell pathway to regulate Rb activity during the immortalisation of primary B-cells. Oncogene 13:1413–1421 [PubMed] [Google Scholar]
- 9. Dave SS, et al. 2006. Molecular diagnosis of Burkitt's lymphoma. N. Engl. J. Med. 354:2431–2442 [DOI] [PubMed] [Google Scholar]
- 10. Demetriades C, Mosialos G. 2009. The LMP1 promoter can be transactivated directly by NF-kappaB. J. Virol. 83:5269–5277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eliopoulos AG, Gallagher NJ, Blake SM, Dawson CW, Young LS. 1999. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J. Biol. Chem. 274:16085–16096 [DOI] [PubMed] [Google Scholar]
- 12. Faumont N, et al. 2009. c-Myc and Rel/NF-kappaB are the two master transcriptional systems activated in the latency III program of Epstein-Barr virus-immortalized B cells. J. Virol. 83:5014–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Forte E, et al. 2012. The Epstein-Barr virus (EBV)-induced tumor suppressor microRNA MiR-34a is growth promoting in EBV-infected B cells. J. Virol. 86:6889–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc. Natl. Acad. Sci. U. S. A. 91:7568–7572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Halder S, et al. 2009. Early events associated with infection of Epstein-Barr virus infection of primary B-cells. PLoS One 4:e7214 doi:10.1371/journal.pone.0007214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Henderson E, Miller G, Robinson J, Heston L. 1977. Efficiency of transformation of lymphocytes by Epstein-Barr virus. Virology 76:152–163 [DOI] [PubMed] [Google Scholar]
- 17. Henkel T, Ling PD, Hayward SD, Peterson MG. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265:92–95 [DOI] [PubMed] [Google Scholar]
- 18. Hollyoake M, Stuhler A, Farrell P, Gordon J, Sinclair A. 1995. The normal cell cycle activation program is exploited during the infection of quiescent B lymphocytes by Epstein-Barr virus. Cancer Res. 55:4784–4787 [PubMed] [Google Scholar]
- 19. Johannsen E, et al. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by J kappa and PU.1. J. Virol. 69:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johansson P, Jansson A, Ruetschi U, Rymo L. 2009. Nuclear factor-kappaB binds to the Epstein-Barr Virus LMP1 promoter and upregulates its expression. J. Virol. 83:1393–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaiser C, et al. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalla M, Hammerschmidt W. 2012. Human B cells on their route to latent infection—early but transient expression of lytic genes of Epstein-Barr virus. Eur. J. Cell Biol. 91:65–69 [DOI] [PubMed] [Google Scholar]
- 23. Keller SA, Schattner EJ, Cesarman E. 2000. Inhibition of NF-kappaB induces apoptosis of KSHV-infected primary effusion lymphoma cells. Blood 96:2537–2542 [PubMed] [Google Scholar]
- 24. Kieff E, Rickinson A. 2007. Epstein-Barr virus and its replication, p 2603–2654 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 25. Klapproth K, Sander S, Marinkovic D, Baumann B, Wirth T. 2009. The IKK2/NF-{kappa}B pathway suppresses MYC-induced lymphomagenesis. Blood 114:2448–2458 [DOI] [PubMed] [Google Scholar]
- 26. Lee JM, et al. 2004. EBNA2 is required for protection of latently Epstein-Barr virus-infected B cells against specific apoptotic stimuli. J. Virol. 78:12694–12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee JM, Lee KH, Weidner M, Osborne BA, Hayward SD. 2002. Epstein-Barr virus EBNA2 blocks Nur77-mediated apoptosis. Proc. Natl. Acad. Sci. U. S. A. 99:11878–11883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu S, Liu P, Borras A, Chatila T, Speck SH. 1997. Cyclosporin A-sensitive induction of the Epstein-Barr virus lytic switch is mediated via a novel pathway involving a MEF2 family member. EMBO J. 16:143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lo AK, et al. 2007. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc. Natl. Acad. Sci. U. S. A. 104:16164–16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luftig M, et al. 2003. Epstein-Barr virus latent membrane protein 1 activation of NF-kappaB through IRAK1 and TRAF6. Proc. Natl. Acad. Sci. U. S. A. 100:15595–15600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maier S, et al. 2006. Cellular target genes of Epstein-Barr virus nuclear antigen 2. J. Virol. 80:9761–9771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maruo S, et al. 2011. Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc. Natl. Acad. Sci. U. S. A. 108:1919–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mattioli I, et al. 2004. Transient and selective NF-kappa B p65 serine 536 phosphorylation induced by T cell costimulation is mediated by I kappa B kinase beta and controls the kinetics of p65 nuclear import. J. Immunol. 172:6336–6344 [DOI] [PubMed] [Google Scholar]
- 34. Mi H, Guo N, Kejariwal A, Thomas PD. 2007. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 35:D247–D252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nikitin PA, et al. 2010. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8:510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reich M, et al. 2006. GenePattern 2.0. Nat. Genet. 38:500–501 [DOI] [PubMed] [Google Scholar]
- 37. Riley KJ, et al. 2012. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 31:2207–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rowe M, et al. 1995. Restoration of endogenous antigen processing in Burkitt's lymphoma cells by Epstein-Barr virus latent membrane protein-1: coordinate up-regulation of peptide transporters and HLA-class I antigen expression. Eur. J. Immunol. 25:1374–1384 [DOI] [PubMed] [Google Scholar]
- 39. Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. 1999. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J. Biol. Chem. 274:30353–30356 [DOI] [PubMed] [Google Scholar]
- 40. Sample J, Kieff E. 1990. Transcription of the Epstein-Barr virus genome during latency in growth-transformed lymphocytes. J. Virol. 64:1667–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seto E, et al. 2010. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 6:e1001063 doi:10.1371/journal.ppat.1001063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shannon-Lowe C, et al. 2005. Epstein-Barr virus-induced B-cell transformation: quantitating events from virus binding to cell outgrowth. J. Gen. Virol. 86:3009–3019 [DOI] [PubMed] [Google Scholar]
- 43. Sinclair AJ, Palmero I, Peters G, Farrell PJ. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 13:3321–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sjoblom A, Nerstedt A, Jansson A, Rymo L. 1995. Domains of the Epstein-Barr virus nuclear antigen 2 (EBNA2) involved in the transactivation of the latent membrane protein 1 and the EBNA Cp promoters. J. Gen. Virol. 76:2669–2678 [DOI] [PubMed] [Google Scholar]
- 45. Skalska L, White RE, Franz M, Ruhmann M, Allday MJ. 2010. Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog. 6:e1000951 doi:10.1371/journal.ppat.1000951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Skalsky RL, et al. 2012. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 8:e1002484 doi:10.1371/journal.ppat.1002484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Spender LC, et al. 1999. Control of cell cycle entry and apoptosis in B lymphocytes infected by Epstein-Barr virus. J. Virol. 73:4678–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spender LC, Cornish GH, Rowland B, Kempkes B, Farrell PJ. 2001. Direct and indirect regulation of cytokine and cell cycle proteins by EBNA-2 during Epstein-Barr virus infection. J. Virol. 75:3537–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Spender LC, Cornish GH, Sullivan A, Farrell PJ. 2002. Expression of transcription factor AML-2 (RUNX3, CBF(alpha)-3) is induced by Epstein-Barr virus EBNA-2 and correlates with the B-cell activation phenotype. J. Virol. 76:4919–4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spender LC, et al. 2006. Cell target genes of Epstein-Barr virus transcription factor EBNA-2: induction of the p55alpha regulatory subunit of PI3-kinase and its role in survival of EREB2.5 cells. J. Gen. Virol. 87:2859–2867 [DOI] [PubMed] [Google Scholar]
- 51. Staege MS, et al. 2002. MYC overexpression imposes a nonimmunogenic phenotype on Epstein-Barr virus-infected B cells. Proc. Natl. Acad. Sci. U. S. A. 99:4550–4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Subramanian A, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tierney RJ, et al. 2000. Methylation of transcription factor binding sites in the Epstein-Barr virus latent cycle promoter Wp coincides with promoter down-regulation during virus-induced B-cell transformation. J. Virol. 74:10468–10479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsang SF, Wang F, Izumi KM, Kieff E. 1991. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J. Virol. 65:6765–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang F, Tsang SF, Kurilla MG, Cohen JI, Kieff E. 1990. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J. Virol. 64:3407–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. White RE, et al. 2010. Extensive co-operation between the Epstein-Barr virus EBNA3 proteins in the manipulation of host gene expression and epigenetic chromatin modification. PLoS One 5:e13979 doi:10.1371/journal.pone.0013979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Woisetschlaeger M, et al. 1991. Role for the Epstein-Barr virus nuclear antigen 2 in viral promoter switching during initial stages of infection. Proc. Natl. Acad. Sci. U. S. A. 88:3942–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Woisetschlaeger M, Yandava CN, Furmanski LA, Strominger JL, Speck SH. 1990. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 87:1725–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu FY, et al. 2004. CCAAT/enhancer binding protein alpha binds to the Epstein-Barr virus (EBV) ZTA protein through oligomeric interactions and contributes to cooperative transcriptional activation of the ZTA promoter through direct binding to the ZII and ZIIIB motifs during induction of the EBV lytic cycle. J. Virol. 78:4847–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xing L, Kieff E. 2007. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J. Virol. 81:9967–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhao B, et al. 2011. Epstein-Barr virus nuclear antigen 3C regulated genes in lymphoblastoid cell lines. Proc. Natl. Acad. Sci. U. S. A. 108:337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao B, et al. 2006. RNAs induced by Epstein-Barr virus nuclear antigen 2 in lymphoblastoid cell lines. Proc. Natl. Acad. Sci. U. S. A. 103:1900–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhao B, et al. 2011. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc. Natl. Acad. Sci. U. S. A. 108:14902–14907 [DOI] [PMC free article] [PubMed] [Google Scholar]