Abstract
Pseudomonas aeruginosa exhibits distinct surface-associated behaviors, including biofilm formation, flagellum-mediated swarming motility, and type IV pilus-driven twitching. Here, we report a role for the minor pilins, PilW and PilX, components of the type IV pilus assembly machinery, in the repression of swarming motility. Mutating either the pilW or pilX gene alleviates the inhibition of swarming motility observed for strains with elevated levels of the intracellular signaling molecule cyclic di-GMP (c-di-GMP) due to loss of BifA, a c-di-GMP-degrading phosphodiesterase. Blocking PilD peptidase-mediated processing of PilW and PilX renders the unprocessed proteins defective for pilus assembly but still functional in c-di-GMP-mediated swarming repression, indicating our ability to separate these functions. Strains with mutations in pilW or pilX also fail to exhibit the increase in c-di-GMP levels observed when wild-type (WT) or bifA mutant cells are grown on a surface. We also provide data showing that c-di-GMP levels are increased upon PilY1 overexpression in surface-grown cells and that this c-di-GMP increase does not occur in the absence of the SadC diguanylate cyclase. Increased levels of endogenous PilY1, PilX, and PilA are observed when cells are grown on a surface compared to liquid growth, linking surface growth and enhanced signaling via SadC. Our data support a model wherein PilW, PilX, and PilY1, in addition to their role(s) in type IV pilus biogenesis, function to repress swarming via modulation of intracellular c-di-GMP levels. By doing so, these pilus assembly proteins contribute to P. aeruginosa's ability to coordinately regulate biofilm formation with its two surface motility systems.
INTRODUCTION
The Gram-negative microbe Pseudomonas aeruginosa is an important model organism for the study of bacterial group behaviors. P. aeruginosa forms surface-attached communities known as biofilms, and this microbe also exhibits surface-associated motile behaviors, including swarming and twitching. It remains unclear how surface-associated P. aeruginosa cells coordinate biofilm formation, twitching, and swarming motility, such that this microbe can readily transition from one type of surface behavior to another when environmental cues, changing surface conditions, etc., dictate that it do so.
One factor that has garnered substantial attention as a key signal governing major lifestyle transitions in bacteria is the second messenger molecule cyclic di-GMP (c-di-GMP) (for a review, see references 25 and 48). Generally speaking, elevated levels of this intracellular signal promote sessile lifestyles such as biofilm formation, and this c-di-GMP-mediated regulation can occur through a variety of mechanisms, including stimulation of exopolysaccharide (EPS) production, cell surface adhesin expression/localization, and/or repression of various forms of motility (26, 37, 38). Conversely, reduced levels of c-di-GMP generally lead to derepression of motile behaviors concomitant with reduced biofilm formation and hence promote the switch to a motile lifestyle (22, 26). Intracellular levels of c-di-GMP are modulated by two opposing enzymatic activities. Diguanylate cyclases (DGC) synthesize c-di-GMP from two molecules of GTP and are noted for the presence of a GGDEF domain (47), whereas phosphodiesterase (PDE) enzymes degrade c-di-GMP and are characterized by the presence of an EAL or HD-GYP domain (11, 50).
Recent studies in P. aeruginosa PA14 have shown that biofilm formation and swarming motility are inversely regulated via a common pathway that is modulated by intracellular levels of c-di-GMP (9, 30, 35). Coordinate regulation of these two surface behaviors depends upon common downstream effectors, such as regulation of flagellar function and production of the Pel-derived EPS. Events during early biofilm formation by P. aeruginosa PA14 require proper control of flagellar function for the reversible to irreversible attachment phase, as well as robust production of the Pel EPS (9, 35, 36). Swarming motility, which occurs on a viscous semisolid agar surface (aided by the production of rhamnolipid surfactant as a surface-wetting agent) is a flagellum-driven process and is therefore sensitive to changes in flagellar function (9, 28, 35, 59). Moreover, strains defective for production of the Pel EPS show enhanced swarming relative to the wild type (WT), indicating that production of this polysaccharide negatively impacts swarming motility in P. aeruginosa PA14 (9).
Genetic and biochemical dissection of the pathway regulating biofilm formation and swarming in P. aeruginosa strain PA14 has revealed distinct regulatory roles for the diguanylate cyclases SadC and RoeA in controlling flagellar function and Pel EPS production, respectively (35, 36). Further studies have shown that the BifA PDE is important for turnover of the c-di-GMP that is synthesized by these two DGCs (30, 35, 36). Strains with mutations in the bifA gene accumulate elevated levels of c-di-GMP, and this accumulation is largely dependent upon the cyclase activities of both SadC and RoeA (30, 36). The resulting excess c-di-GMP produced in the bifA mutant leads to hyper-biofilm formation and repression of swarming motility. Moreover, it was shown that enhanced production of the Pel EPS is required for the hyper-biofilm-forming phenotype but contributes only marginally to the swarming defect of the bifA mutant (30).
The finding described above prompted us to ask what factors are required for repression of swarming motility when c-di-GMP levels are elevated. To address this question, we performed a genetic screen to identify suppressors in the bifA background that restored the ability to swarm (29). Through this analysis, we identified a role for the pilY1 gene in c-di-GMP-mediated repression of swarming motility. Strains with mutations in the pilY1 gene show robust suppression of both the swarming deficiency as well as the hyper-biofilm-forming phenotype exhibited by the bifA mutant (29). The PilY1 protein is involved in biogenesis, stability, and function of the type IV pilus required for twitching motility, a form of movement on solid surfaces that involves cycles of pilus extension, tethering, and retraction (2, 8, 21, 34, 43). However, mutation of the pilA gene, encoding the major subunit of the type IV pilus, showed only weak suppression of the bifA swarming and biofilm defects, indicating that it is loss of PilY1 specifically, and not loss of pili, that fully suppresses the bifA mutant defects (29).
Taken together, data from our studies outlined above suggest that the PilY1 protein plays two distinct cellular roles: one role promoting pilus assembly and a second role repressing swarming motility. In this report, we demonstrate that this duality of functions extends to a limited subset of components of the pilus assembly machinery. Specifically, we provide evidence that the minor pilins, PilW and PilX, impact both pilus assembly and swarming motility, and we propose a model wherein this subset of pilus assembly proteins functions to allow P. aeruginosa to coordinately regulate two distinct motility behaviors, swarming and twitching, when associated with a surface. These findings may provide new insights into how P. aeruginosa transitions between surface behaviors.
MATERIALS AND METHODS
Strains and media.
Strains used in this study are listed in Table 1. P. aeruginosa PA14 and Escherichia coli DH5α and S17-1 λpir strains were routinely cultured in lysogeny broth (LB) medium, solidified with 1.5% agar when necessary. Gentamicin (Gm) was used at 25 μg/ml for P. aeruginosa and at 10 μg/ml for E. coli. Ampicillin (Ap) was used at 150 μg/ml and nalidixic acid (NA) at 20 μg/ml for E. coli. For phenotypic assays with P. aeruginosa, either M63 or M8 minimal salts medium (as indicated below) was supplemented with MgSO4 (1 mM), glucose (0.2%), and Casamino Acids (CAA; 0.5%). For expression plasmids harboring the PBAD promoter, arabinose was added to cultures at a 0.2% final concentration, unless noted otherwise.
Table 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype, description, or sequence | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | l− f80dlacZDM15D(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Life Technologies |
| S17-1 (λpir) | thi pro hsdR- hsdM+ ΔrecA RP4-2::TcMu-Km::Tn7 | 55 |
| P. aeruginosa strains | ||
| PA14 | Wild type | |
| bifA strain | Unmarked in-frame deletion of the bifA gene | 30 |
| bifA pilY1 strain | Unmarked in-frame deletions of bifA and pilY1 | 29 |
| bifA pilA strain | Unmarked in-frame deletions of bifA and pilA | 29 |
| bifA fimU strain | Unmarked in-frame deletions of bifA and fimU | This study |
| bifA pilV strain | Unmarked in-frame deletions of bifA and pilV | This study |
| bifA pilW strain | Unmarked in-frame deletions of bifA and pilW | This study |
| bifA pilX strain | Unmarked in-frame deletions of bifA and pilX | This study |
| bifA pilE strain | Unmarked in-frame deletions of bifA and pilE | This study |
| bifA fimU-pilVWXY1Y2E strain | Unmarked deletions of bifA and the fimU-pilVWXY1Y2E operon | This study |
| bifA pilD strain | Unmarked in-frame deletions of bifA and pilD | This study |
| pilX+::His strain | PA14 with chromosomal insertion of a C-terminal His6 tag in the pilX gene | This study |
| pilX(G−1F)::His strain | PA14 with chromosomal insertion of a His6 tag and mutation of G−1 to F in the pilX gene | This study |
| pilXΔLP::His strain | PA14 with chromosomal deletion of the pilX leader peptide sequence and insertion of a His6 epitope | This study |
| bifA pilX+::His strain | bifA mutant with chromosomal insertion of a C-terminal His6 tag in the pilX gene | This study |
| bifA pilX(G−1F)::His strain | bifA mutant with chromosomal insertion of a His6 tag and mutation of G−1 to F in the pilX gene | This study |
| bifA pilXΔLP::His strain | bifA mutant with chromosomal deletion of the pilX leader peptide sequence and insertion of a His6 epitope | This study |
| pilD pilX+::His strain | pilD mutant with chromosomal insertion of a C-terminal His6 tag in the pilX gene | This study |
| pilD pilXΔLP::His strain | pilD mutant with chromosomal deletion of the pilX leader peptide sequence and insertion of a His6 epitope | This study |
| Plasmids | ||
| pMQ72 | Shuttle vector for cloning in yeast and for arabinose-inducible gene expression; Gmr | 52 |
| pMQ30 | Shuttle vector for yeast cloning and Gram-negative allelic replacement; Gmr | 52 |
| pMQ30-fimU | Plasmid for in-frame deletion of fimU gene; Gmr | This study |
| pMQ30-pilV | Plasmid for in-frame deletion of pilV gene; Gmr | This study |
| pMQ30-pilW | Plasmid for in-frame deletion of pilW gene; Gmr | This study |
| pMQ30-pilX | Plasmid for in-frame deletion of pilX gene; Gmr | This study |
| pMQ30-pilE | Plasmid for in-frame deletion of pilE gene; Gmr | This study |
| pMQ30-pilD | Plasmid for in-frame deletion of pilD gene; Gmr | This study |
| pMQ30-fimUpilVWXY1Y2E | Plasmid for in-frame deletion of the fimU-pilVWXY1Y2E operon; Gmr | This study |
| pMQ30-pilX+::His | Plasmid for chromosomal insertion of a His6 tag in the pilX gene | This study |
| pMQ30-pilX(G−1F)::His | Plasmid for chromosomal insertion of a His6 epitope and an amino acid substitution of G−1 to F in pilX | This study |
| pMQ30-pilXΔLP::His | Plasmid for chromosomal deletion of the pilX leader peptide sequence and insertion of a His6 epitope | |
| pPilX-His | His-tagged pilX gene in pMQ72; Gmr | This study |
| pPilX(G−1A)-His | His-tagged pilX gene with the G−1A substitution cloned in pMQ72; Gmr | This study |
| pPilX(G−1D)-His | His-tagged pilX with the G−1D substitution cloned in pMQ72; Gmr | This study |
| pPilX(G−1F)-His | His-tagged pilX gene with the G−1F substitution cloned in pMQ72; Gmr | This study |
| pPilW-His | His-tagged pilW gene in pMQ72; Gmr | This study |
| pPilW(G−1F)-His | His-tagged pilW gene with the G−1F substitution cloned in pMQ72; Gmr | This study |
| pPilY1-His | His-tagged pilY1 gene cloned in the pMQ80 expression vector | 29 |
Saccharomyces cerevisiae strain InvSc1 (Invitrogen), used for plasmid construction via in vivo homologous recombination, was grown with yeast extract-peptone-dextrose (1% Bacto yeast extract, 2% Bacto peptone, and 2% dextrose), as reported previously (52). Selections with InvSc1 were performed using synthetic defined agar-uracil (4813-065; Qbiogene).
Construction of mutant strains and plasmids.
Table 1 lists all plasmids constructed and/or used in this study. Primers used in plasmid construction are listed in Table S1 in the supplemental material. In-frame gene deletions, chromosomal knock-in epitope tagging, and mutagenesis were performed via allelic exchange. Plasmids for these purposes were constructed via cloning by homologous recombination of relevant PCR products into the pMQ30 vector in yeast (52). In-frame deletions of genes at the fimU-pilE locus were generated in both the WT and bifA mutant backgrounds using methods previously described (29, 30). The resulting internal gene deletions remove the following amino acids: fimU (amino acids [aa] 8 to 149), pilV (aa 15 to 158), pilW (aa 36 to 241), pilX (aa 41 to 171), pilY2 (aa 13 to 100), pilE (aa 2 to 127), and pilD (aa 22 to 280). For the fimU-pilVWXY1Y2E operon deletion, the deleted region begins 645 bp upstream of the fimU translational start and ends within pilE at aa 127, as for the pilE single mutant. All deletions were confirmed by PCR, and all chromosomal modifications were confirmed by PCR and sequencing.
A chromosomal knock-in construct with a deletion of the leader peptide (LP) in pilX and insertion of a His6 epitope (pilXΔLP::His) is expected to yield the N-terminal amino acid sequence MSTLL in contrast to the predicted sequence STLL formed by PilD cleavage at the G−1 (gylcine residue at the −1 position with respect to the cleavage site) and methylation of the newly formed N-terminal serine. The methionine start codon of pilXΔLP::His was included for proper translation initiation. Thus, our version of processed PilX (PilXΔLP::His) is not identical to the expected PilD-processed version of PilX. A similar construct was used previously to test the requirement for the leader peptide of PilA in pilus assembly (56).
Plasmids for complementation and overexpression were generated using the pMQ72 vector via homologous recombination in yeast. For primers used see Table S1 in the supplemental material. For all complementation and mutant expression constructs for the pilX gene, we designed primers to include six additional base pairs upstream of the predicted translational start codon for the pilX gene of strain P. aeruginosa PA14 denoted in the Pseudomonas Genome Database (www.pseudomonas.com). The database indicates that the start of translation is a CTG; however, there is an in-frame ATG two codons upstream of the CTG. Since it seemed plausible that the upstream in-frame ATG might be a bona fide start site, we chose to include this sequence in the design of our pilX expression constructs.
Motility assays.
Swim (0.3% agar) and twitch (1.5% agar) motility plates consisted of M8 medium supplemented with glucose, MgSO4, CAA, and arabinose (0.2%) where indicated in the figure legends. Swim and twitch assays were performed as previously reported (45, 60). For quantification of twitch zones, agar was removed from the petri plate, twitch zones were stained with crystal violet, and digital images of the zones were then analyzed using ImageJ. Swarm motility plates (0.5% agar) were comprised of M8 medium supplemented with MgSO4, glucose, CAA, and arabinose (0.2%) where indicated in the figure legends. Swarm assays were performed as previously described (29, 30).
Biofilm formation assay.
Biofilm formation in 96-well microtiter plates was assayed and quantified as previously described (10, 44). All biofilm assays were performed using M63 minimal medium supplemented with glucose, MgSO4, CAA, and arabinose where indicated.
CR binding assays.
Congo red (CR) binding assays were performed as reported previously (14, 15, 29).
Expression analysis of PilX-His/PilW-His proteins.
Cultures grown in LB medium overnight were diluted (1:100) into M8 (250 ml volume) supplemented with glucose, MgSO4, CAA, and 0.2% arabinose, where indicated in the figure legends, and grown to an optical density at 600 nm (OD600) of ∼0.5 at 37°C with shaking (250 rpm). Bacterial cells were harvested by centrifugation at 5,520 × g for 15 min at 4°C and stored at −80°C. Cell pellets were resuspended in cell lysis buffer (200 mM Tris-HCl [pH 7.5], 1 mM EDTA, 2 mM MgCl2, and complete protease inhibitors [Roche Diagnostics Corp., Indianapolis, IN]). Benzonase nuclease (Novagen, San Diego, CA) was added to a final concentration of ∼50 units/ml, and bacterial cells were lysed in a French pressure cell. Samples were centrifuged at 9,300 × g for 10 min at 4°C to remove unbroken cells, and supernatants were collected as whole-cell lysates (WCL).
For Western blotting of whole-cell lysates, samples were normalized to the same protein concentrations, and equivalent amounts of total protein were mixed with 2× SDS loading buffer containing freshly added dithiothreitol ([DTT] 200 mM). The concentration of total protein in lysates was determined using a Bio-Rad detergent-compatible (DC) protein assay kit (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. Samples were boiled for 10 min and resolved by SDS-PAGE using 15% polyacrylamide gels (Bio-Rad). Proteins were transferred to a nitrocellulose membrane and probed with an anti-penta-His antibody (Qiagen, Valencia, CA) to detect His-tagged PilX and PilW and mutant variants of these proteins. Western blots were developed with a Western Lightning ECL detection kit (Perkin-Elmer, Boston, MA) according to the manufacturer's instructions.
Cellular localization of PilX.
For cellular fractionation of strains expressing the chromosomally modified genes pilX::His and pilX::His with a mutation of the G−1 residue to F [pilX(G−1F)::His], we cultured strains on soft agar (0.5%) swarm plates where cells were allowed to swarm for 16 h at 37°C; cells were then harvested by gentle scraping with an ethanol-washed plastic coverslip into microcentrifuge tubes. Samples were centrifuged for 2 min at room temperature (RT), supernatants were removed, and cell pellets were frozen at −80°C until further fractionation (see below).
For fractionation, cell pellets were then resuspended in cell lysis buffer (see above), and Benzonase nuclease (Novagen, San Diego, CA) was added to a final concentration of ∼50 units/ml. Bacterial cells were lysed in a French pressure cell, and samples were centrifuged at 9,300 × g for 10 min at 4°C to remove unbroken cells. Supernatants were collected as whole-cell lysates (WCL), and samples were further fractionated to yield cytoplasmic and inner and outer membrane fractions (IM and OM, respectively), as described previously (29, 30, 41).
For surface protein (SP) preparations, cultures grown overnight in LB medium were struck in a crosshatched pattern to M8 minimal salts medium plates supplemented with glucose, MgSO4, and CAA, and SP fractions were isolated as described previously (16).
For Western blotting of fractions, samples within a fraction were normalized to the same protein concentrations, and equivalent amounts of total protein were mixed with 2× SDS loading buffer containing dithiothreitol (200 mM). The concentration of total protein in fractions was determined as described above. Samples were boiled for 10 min and resolved by SDS-PAGE as follows: using 15% gels for resolution of PilX-His, 7.5% gels for resolution of the inner membrane marker SecY, and 4 to 15% gels for resolution of the outer membrane marker OprF. Proteins were transferred to a nitrocellulose membrane and probed with one of the following antibodies: (i) an anti-penta-His antibody to detect His-tagged PilX and PilX(G−1F), (ii) an anti-SecY antibody (1, 24), and (iii) an anti-OprF antibody (6, 20, 32). Western blots were developed as described above.
In vivo quantification of c-di-GMP. (i) Quantification of c-diGMP levels in liquid-grown cells.
Overnight LB-grown cultures were diluted to an OD600 of 0.04 in M8 medium supplemented with MgSO4, glucose, and Casamino Acids and grown at 37°C (250 rpm) for approximately 3 h, whereupon cultures were normalized to an OD600 of 0.4 prior to nucleotide extraction. Subsequent extraction of nucleotides and quantification of c-di-GMP by liquid chromatography-mass spectrometry (LC-MS) were performed as described previously (39) with the exception that extractions were incubated at −20°C for 1 h. Following LC-MS analysis, samples were compared to a standard curve derived from measurements of known concentrations of pure c-di-GMP to determine the concentration (in nM) of c-di-GMP in the samples. The data were then normalized to the dry weight of the cell pellet from which the c-di-GMP was extracted and presented as pmol of c-di-GMP/mg of dry weight.
(ii) Analysis of c-diGMP levels in swarm-grown cells.
Overnight LB-grown cells were inoculated onto the surface of swarm plates and grown overnight for 16 h at 37°C. Swarm cells were then scraped from the surface using an ethanol-washed plastic coverslip, deposited into a 1.5-ml microcentrifuge tube, and centrifuged for 1 min at RT. Supernatants were removed, and cell pellets were resuspended in nucleotide extraction buffer and further processed as described above.
Analysis of protein expression in liquid- versus surface-grown cells.
Strains were cultured under the same liquid and swarm plate conditions used to prepare cells for c-di-GMP measurements (see above) with the exception that cells harvested after the growth period were centrifuged, and pellets were stored at −80°C after removal of the supernatant. Whole-cell lysates were then prepared for Western blot analysis as described above (see “Expression analysis of PilX-His/PilW-His proteins”). Western blotting was performed as described above. Samples were resolved by SDS-PAGE using 15% gels for resolution of PilX-His and PilA and 7.5% gels for resolution of PilY1. Proteins were transferred to a nitrocellulose membrane and probed with one of the following antibodies: (i) an anti-PilY1 antibody (43), (ii) an anti-PilA antibody (29), or (iii) an anti-penta-His antibody to detect PilX-His. Following detection of proteins as described above, autoradiograph films were scanned, and protein species were quantified using ImageJ software (http://rsbweb.nih.gov/ij/).
Rhamnolipid production.
To detect rhamnolipid production, plate-based assays were performed as described previously (54), using M8 minimal salts medium plates supplemented with glucose, MgSO4, and CAA and solidified with 1.5% agar. Plates were incubated overnight at 37°C followed by 6 days at room temperature.
Analysis of flagellin expression.
Cells were harvested from swarm plates by gentle scraping with an ethanol-washed plastic coverslip. Samples were normalized by OD600, and cells were harvested by centrifugation. Cell pellets were resuspended in 1× SDS loading buffer containing 100 mM DTT, boiled for 10 min, and resolved by SDS-PAGE using 7.5% polyacrylamide gels. Proteins were transferred to a nitrocellulose membrane and probed with a polyclonal antibody to FliC (51), and detection was performed as described above.
RESULTS
The minor pilins, PilW and PilX, participate in c-di-GMP-mediated repression of swarming motility.
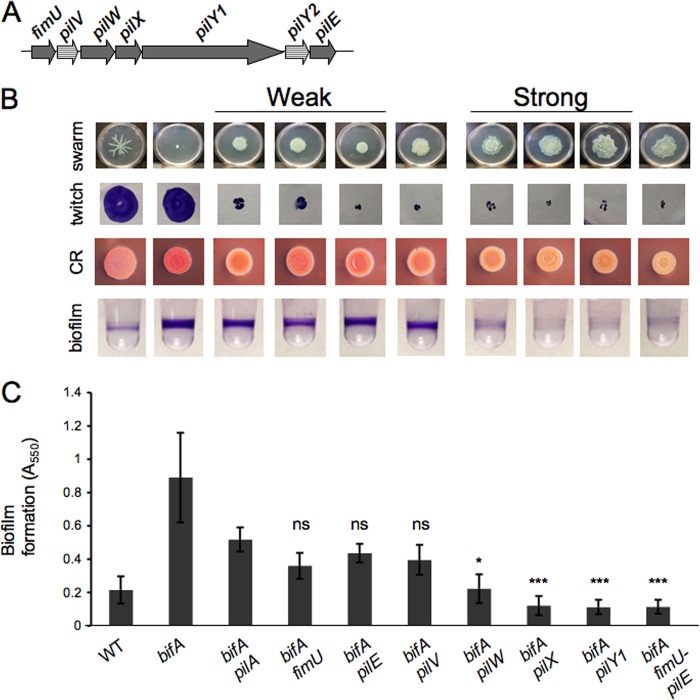
To address the question of whether additional proteins involved in pilus assembly might also function with PilY1 in c-di-GMP-mediated swarming repression, we first looked to the pilY1 genomic locus for candidate genes (Fig. 1A). Previous work had shown that the fimU-pilVWXY1Y2E locus is expressed as a polycistronic operon in Pseudomonas aeruginosa (5). The fimU, pilV, pilW, pilX, and pilE genes encode pilin-like proteins containing a predicted hydrophobic N-terminal α-helical region that is a hallmark of pilins such as the major pilin subunit PilA. While the precise role of these so-called “minor” pilins (minor refers to their low abundance relative to PilA) in pilus biogenesis is not clear, the minor pilins were recently reported to be incorporated into pilus fibers; and, furthermore, while pilus assembly is not absolutely dependent upon the minor pilins, assembly is suboptimal in their absence (16). It is worth noting that the genome annotations at the pilY1 locus in P. aeruginosa strain PA14 do not include the pilV and pilY2 genes (www.pseudomonas.com), despite the fact that the coding sequences of these genes appear to be intact in the genomic sequence, indicating that the pilV and pilY2 genes may represent bona fide open reading frames (ORFs). Indeed, Giltner et al. (17) have shown that the pilV gene from strain P. aeruginosa PA14, when expressed in a P. aeruginosa PAO1 pilV mutant, is able to complement its twitching defect, indicating that the P. aeruginosa PA14 pilV gene encodes a functional homolog. Therefore, we included these ORFs in our initial analysis.
Fig 1.
Genetic analysis of the pilY1 locus. (A) Schematic diagram of the pilY1 genomic locus. The annotation for the P. aeruginosa PA14 genome (www.pseudomonas.com) does not include the pilV and pilY2 open reading frames (striped). (B) Phenotypes of the bifA mutant suppressor analysis. The strain genotypes for each image in this panel are shown at the bottom of panel C. The first row shows representative swarm images for the WT and bifA single mutant, followed by the weak-suppressor class consisting of the bifA pilA, bifA fimU, and bifA pilE double mutants. The bifA pilV mutant has an intermediate phenotype and is followed by the strong-suppressor class consisting of the bifA pilW, bifA pilX, and bifA pilY1 mutants. Note that suppression of the bifA mutant swarming defect by mutation of pilW, pilX, or pilY1 does not restore WT tendril formation. The rightmost image shows the bifA fimU-pilVWXY1Y2E (fimU-pilE) mutant. Swarm plates were incubated at 37°C for 16 h. Images of twitch zones stained with crystal violet to aid visualization are shown in the second row. Twitch plates were incubated overnight at 37°C followed by 6 days at RT, at which time agar was removed from the petri plate for subsequent crystal violet staining. Images of Congo red (CR) binding by each of the strains are shown in the third row. CR plates were incubated overnight at 37°C followed by an additional 48 h at RT. The fourth row shows representative wells of a 96-well dish from a biofilm assay for each strain. Strains were grown in M63 medium with MgSO4, glucose, and CAA for 24 h at 37°C prior to crystal violet staining. (C) Quantification of crystal violet-stained biofilms. The strain genotypes are shown on the x axis. Crystal violet was solubilized with 30% glacial acetic acid, and the absorbance was measured at 550 nm. Error bars represent standard deviations of the averages of four experiments with four replicates per experiment. Data were analyzed by one-way analysis of variance followed by a Tukey's posttest comparison. ns, not significantly different; *, P < 0.05; ***, P < 0.001 (all relative to the bifA pilA double mutant).
To determine whether any of the minor pilins, like PilY1, play a role in swarming repression, we constructed in-frame deletions in each of the minor pilin genes in the bifA mutant background and then assessed suppression of the bifA mutant phenotypes. We found that the double mutant strains could be grouped into four classes based on suppression of the bifA phenotypes: a weak-suppressor class, a strong-suppressor class, an intermediate suppressor class, as well as a fourth class of one mutant that displayed no detectable phenotype.
Weak-suppressor strains showed only partial suppression of the swarming defect, (typically characterized by a small circular zone of expansion surrounded by a ruffled border), the enhanced binding of the dye Congo red, and the hyper-biofilm-formation phenotype of the bifA mutant (Fig. 1B, first, third, and fourth rows, and C, respectively). CR binding provides a visual assessment of the polysaccharide produced by the bifA mutant, and we have previously shown that this enhanced polysaccharide production is required for the hyper-biofilm-forming phenotype of this mutant and requires the Pel locus (30).
Strains in the weak-suppressor class include the bifA fimU and bifA pilE double mutants. Phenotypes of the weak suppressors closely resemble those of the bifA pilA double mutant in all assays (Fig. 1B, third from left). Quantification of biofilm formation indicates that the biofilms formed by the bifA fimU and bifA pilE mutants do not differ significantly from the biofilm of the bifA pilA double mutant (Fig. 1C). Furthermore, mutation of fimU or pilE in the bifA background leads to loss of twitching motility (Fig. 1B, second row). These results agree with our previous observations of the bifA pilA double mutant (29) and indicate that loss of pilus assembly/twitching motility leads to weak suppression of the bifA swarming defect.
Strains in the strong-suppressor class include the bifA pilW and bifA pilX double mutants. The bifA pilX and bifA pilW double mutants exhibit phenotypes that closely resemble the bifA pilY1 double mutant, showing both robust rescue of the swarming defect and strong reduction of the hyper-biofilm formation and CR-binding of the bifA mutant (Fig. 1B). Quantification of biofilm formation indicates that biofilms formed by the bifA pilX and bifA pilW mutants are not significantly different from the biofilm of the bifA pilY1 mutant, but, in contrast, are significantly reduced relative to the biofilm of the bifA pilA double mutant. It should also be noted that while swarming is restored, the classic pattern (i.e., tendril formation) exhibited by the WT is not restored, similar to what was reported for the bifA pilY1 double mutant (29).
Mutations in either pilW or pilX render those strains defective for twitching motility (Fig. 1B), consistent with previous observations by others that the PilW and PilX minor pilins are involved in pilus assembly and twitching motility (2, 16). Deletion of the entire fimU-pilVWXY1Y2E locus in the bifA mutant background results in phenotypes that are comparable to the bifA pilX, bifA pilW, and bifA pilY1 double mutants (Fig. 1A to C, rightmost columns). Thus, with no obvious additive effects, we infer that PilY1 and the minor pilins PilX and PilW participate in the regulation of swarming motility in a bifA mutant. Furthermore, these data support the notion that PilX and PilW play dual roles in the cell by participating in both pilus assembly and repression of swarming motility.
The bifA pilV double mutant exhibited phenotypes akin to both suppressor classes, being a weak suppressor of the hyper-biofilm formation and CR binding but a strong suppressor of swarming motility. While we have been able to partially complement the biofilm and twitching motility defects of the bifA pilV mutant by arabinose-inducible expression of a His-tagged version of pilV in multicopy, we have been unable to complement the swarming phenotype using a range of arabinose concentrations (data not shown). In light of these data, we cannot make any strong conclusions regarding the role of pilV in swarming repression and, thus, have not further pursued analysis of this gene at this time.
Finally, the bifA pilY2 double mutant was indistinguishable from the bifA single mutant in all assays, including the twitching motility assay (data not shown), suggesting that the pilY2 gene does not play an obvious role in pilus assembly or swarming repression; therefore, we did not include this gene in further studies. These data are consistent with the absence of pilY2 from the genome annotation at this locus in the P. aeruginosa PA14 strain and with the recent suggestion that pilY2 may be a pseudogene (16).
Expression of His-tagged PilX in trans complements the bifA pilX double mutant phenotypes.
To ensure that suppression of the bifA mutant phenotypes and loss of twitching motility observed for the bifA pilX mutant are due to inactivation of pilX and not caused by polar effects on expression of the pilY1 gene downstream or secondary mutations, we introduced an arabinose-inducible expression plasmid bearing a His6-tagged version of the pilX gene (pPilX-His) into the bifA pilX double mutant and assessed the ability of this construct to complement the mutant phenotypes. As shown in Fig. 2A, expression of PilX-His in the bifA pilX double mutant fully restores the swarming defect observed for the bifA single mutant relative to the vector control.
Fig 2.
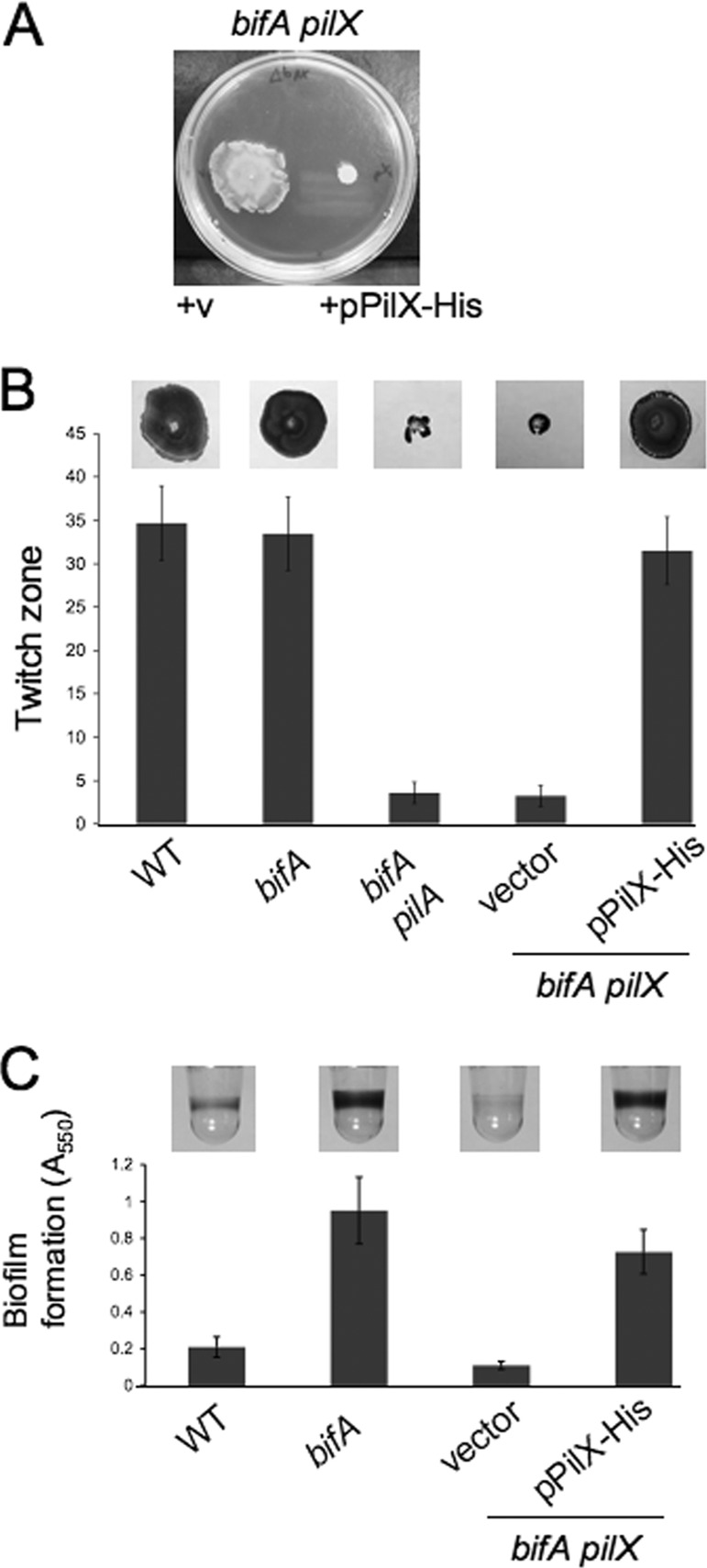
Complementation of the bifA pilX double mutant phenotypes. (A) Representative swarm plate showing the bifA pilX double mutant carrying either the empty vector (+v) on the left or the pPilX-His plasmid on the right. Swarm plates were supplemented with 0.2% arabinose and were incubated at 37°C for 16 h. (B) Crystal violet-stained twitching zones for the WT, bifA mutant, bifA pilA mutant, and bifA pilX mutant carrying either the vector or the pPilX-His plasmid are shown at the top. Twitch plates supplemented with arabinose were incubated overnight at 37°C followed by 6 days at RT, when agar was removed from petri plates for subsequent crystal violet staining. The graph shows corresponding quantification of twitch zones using the ImageJ program with units in pixels (×1,000). Error bars represent standard deviations of the averages from four twitch zones. (C)Representative wells of a 96-well microtiter dish from a biofilm assay for the WT, bifA mutant, and the bifA pilX double mutant carrying either the vector or the pPilX-His plasmid are shown at the top. Strains were grown in M63 medium with MgSO4, glucose, CAA, and arabinose for 24 h at 37°C prior to crystal violet staining. The graph shows quantification of the biofilms by solubilization of crystal violet with 30% glacial acetic acid and measurement of absorbance at 550 nm. Error bars represent standard deviations of averages from three independent experiments with four wells per experiment.
In twitching motility assays, we observed that expression of PilX-His is able to complement the twitching defect of the bifA pilX double mutant (Fig. 2B). Furthermore, PilX-His expression restored hyper-biofilm formation to levels comparable to the bifA single mutant (Fig. 2C). Collectively, these results confirm that it is inactivation of pilX alone that is responsible for the observed suppression of the bifA mutant phenotypes.
Separation of functions by mutation: cleavage of the PilX N-terminal leader sequence is required for function in twitching motility but not for repression of swarming motility.
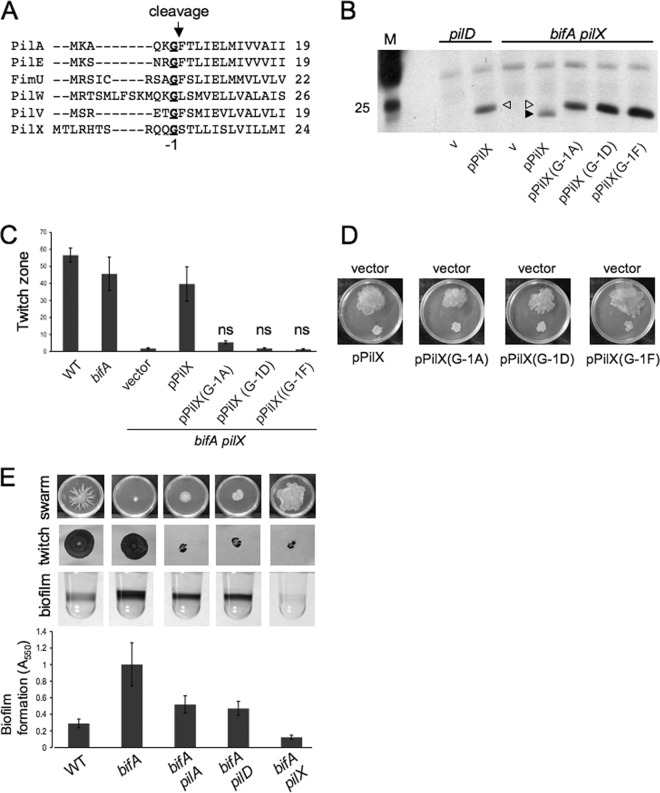
To further support our hypothesis that the minor pilins play two distinct roles in the cell, we sought to separate these functions by mutational analysis. Shown in Fig. 3A is an alignment of the N termini of PilA and the minor pilins of P. aeruginosa strain PA14. Previous studies of the major pilin subunit, PilA, have shown that PilA undergoes cleavage of a leader peptide just after the glycine residue (in the −1 position with respect to the cleavage site), followed by methylation of the newly formed N-terminal amino acid, phenylalanine (46, 56). Both the proteolysis and methylation activities are performed by the PilD peptidase, and it has been shown that the cleavage event is required for proper pilus assembly while the requirement for methylation is less clear (40, 42, 56, 58). Recently, it was shown that the minor pilins of P. aeruginosa are also cleaved by PilD and that they are incorporated into the mature pilus (16) although it has not been established whether cleavage of the minor pilins is required for incorporation into the pilus and for proper pilus assembly.
Fig 3.
Separation of PilX functions by mutation. (A) Alignment of the N termini of PilA and the minor pilins PilE, FimU, PilW, PilV, and PilX from P. aeruginosa PA14 using the ClustalW alignment software (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Sequences were obtained from the Pseudomonas Genome Database (www.pseudomonas.com). For the PilX protein, the amino acid sequence shown differs from the sequence predicted by the genome annotation (MRHTS…) by including an in-frame methionine two codons upstream of the predicted coding sequence (see Materials and Methods for further information). The conserved glycine residue (G) is indicated in bold and underlined and is in the −1 position with respect to the indicated cleavage site. (B) Western blot showing arabinose-induced expression of PilX-His and the G−1 mutant variants in the pilD and bifA pilX mutant backgrounds. Equal amounts of total protein from whole-cell lysates for each strain were resolved by SDS-PAGE using a 15% polyacrylamide gel. The PilX-His protein and its mutant variants were detected using an anti-penta-His antibody. The marker lane (M) indicates the position of the 25-kDa marker. The filled arrowhead indicates the processed form of PilX-His (∼21 kDa), and the open arrowhead indicates the unprocessed form. v, vector control. (C) Graph depicting quantification of twitch zones using the ImageJ program with units in pixels (×1,000). Twitch plates were supplemented with 0.2% arabinose. Error bars represent standard deviations of averages from four independent experiments with four twitch zones per experiment. Data were analyzed by one-way analysis of variance followed by a Tukey's posttest comparison. ns, not significantly different from the bifA pilX strain carrying the vector alone. (D) Images of swarms for the bifA pilX mutant carrying the vector (top of the plate) and the pPilX-His plasmid or its mutant variants (bottom of the plate). The medium was supplemented with 0.2% arabinose and incubated at 37°C for 16 h. (E) Representative images of swarms, twitch zones, and biofilm wells for the indicated strains. The strain genotypes for each image in this panel are shown below the graph depicting quantification of biofilm formation. Error bars represent standard deviations of averages from three independent experiments with four wells per experiment.
Based on the studies with PilA, we predicted that cleavage of the PilX N-terminal leader peptide would be required for assembly of functional pili and asked whether cleavage was also required for the role of PilX in repression of swarming. To address this question, we made amino acid substitutions of the PilX G−1 residue to prevent processing by PilD and assessed whether the resulting mutant versions of PilX were functional in both pilus assembly and swarming repression. For the amino acid substitutions, we selected a subset of those generated in previous studies examining PilD-mediated processing of PilA (56). We chose to replace the G−1 residue of PilX with amino acids that fully blocked cleavage of PilA (D and F) as well as the single amino acid tested (A) that allowed for partial cleavage of PilA (56).
First, we examined whether the amino acid substitutions of the PilX G−1 led to inhibition of PilD-mediated cleavage. We expressed the wild-type PilX-His and the PilX(G−1A)-, PilX(G−1D)-, and PilX(G−1F)-His mutant proteins from a plasmid in the bifA pilX (pilD+) background and observed that the mutant versions of PilX migrate at a higher molecular weight than the wild-type protein, suggesting that the mutant proteins are unprocessed, as expected (Fig. 3B, right side). Indeed, we observe a similar size shift when comparing migration of the wild-type PilX protein in a pilD mutant strain to a PilD+ strain (Fig. 3B, left side), indicating that the size increase in the pilD mutant strain is due to retention of the leader peptide. Thus, we conclude that these mutations prevent cleavage of the leader peptide by PilD. For the PilX(G−1A)-His mutant protein, we were unable to detect the processed form of this protein as had been reported for the non-His-tagged PilA(G−1A) protein (56).
Next, we assessed the ability of these constructs to complement the twitching phenotype of the bifA pilX double mutant. As shown in Fig. 3C, expression of wild-type pPilX-His in the bifA pilX strain restores twitching motility to levels comparable to those observed for the bifA single mutant. In contrast, expression of either pPilX(G−1D)-His or pPilX(G−1F)-His is unable to restore twitching motility as strains carrying these constructs are indistinguishable from the bifA pilX mutant carrying the vector control. Expression of pPilX(G−1A)-His leads to a small recovery in twitching ability; however, this increase in motility is not statistically significant relative to the vector control (Fig. 3C). Collectively, our results demonstrate that mutations in the PilX G−1 residue that inhibit cleavage by PilD compromise the ability of the mutant PilX proteins to function in pilus assembly and therefore indicate that cleavage of PilX is required for its function in pilus assembly and twitching motility.
To determine whether cleavage of PilX is required for its role in swarming repression, we tested the pPilX-His mutant constructs for the ability to complement the bifA pilX double mutant by restoring the swarming defect observed for the bifA single mutant. The results in Fig. 3D show that expression of each of the pPilX-His mutant constructs in the bifA pilX mutant is able to repress swarming motility in a manner indistinguishable from that of the wild-type pPilX-His construct. These data indicate that repression of swarming motility by PilX does not require cleavage by PilD.
As further verification of these findings, we generated a pilD mutation in the bifA mutant background and assessed suppression of the bifA mutant phenotypes. If cleavage of PilX were required for its role in swarming repression, then we would expect a pilD mutation in the bifA background to strongly suppress the swarming defect, similar to what we observe for a pilX mutation. However, that is not what we observe. Instead, we find that the bifA pilD double mutant exhibits only weak suppression of the bifA phenotypes and is virtually indistinguishable from the bifA pilA double mutant (Fig. 3E). Thus, these genetic data agree with our mutational analysis of PilX and support the notion that cleavage of PilX, while required for its role in pilus assembly, is not required for repression of swarming motility.
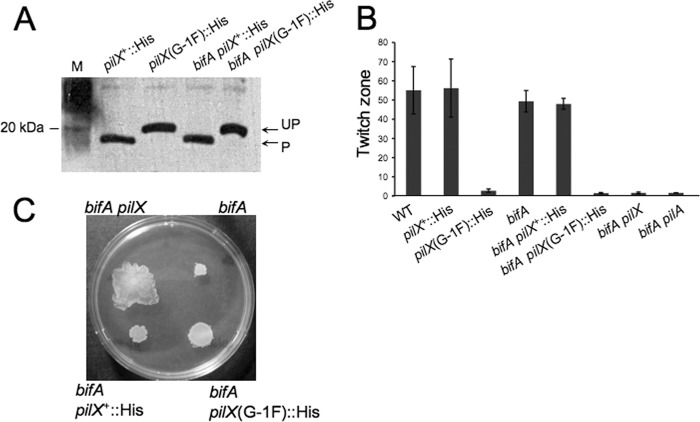
Thus far, our assessment of the function of unprocessed PilX mutant proteins in pilus assembly and swarming repression is based upon complementation of the bifA pilX mutant, relying on overexpression of these proteins in this strain. To examine the role of cleavage on function of PilX when it is expressed from its native locus, we constructed the G−1F amino acid substitution in the pilX gene on the chromosome in both the WT and bifA mutant backgrounds. To monitor expression of both the wild-type PilX and PilX(G−1F) proteins in these strains, we inserted a His6 tag at the C terminus of the pilX gene. As shown in Fig. 4A, the PilX::His and PilX(G−1F)::His proteins are expressed at similar levels in both the WT and bifA mutant backgrounds. We also observe the expected size shift in the PilX(G−1F)::His proteins indicating lack of cleavage by PilD. We next performed twitching motility assays and found that insertion of the His tag alone had no impact on twitching abilities of either the WT or the bifA mutant strain (Fig. 4B). In contrast, WT and bifA cells expressing the PilX(G−1F)::His mutant protein were as defective in twitching motility as pilX and pilA null mutants. These data are consistent with the results obtained in the complementation experiments when the PilX(G−1F) mutant protein was expressed in multicopy and further support the idea that cleavage of PilX is required for its function in pilus assembly and twitching motility.
Fig 4.
Analysis of PilX. (A) Assessment of chromosomally expressed PilX-His and PilX(G−1F)-His protein levels by Western blot analysis. Equal amounts of total protein from whole-cell lysates prepared from each strain were resolved by SDS-PAGE on a 15% polyacrylamide gel. PilX-His and the PilX(G−1F)-His mutant variant were detected using an anti-penta-His antibody. P, processed form; UP, unprocessed form. (B) Graph showing quantification of twitch zones using ImageJ with units in pixels (×1,000). Error bars represent standard deviations of averages from three twitch zones per strain. (C) Representative images of swarm plates for the indicated strains. Plates were incubated at 37°C for 16 h.
We next examined the ability of these chromosomal knock-in strains to swarm. In the bifA mutant background, insertion of the His epitope tag alone had no discernible impact on the swarming defect, indicating that the His epitope tag did not disrupt the ability of PilX to repress swarming in this background (Fig. 4C). In contrast, the bifA pilX(G−1F) mutant exhibited weak suppression of the swarming defect similar to what is observed for the bifA pilA mutant. This is not surprising, given that the pilX(G−1F) mutation renders cells defective for pilus assembly and twitching motility, and our previous observations of the bifA pilA mutant suggest that loss of pilus assembly does result in some loss of swarming repression (Fig. 1). Notably, however, we did not observe full suppression of the bifA swarming defect, which is the result expected if cleavage of PilX was absolutely required for its role in swarming repression.
Our genetic analyses of the bifA pilX and bifA pilW mutants suggest that PilW behaves similarly to PilX in influencing both pilus assembly and swarming repression (Fig. 1B and C). Therefore, we further investigated the involvement of PilW in pilus assembly and swarming repression. Based on our results with PilX, we predicted that the PilW(G−1F) mutant protein would fail to be processed by PilD and would be unable to rescue the twitching defect when expressed in the bifA pilW mutant strain, in contrast to expression of the wild-type PilW-His protein. And this is, indeed, what we observe (see Fig. S1A and B in the supplemental material), indicating that processing of PilW is required for its function in pilus assembly. Furthermore, expression of either the wild-type PilW-His or the PilW(G−1F) mutant protein in the bifA pilW mutant restores the bifA swarming defect (see Fig. S1C), indicating that cleavage of the PilW N terminus is not required for its function in swarming repression, as expected based on our PilX studies.
Collectively, the data presented above support our hypothesis that the minor pilins, PilX and PilW, play two roles in the cell, both involving surface-associated motile behaviors. In one role, PilX/PilW promote pilus assembly, and this function requires PilD-mediated cleavage of the leader peptide; in the second role, PilX/PilW repress swarming motility, and this is independent of PilD-mediated cleavage.
Unprocessed PilX is not required for swarming repression.
The findings presented thus far are consistent with the notion that the processed and unprocessed forms of the minor pilins PilW and PilX have distinct cellular functions. We presume that the PilD-processed versions of these proteins proceed directly to their role in pilus assembly. Given that PilD-mediated processing is not required for the role of PilX/PilW in swarming repression, we hypothesized that some fractions of PilW and PilX proteins might remain in the unprocessed form and that this unprocessed pool of proteins participates in the repression of swarming motility.
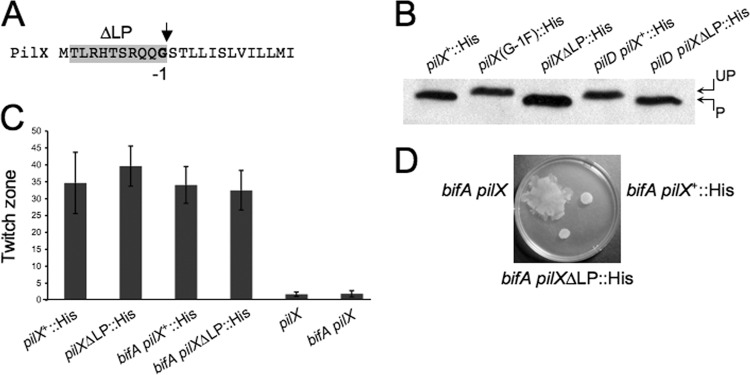
To test this hypothesis directly, we sought to generate a processed version of PilX without requiring synthesis of unprocessed PilX. To this end, we deleted the DNA sequence encoding the leader peptide normally cleaved by PilD in the pilX gene (denoted pilXΔLP) on the chromosome and inserted a C-terminal His6 epitope tag for detection of the PilXΔLP protein in the WT and bifA strain backgrounds (see Materials and Methods for details) (Fig. 5A). As shown in Fig. 5B, the PilXΔLP-His protein (lane 3) migrates at the same size as the processed PilX-His (lane 1) in the WT strain unlike the larger unprocessed PilX(G−1F)-His protein (lane 2). We observe the same migration pattern when these proteins are expressed in the bifA mutant background (data not shown).
Fig 5.
Deletion of the PilX leader peptide at the native locus and impact on pilus assembly and repression of swarming. (A) Sequence of the PilX N terminus with the leader peptide deletion (ΔLP) shaded in gray and the conserved glycine in the −1 position relative to the PilD cleavage site indicated by the arrow (See Materials and Methods for details of this mutation). (B) Western blot showing cleavage status of PilX-His, PilX(G−1F)-His, and PilXΔLP-His proteins in the WT strain compared to cleavage status of PilX-His and PilXΔLP-His in the pilD mutant strain. UP, unprocessed; P, processed. Western blots were probed with an anti-His antibody. (C) Graph showing quantification of twitch zones using the ImageJ program with units in pixels (×1,000). Error bars represent standard deviations of averages from three twitch zones per strain. (D) Representative image of a swarm plate with the indicated strains. Plates were incubated at 37°C for 16 h.
To verify that this deletion construct was behaving as expected, we also generated the leader peptide deletion in the pilD mutant background. Expression of the wild-type pilX+::His gene in the pilD mutant background yields the unprocessed form of PilX-His, as expected given the absence of the PilD peptidase (Fig. 5B, lane 4). In contrast, expression of pilXΔLP::His results in synthesis of a protein consistent with the size of the processed PilX, despite the absence of PilD (Fig. 5B, lane 5), and thus confirms that the pilXΔLP deletion mutation eliminates synthesis of unprocessed PilX-His.
We then assessed the ability of the PilXΔLP protein to function in pilus assembly by performing twitching assays with these strains. Interestingly, both the pilXΔLP::His and the bifA pilXΔLP::His strains exhibited twitching motility that was comparable to strains expressing the wild-type PilX-His protein (Fig. 5C). These data suggest that interaction with PilD and the subsequent processing event itself are not absolutely required for the function of PilX in pilus assembly, provided that a processed version of PilX is present in cells. This is in contrast to what has been observed for the major pilin subunit, PilA, where expression of a leaderless PilA protein resulted in the absence of surface piliation (56), indicating that processing of the major pilus subunit by PilD is required for proper pilus assembly.
We next evaluated the swarming ability of the bifA pilXΔLP::His strain. If unprocessed PilX is required for repression of swarming in the bifA mutant, then we would expect the bifA pilXΔLP::His strain to regain the ability to swarm. What we observe, however, is that swarming by the bifA pilXΔLP::His strain remains defective and is indistinguishable from that of the bifA pilX+::His strain (Fig. 5D). These data suggest that the unprocessed version of PilX per se is not required to repress swarming in the bifA mutant and, thus, argue against our hypothesis regarding a distinct function for unprocessed PilX. Together, these data are consistent with a model in which the processed version of PilX is required for pilus assembly, whereas either form of PilX, processed or unprocessed, appears to be functional in repression of swarming.
Cellular localization of PilX-His.
Our data indicate that the PilX protein is involved in c-di-GMP-mediated repression of swarming motility. Thus, we postulated that levels of c-di-GMP might influence the expression and/or localization of this protein. To address these possibilities, we fractionated (swarm) surface-grown cells of both the WT and bifA mutant carrying the chromosomally encoded His epitope-tagged versions of PilX at the native locus [pilX+::His and pilX(G−1F)::His] and probed for the presence of these proteins in various cellular fractions. In both the WT and bifA strain backgrounds, we detect wild-type PilX-His and the larger variant PilX(G−1F)-His in the whole-cell (WC), cytosolic (Cyt), total membrane (TM), and inner membrane (IM) fractions but not in the outer membrane (OM) fractions (see Fig. S2 in the supplemental material). Our data indicate that PilX-His is predominantly found in the IM fraction, which is consistent with an earlier report showing PilX primarily in the insoluble membrane fraction; however, the specific membrane compartment was not further delineated in those studies (2).
In the WT strain background, we did not observe any significant changes in the localization pattern of PilX(G−1F)-His compared to PilX-His, suggesting that lack of processing by the PilD peptidase (and thus retention of the leader peptide) has no impact on cellular localization. In comparing the WT and bifA mutant strains, we did not observe any significant alterations in the PilX fractionation pattern. Thus, levels of c-di-GMP do not appear to grossly alter the localization or abundance of the PilX protein.
The integrity of the inner and outer membrane fractions was confirmed by Western blotting using antibodies against SecY, an inner membrane protein (1, 24), and OprF, an outer membrane protein (6, 20, 32) (see Fig. S2 in the supplemental material).
Given that the PilX protein was recently shown to be associated with the pilus fiber (16), we wanted to examine the cell surface-associated proteins for the presence of PilX and to assess any differences that might be attributable to differences in c-di-GMP levels. To this end, we prepared surface protein (SP) fractions according to the protocol used by Giltner et al. (16) in which cells are grown overnight on agar plates and harvested directly for isolation of cell surface proteins. However, we were unable to detect the PilX-His protein in SP fractions prepared from strains carrying either the chromosomally expressed version of PilX-His or the multicopy plasmid-based version (data not shown).
It is worth noting that we did not detect unprocessed PilX in any of the fractions we examined from strains expressing the pilX+::His gene. The lack of detectable cellular pools of unprocessed PilX suggests that this version of PilX is indeed transient. Taken together with our results showing that the PilXΔLP protein is sufficient for function in both pilus assembly and swarming repression, these data strongly argue against the notion that unprocessed PilX has a distinct cellular role.
Given that the leader peptide of PilX does not appear to play an important role in either pilus assembly or swarming repression, we wondered whether it was also dispensable for cellular localization of PilX. Previous work with the PilA protein suggests that the leader peptide is not involved in membrane association but, rather, the maturation of pilin subunits into pilin fibers (56, 57). However, it is possible that the leader peptide of the minor pilins behaves differently and may influence cellular localization. If so, then any changes in cellular localization of the PilXΔLP protein could indicate that proper localization of PilX is not absolutely required for function in either pilus assembly or swarming repression.
To evaluate the role of the PilX leader peptide in cellular localization, we fractionated WT and bifA mutant cells expressing the PilXΔLP-His protein after growth on swarm plates and found no detectable alterations in the cellular localization profile relative to the PilX-His protein (see Fig. S3 in the supplemental material), suggesting that the leader peptide is not a cellular localization determinant for PilX. However, we did observe that the overall levels of PilXΔLP-His protein appear to be somewhat elevated relative to PilX-His in all fractions where these proteins are detected although the biological significance of this difference is not clear.
PilY1 and the minor pilins, PilX/PilW, impact cellular c-di-GMP levels in a surface-specific manner.
Having established that PilY1, PilX, and PilW participate in c-di-GMP-mediated repression of swarming motility, we next explored the mechanism by which this repression might occur. One hypothesis is that the minor pilins impact intracellular c-di-GMP levels by modulating some aspect of c-di-GMP metabolism, such as synthesis or degradation. An alternative hypothesis is that the minor pilins influence factors important for swarming function or regulation that are downstream of c-di-GMP metabolism, such as rhamnolipid production or flagellar synthesis. We tested the latter hypothesis and found that strains with a mutation in pilY1, pilX, or pilW do not exhibit any detectable changes in rhamnolipid production or levels of the flagellar subunit, FliC, relative to the WT parent strain (data not shown).
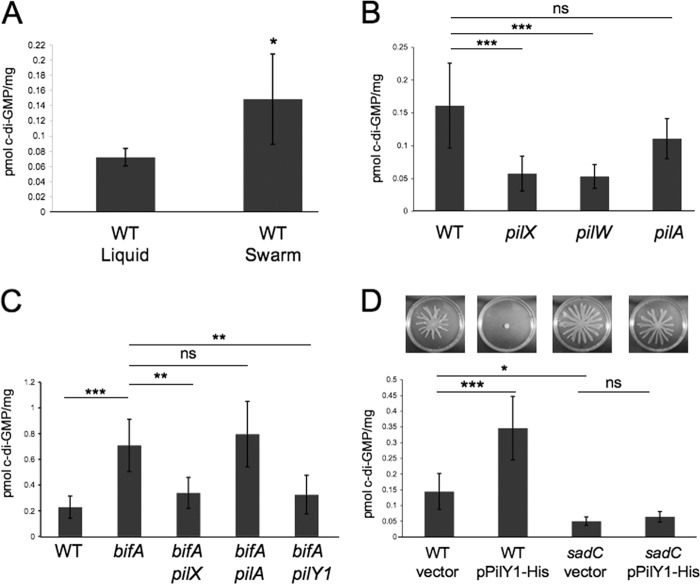
Thus, we turned our attention to the hypothesis that the minor pilins impact c-di-GMP metabolism. In our previous studies of the pilY1 gene and its involvement in c-di-GMP-mediated repression of swarming motility, we observed that mutating pilY1 had no impact on c-di-GMP levels in either WT or bifA mutant cells, despite all phenotypic evidence indicating some role for PilY1 in these processes (29). However, c-di-GMP measurements for those studies were taken from cells grown in liquid culture. These results prompted us to consider the possibility that pilY1 and the minor pilins might exhibit surface-specific impacts on c-di-GMP levels. To begin to test this notion, we first extracted cellular nucleotides from WT cells grown in liquid culture and on swarm plates and measured levels of the c-di-GMP species using LC-MS (see Materials and Methods for details). As shown in Fig. 6A, we observe that c-di-GMP levels are significantly elevated ∼2.5-fold in WT cells grown on swarm plates compared to liquid growth, suggesting that growth on a surface stimulates c-di-GMP production.
Fig 6.
Quantification of intracellular c-di-GMP levels by LC-MS. (A) Graph depicting quantification of global pools of c-di-GMP in WT cells grown either in liquid culture or on swarm plates. Data are expressed as picomoles of c-di-GMP per mg (dry weight) of the cell pellets from which the nucleotides were extracted (see Materials and Methods for details). Error bars represent standard deviation of the mean with four replicates per experiment. *, P < 0.02 (t test). (B) Quantification of c-di-GMP levels in the indicated strains grown on swarm plates. Error bars represent standard deviations of the averages of two experiments with three replicates per experiment. Data were analyzed by analysis of variance followed by a Tukey's posttest comparison. ns, not significant; ***, P < 0.001. (C) Measurements of c-di-GMP in swarm-grown cells of the indicated strains. Error bars represent standard deviation of the averages of two experiments with three replicates per experiment. Data were analyzed by analysis of variance with a Tukey's posttest comparison. ns, not significant; **, P < 0.01; ***, P < 0.001. (D) The top panel shows images of swarm plates for the indicated strains (see strain labels below the graph). The strains carried either the vector control or the pPilY1 plasmid. The medium was supplemented with 0.2% arabinose, and plates were incubated at 37°C for 16 h, after which cells were harvested for c-di-GMP quantification by LC-MS. The graph shows c-di-GMP measurements for each strain taken from swarm plates. Error bars represent standard deviations of the averages of two experiments with four replicates per experiment. Data were analyzed by analysis of variance and Tukey's posttest comparison. *, P < 0.05; ***, P < 0.001.
Given that c-di-GMP levels increase when WT cells are grown on the surface of a swarm plate, we next asked whether the pilX or pilW mutant is defective in this surface-induced c-di-GMP response. Indeed, surface growth of pilX and pilW mutant strains resulted in cellular levels of c-di-GMP that were nearly 3-fold lower than the level of the WT (Fig. 6B), approaching levels seen for the WT grown in liquid, indicating that the pilX and pilW genes likely participate in the surface stimulation of c-di-GMP synthesis. In contrast, a surface-grown pilA mutant showed a small, nonsignificant decrease in c-di-GMP levels relative to WT levels, indicating that the c-di-GMP surface response we observe is independent of pilus assembly.
We next assessed whether the minor pilins might impact cellular pools of c-di-GMP in the bifA mutant grown on swarming agar. Our genetic analyses of the bifA pilX and bifA pilW strains would indicate that this is indeed the case. Specifically, mutation of either the pilX or pilW gene in the bifA mutant background leads to suppression of all of the c-di-GMP-related phenotypes of this mutant (Fig. 1B). Results of c-di-GMP measurements from surface-grown cells (Fig. 6C) show that the bifA mutant accumulates ∼3-fold higher levels of c-di-GMP than the WT, as expected (30), and that mutation of pilX in the bifA mutant background leads to a reduction in c-di-GMP to levels comparable to the WT level, implying that pilX is required for the observed c-di-GMP elevation in the bifA mutant. In contrast, c-di-GMP measurements from liquid-grown cultures show no differences between the bifA and bifA pilX strains (data not shown). Furthermore, mutation of the pilA gene in the bifA mutant background has no impact on c-di-GMP levels from surface-grown cells, further supporting our observations that defects in pilus assembly alone do not impact c-di-GMP levels. Thus, the weak suppression of the bifA swarming defect that we observe in the bifA pilA double mutant does not appear to result from c-di-GMP alterations.
Upon revisiting the question of whether the pilY1 gene also impacts c-di-GMP levels in the bifA mutant specifically when cells are surface grown, we found that c-di-GMP levels are indeed reduced in the bifA pilY1 double mutant relative to the bifA mutant alone and that the c-di-GMP levels of the bifA pilY1 and bifA pilX double mutants are indistinguishable from one another (Fig. 6C). As mentioned above, this is in contrast to our previous observations where in liquid we did not see any difference in c-di-GMP levels between either the WT and the pilY1 mutant or between the bifA strain and the bifA pilY1 double mutant (29). Taken together, we find that PilX, PilW, and PilY1 are involved in a surface growth-dependent elevation of c-di-GMP levels.
Enhanced pilY1 gene expression leads to elevated c-di-GMP levels in the WT but not in a sadC diguanylate cyclase mutant.
Previously, we showed that overexpression of the pilY1 gene in the WT led to repression of swarming and that this repression required the function of the SadC diguanylate cyclase (29) (Fig. 6D, top panel). Based on those studies and the results described above, we predicted that pilY1 overexpression would lead to increased c-di-GMP levels in the WT but not in the sadC mutant background. In other words, we hypothesized that pilY1 overexpression stimulates c-di-GMP synthesis and that this stimulation requires the SadC diguanylate cyclase. To test this hypothesis, we measured intracellular c-di-GMP levels in swarm surface-grown cells of either the WT or the sadC mutant carrying a vector control or an arabinose-inducible pilY1 expression vector (pPilY1-His). We showed previously that this PilY1 expression construct does result in PilY1 overexpression (29). First, we observed that c-di-GMP levels are reduced nearly 3-fold in the sadC mutant relative to the WT (Fig. 6D, both carrying the vector control) which is consistent with data from a previous report (36). We also found that pilY1 overexpression leads to a nearly 2.5-fold increase in c-di-GMP levels in the WT strain; however, we did not observe a significant change in c-di-GMP levels upon overexpression of PilY1 in the sadC mutant background. These results support our hypothesis that PilY1 overexpression in surface-grown cells leads to a SadC-dependent increase in intracellular c-di-GMP levels and further bolster a link between PilY1, a pilus assembly protein, and SadC, a diguanylate cyclase, in the c-di-GMP-mediated repression of swarming motility.
In contrast to PilY1, multicopy plasmid-based expression of PilX or PilW does not lead to repression of swarming by the WT (data not shown). Therefore, we did not assess whether any changes in c-di-GMP levels occurred in strains carrying pilX or pilW overexpression constructs. Interestingly, we also observed that expression of multicopy pPilY1-His in either a pilX or pilW mutant background is still able to repress swarming motility (data not shown), suggesting that multicopy expression of PilY1 likely diminishes the requirement for PilX and PilW in the negative regulation of swarming motility.
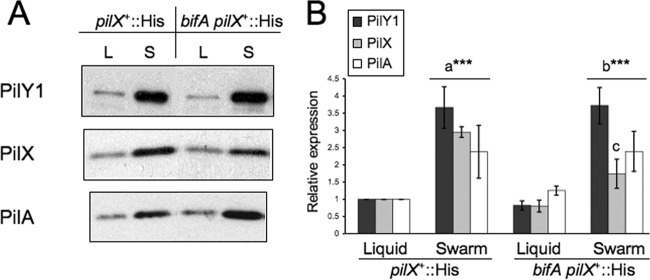
Surface growth results in increased levels of PilY1, PilX, and PilA.
Thus far, our data suggest that surface growth increases cellular c-di-GMP levels and that this increase involves the functions of PilY1 and SadC, as well as of PilX and PilW. Based on the data presented above showing that overexpression of PilY1 leads to stimulation of c-di-GMP synthesis, we reasoned that surface growth might alter PilY1 levels and that this, in turn, would lead to an increase in c-di-GMP synthesis via the SadC diguanylate cyclase.
To test this hypothesis, we examined PilY1 protein levels in lysates from strains grown either in liquid or on the surface of swarm plates. For these experiments, we used the pilX+::His variants of the WT and bifA mutant strains so that we could also examine PilX-His levels under these conditions as well. Recall that strains expressing PilX-His behave like their parental counterparts in all assays tested. We observed that levels of PilY1 are increased approximately 3.7-fold when pilX+::His cells are cultured on the surface of swarm plates relative to growth in liquid (Fig. 7A, top panel, left two lanes, and B, black bars). We observed a similar increase (∼4.5-fold) in PilY1 levels in surface-grown versus liquid-grown cells of the bifA pilX+::His mutant (Fig. 7A, right two lanes, and B, black bars). We detected no significant differences in PilY1 levels between the WT and bifA mutant strains when they were grown under the same conditions (either liquid or surface growth), indicating that the level of c-di-GMP (i.e., relatively low versus high) does not have a detectable impact on PilY1 expression under either growth condition. These data suggest that there is no apparent feedback regulation of PilY1 expression by c-di-GMP. Overall, these results support our hypothesis that surface growth increases PilY1 levels and, furthermore, are consistent with PilY1 playing a role upstream of c-di-GMP surface-induced synthesis.
Fig 7.
Expression of pilus assembly proteins under liquid versus swarm surface growth conditions. (A) Representative Western blot images showing expression of PilY1, PilX-His, and PilA in the WT or bifA strain carrying the His epitope insertion in the pilX gene (pilX+::His) grown either in liquid cultures (L lanes) or on swarm plates (S lanes). The pilX+::His strain behaves like its parental strain under all conditions tested. (B) Graph depicting quantification of relative protein levels from Western blots. Protein levels were quantified from scanned autoradiographs using ImageJ, and levels of PilY1, PilX, and PilA were set equal to 1 for the pilX+::His strain grown in liquid. Error bars indicate standard deviations of the averages of two experiments with three replicates per strain under each condition of growth. Significance was determined by a t test as follows: a***, P < 0.001, for the difference in protein levels between the pilX+::His strain grown in liquid versus growth on swarm plates; b***, P < 0.001, for the difference in protein levels between bifA pilX+::His grown in liquid versus growth on swarm plates; c, P < 0.001, for the difference in PilX levels between the pilX+::His and bifA pilX+::His strains grown on swarm plates. Besides this exception, there are no other significant differences in levels of PilY1, PilX, or PilA between the pilX+::His and bifA pilX+::His strains when they are grown under the same conditions.
Moreover, these data are reminiscent of recent work by Cowles and Gitai showing that synthesis of the pilus assembly proteins, PilA and PilT, is induced when cells are grown on surfaces (12). Thus, we next assessed whether PilX and PilA levels were also elevated upon surface growth. Indeed, we found that both PilX and PilA protein levels are elevated in the pilX+::His (WT) and bifA pilX+::His (bifA) strains when they are grown on the surface of swarm plates relative to growth in liquid (Fig. 7A, middle and bottom panels, and B, gray and white bars, respectively).
For PilA, we observed a ∼2.4-fold increase in PilA levels when WT cells were grown under solid versus liquid conditions and a ∼1.9-fold increase in PilA for the bifA mutant grown under solid versus liquid conditions. We did not observe a statistically significant difference in PilA levels between WT and the bifA mutant when both strains were grown in liquid, nor was there a significant difference when both were cultured on swarm plates.
For PilX, we observed a ∼3-fold increase in PilX levels in the WT and a ∼2.2-fold increase in the bifA mutant when they were grown under solid versus liquid conditions. While PilX levels are not significantly different in the WT versus the bifA mutant when the strains were grown in liquid, there is a significant difference when these strains are grown on swarm plates. Specifically, in the bifA mutant, PilX accumulates to only ∼60% of the level observed in the WT strain, indicating a less robust surface-induced increase in PilX when the bifA gene is mutated; however, the biological significance of this difference remains unclear.
DISCUSSION
Pseudomonas aeruginosa exhibits several surface-associated group behaviors, including biofilm formation, and two distinct surface-associated motile behaviors, namely, flagellum-propelled swarming motility and type IV pilus-mediated twitching motility. An open question is how cells transition from one surface behavior to another when changing environmental cues and/or surface conditions necessitate such a switch.
The work presented here illustrates one connection among surface-associated behaviors in this organism; that is, we show that components of the pilus assembly machinery apparently repress swarming motility. For this study, we focused on the genes encoding the minor pilins (fimU, pilV, pilW, pilX, pilY2, and pilE) because they are expressed in an operon containing the pilY1 gene (5). In previous work, we showed that mutating the pilY1 gene relieves the inhibition of swarming motility conferred on strains producing high levels of c-di-GMP due to loss of the BifA c-di-GMP phosphodiesterase (29). The minor pilins have been shown genetically to be important for optimal pilus assembly and were recently shown to be incorporated into the pilus fiber; however, the significance of this incorporation is not yet understood (2–4, 16, 49).
In the experiments here, we utilized the bifA mutant as a tool to assess the involvement of the minor pilins in repression of swarming motility. By mutating each of the minor pilin genes in the bifA background and assessing the suppression of the swarming defect, we determined that mutation of either the pilW or pilX gene showed robust suppression of the swarming defect, resembling the suppression we reported for the pilY1 mutation in the bifA strain (29). Phenotypic analysis of the fimU, pilE, and pilY2 mutants revealed weak-suppression phenotypes (akin to mutating the PilA pilin) or no phenotype, while a pilV mutant yielded an inconsistent and therefore inconclusive phenotype. Thus, we conclude that PilW and PilX indeed participate, along with PilY1, in swarming repression.
Our studies indicate that PilW and PilX (as well as the previously reported PilY1) play two roles in the cell: in one role these minor pilins promote assembly of the pilus, and in a second role they repress swarming when c-di-GMP levels are elevated. Here, we show that, for PilW and PilX, these two roles represent distinct functions of these proteins such that they can be separated by mutation. Mutating the invariant glycine (G−1) to a phenylalanine (F) in the N-terminal leader peptide of both PilW and PilX inhibits cleavage of this peptide by the PilD peptidase. With cleavage blocked, the PilW(G−1F) and PilX(G−1F) mutant proteins are unable to function in pilus assembly, indicating that processing by PilD is required for this role, a finding that is not unexpected given that PilD-mediated processing of the major pilin, PilA, is also required for proper pilus assembly (56). In contrast, the PilW(G−1F) and PilX (G−1F) mutant proteins are still able to function in swarm repression, indicating that processing of the minor pilins is not required for this second function. This conclusion is further supported by genetic evidence, whereby deletion of the pilD gene (which would inhibit processing of PilA and all of the minor pilins) in the bifA mutant background fails to fully suppress the swarming defect of a bifA mutant and instead yields a weak-suppression phenotype that is indistinguishable from the bifA pilA mutant. This finding indicates that, despite lack of N-terminal processing of PilA, PilW, and PilX, repression of swarming still occurs.
An important goal moving forward is to understand the mechanism by which the minor pilins repress swarming. Here, we present findings supporting a mechanism whereby PilX/PilW and PilY1 repress swarming by modulating levels of the intracellular signaling molecule c-di-GMP, specifically when cells are grown on a surface. These findings are consistent with the model depicted in Fig. 8. We show that surface growth of P. aeruginosa cells increases the level of pilus assembly proteins, including PilY1 and PilX as well as PilA. These data are consistent with a recent report in which Cowles and Gitai demonstrate that surface growth stimulates pilus production, in part by increasing levels of PilA (12). Furthermore, these investigations showed that the level of PilA increases with increasing surface hardness (i.e., agar concentration), suggesting a response that may be fine-tuned, depending upon specific surface conditions (12). From these studies, the authors surmise that surface induction of pilus biogenesis proteins allows cells to assemble pili when conditions favor twitching motility.
Fig 8.
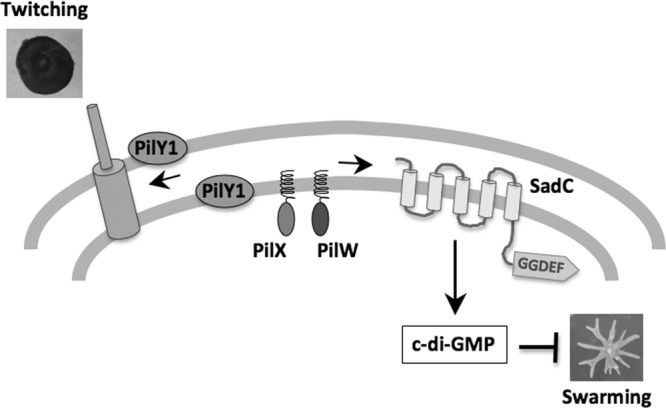
Model for coordinate regulation of surface-based swarming and twitching motility. Surface growth induces expression of pilus assembly proteins, including PilY1, PilX, and PilA, likely to prepare cells for twitching motility. Recent evidence indicates that the level of this induction may be tuned to surface conditions, such as surface hardness (12), and one could postulate that additional surface cues such as surface wetness or relative humidity might also impact this induction. Surface induction of PilY1 not only facilitates pilus biogenesis but also stimulates synthesis of the intracellular signaling molecule, c-di-GMP, via an as yet uncharacterized interaction with the SadC diguanylate cyclase. Our data are consistent with the minor pilins, PilX and PilW, also participating in the modulation of c-di-GMP levels, either in conjunction with PilY1 or possibly via a separate pathway. Increased synthesis of c-di-GMP then leads to repression of swarming motility, most likely by influencing flagellar function as previously proposed (9). Elevated c-di-GMP levels also stimulate biofilm formation (9, 30, 35), allowing P. aeruginosa cells to coordinately regulate all three of these distinct surface behaviors. Note that depictions in the figure are not to scale.
In our model, we propose that a surface-induced increase in PilY1 facilitates not only pilus assembly but also suppression of swarming via stimulation of c-di-GMP synthesis by a direct or indirect interaction with the SadC diguanylate cyclase. Under surface growth conditions that favor swarming (i.e., 0.5% agar), where we observe an increase in PilY1 levels, we also observe increased c-di-GMP levels. Furthermore, we show that overexpression of PilY1 leads to increased c-di-GMP levels and repression of swarming, both of which are dependent upon the SadC diguanylate cyclase. We postulate that the increase in c-di-GMP levels when cells are grown on 0.5% agar acts as a signal for the cell to undergo swarming motility versus the swimming motility utilized in liquid medium. However, if c-di-GMP levels are further increased, for example, when we overexpress PilY1 or presumably when cells are grown on a hard surface (1.5% agar), swarming motility is completely repressed.
Additional evidence supporting the notion that surface growth can lead to enhancement of diguanylate cyclase activity has been reported by Güvener and Harwood (19). These studies revealed that surface growth of P. aeruginosa stimulates clustering of the phosphorylated (i.e., active) form of the WspR diguanylate cyclase, leading to enhanced signal transduction through the Wsp chemosensory system. While this study did not directly measure c-di-GMP levels in surface-grown cells, it has been well established that signal transduction through the Wsp system results in WspR-mediated c-di-GMP synthesis (18, 23, 33).
Together with previous studies, we have shown that PilX, PilY1, and SadC localize to the inner membrane of cells (29, 35). While this is a broad brushstroke in terms of compartmentalizing these proteins, such observations allow for the possibility of interaction among these proteins. However, previous reports have found PilY1 present in sheared surface protein fractions, indicating that PilY1 is a cell surface-localized protein, possibly associated with the pilus (7, 21). Thus, we cannot rule out the possibility that signaling by PilY1 occurs through a more complex mechanism involving communication between the outer cell surface and inner membrane compartment.
Given that mutation of pilY1, pilX, or pilW leads to similar impacts on c-di-GMP levels in both the WT and bifA strain backgrounds, it is reasonable to propose that PilX/PilW participate together with PilY1 in the surface-induced stimulation of c-di-GMP production. PilY1 is likely to be the main actor in this scenario, given the additional observations here that multicopy plasmid-based expression of pilY1 but not pilX or pilW is able to repress swarming motility. However, we have also observed that overexpression of pilY1 does not require the functions of either PilX or PilW (data not shown). One interpretation of these data is that PilX/PilW are accessory factors whose role in swarming repression can be bypassed by excess PilY1 expression. An alternative possibility is that PilX/PilW function to modulate c-di-GMP in a pathway distinct from that of PilY1, providing a potentially intriguing depth to the regulation of swarming motility by these pilus assembly proteins. These questions as well as the nature of potential interactions of these proteins with SadC remain to be investigated.
Additional questions remain regarding the specific signals that are sensed when these microbes encounter or grow on a surface and how such signals are perceived by cells. The investigations by Cowles and Gitai suggest that surface hardness is a potential signal, with the notion that cells may be able to sense surface hardness over a broad spectrum and respond by building pili incrementally across this spectrum (12). Our model based on the findings presented here fits within this framework. We present evidence that cells respond to growth on a soft (swarm) agar surface by increasing levels of PilY1, PilX, PilW, and PilA, followed by induction of c-di-GMP synthesis relative to growth in liquid cultures. We would predict that further increases in PilY1 levels and c-di-GMP induction would lead to incremental decreases in swarming motility as cells transition from soft to hard agar surfaces where full repression would be achieved. However, additional factors (that may or may not be controlled by c-di-GMP) may contribute to full repression of swarming motility on hard surfaces, a notion supported by recent studies showing that growth on hard agar surfaces inhibits production of the rhamnolipid surfactant required for swarming motility (27). Alternatively, surface hardness may represent only a surrogate measure of a more complex set of surface conditions involving changes in relative humidity or water potential of the surface, as described recently (13). In any case, the precise nature of surface signals and how such signals are perceived by cells remain to be fully explored.
More broadly, our findings suggest that P. aeruginosa has evolved mechanisms that allow this organism to suppress swarming motility when twitching motility is favored. Consistent with this idea, both the extent and patterning of swarming motility are altered when pilus biogenesis is disrupted (29, 31, 53) (Fig. 1B), implying an interaction between swarming and twitching functions in P. aeruginosa. Potentially, the reverse may be true as well; that is, we postulate that there may be regulatory mechanisms allowing this microbe to modulate twitching when swarming is favored. Together with our previous findings showing that biofilm formation and swarming motility are inversely regulated (9, 30, 35), a picture is emerging whereby P. aeruginosa can effectively coordinate its surface-associated behaviors and where, furthermore, at least some aspects of this regulation are mediated via the intracellular signaling molecule c-di-GMP.
Supplementary Material
ACKNOWLEDGMENTS
We thank Beverly Chamberlin and Lijun Chen at the Michigan State University Mass Spectrometry Facility for the quantitative analysis of c-di-GMP.
This work was supported by NIH grant R01AI083256 to G.A.O.
Footnotes
Published ahead of print 3 August 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Akiyama Y, Ito K. 1985. The SecY membrane component of the bacterial protein export machinery: analysis by new electrophoretic methods for integral membrane proteins. EMBO J. 4:3351–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alm RA, Hallinan JP, Watson AA, Mattick JS. 1996. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol. Microbiol. 22:161–173 [DOI] [PubMed] [Google Scholar]
- 3. Alm RA, Mattick JS. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16:485–496 [DOI] [PubMed] [Google Scholar]
- 4. Alm RA, Mattick JS. 1996. Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178:3809–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belete B, Lu H, Wozniak DJ. 2008. Pseudomonas aeruginosa AlgR regulates type IV pilus biosynthesis by activating transcription of the fimU-pilVWXY1Y2E operon. J. Bacteriol. 190:2023–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benz R, Hancock RE. 1981. Properties of the large ion-permeable pores formed from protein F of Pseudomonas aeruginosa in lipid bilayer membranes. Biochim. Biophys. Acta 646:298–308 [DOI] [PubMed] [Google Scholar]
- 7. Bohn YST, et al. 2009. Multiple roles of Pseudomonas aeruginosa TBCF10839 PilY1 in motility, transport and infection. Molecular Microbiology. 71:730–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burrows LL. 2005. Weapons of mass retraction. Mol. Microbiol. 57:878–888 [DOI] [PubMed] [Google Scholar]
- 9. Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:3603–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caiazza NC, O'Toole GA. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 186:4476–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280:30829–30837 [DOI] [PubMed] [Google Scholar]
- 12. Cowles KN, Gitai Z. 2010. Surface association and the MreB cytoskeleton regulate pilus production, localization and function in Pseudomonas aeruginosa. Mol. Microbiol. 76:1411–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dechesne A, Smets BF. 2012. Pseudomonad swarming motility is restricted to a narrow range of high matric water potentials. Appl. Environ. Microbiol. 78:2936–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690 [DOI] [PubMed] [Google Scholar]
- 15. Friedman L, Kolter R. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giltner CL, Habash M, Burrows LL. 2010. Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J. Mol. Biol. 398:444–461 [DOI] [PubMed] [Google Scholar]
- 17. Giltner CL, Rana N, Lunardo MN, Hussain AQ, Burrows LL. 2011. Evolutionary and functional diversity of the Pseudomonas type IVa pilin island. Environ. Microbiol. 13:250–264 [DOI] [PubMed] [Google Scholar]
- 18. Goymer P, et al. 2006. Adaptive divergence in experimental populations of Pseudomonas fluorescens. II. Role of the GGDEF regulator WspR in evolution and development of the wrinkly spreader phenotype. Genetics 173:515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Güvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 66:1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hancock RE, Decad GM, Nikaido H. 1979. Identification of the protein producing transmembrane diffusion pores in the outer membrane of Pseudomonas aeruginosa PAO1. Biochim. Biophys. Acta 554:323–331 [DOI] [PubMed] [Google Scholar]
- 21. Heiniger RW, Winther-Larsen HC, Pickles RJ, Koomey M, Wolfgang MC. 2010. Infection of human mucosal tissue by Pseudomonas aeruginosa requires sequential and mutually dependent virulence factors and a novel pilus-associated adhesin. Cell. Microbiol. 12:1158–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 23. Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hinsa SM, O'Toole GA. 2006. Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology 152:1375–1383 [DOI] [PubMed] [Google Scholar]
- 25. Jenal U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185–191 [DOI] [PubMed] [Google Scholar]
- 26. Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385–407 [DOI] [PubMed] [Google Scholar]
- 27. Kamatkar NG, Shrout JD. 2011. Surface hardness impairment of quorum sensing and swarming for Pseudomonas aeruginosa. PLoS One 6:e20888 doi:10.1371/journal.pone.0020888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kohler T, Curty LK, Barja F, van Delden C, Pechere JC. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuchma SL, et al. 2010. Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J. Bacteriol. 192:2950–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuchma SL, et al. 2007. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:8165–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leech AJ, Mattick JS. 2006. Effect of site-specific mutations in different phosphotransfer domains of the chemosensory protein ChpA on Pseudomonas aeruginosa motility. J. Bacteriol. 188:8479–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. MacEachran DP, et al. 2007. The Pseudomonas aeruginosa secreted protein, PA2934, decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect. Immun. 75:3902–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malone J, et al. 2007. The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology 153:980–994 [DOI] [PubMed] [Google Scholar]
- 34. Mattick JS. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289–314 [DOI] [PubMed] [Google Scholar]
- 35. Merritt JH, Brothers KM, Kuchma SL, O'Toole GA. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189:8154–8164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Merritt JH, et al. 2010. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1:e00183–10 doi:10.1128/mBio.00183–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Monds RD, Newell PD, Gross RH, O'Toole GA. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63:656–679 [DOI] [PubMed] [Google Scholar]
- 38. Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3,5)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. U. S. A. 106:3461–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O'Toole GA. 2011. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J. Bacteriol. 193:4685–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nunn D, Bergman S, Lory S. 1990. Products of three accessory genes, pilB, pilC, and pilD are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 172:2911–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nunn D, Lory S. 1993. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J. Bacteriol. 175:4375–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nunn DN, Lory S. 1991. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. U. S. A. 88:3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Orans J, et al. 2010. Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility. Proc. Natl. Acad. Sci. U. S. A. 107:1065–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 45. O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 46. Pasloske B, Paranchych W. 1988. The expression of mutant pilins in Pseudomonas aeruginosa: fifth position glutamate affects pilin methylation. Mol. Microbiol. 2:489–495 [DOI] [PubMed] [Google Scholar]
- 47. Paul R, et al. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Römling U, Gomelsky M, Galperin MY. 2005. c-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629–639 [DOI] [PubMed] [Google Scholar]
- 49. Russell MA, Darzins A. 1994. The pilE gene product of Pseudomonas aeruginosa, required for pilus biogenesis, shares amino acid sequence identity with the N-termini of type 4 prepilin proteins. Mol. Microbiol. 13:973–985 [DOI] [PubMed] [Google Scholar]
- 50. Ryan RP, et al. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U. S. A. 103:6712–6717 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51. Sauer K, Camper AK. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shrout JD, et al. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264–1277 [DOI] [PubMed] [Google Scholar]
- 54. Siegmund I, Wagner F. 1991. New method for detecting rhamnolipids excreted by Pseudomonas aeruginosa species during growth on minimal agar. Biotechnol. Technol. 5:265–268 [Google Scholar]
- 55. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 56. Strom MS, Lory S. 1991. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J. Biol. Chem. 266:1656–1664 [PubMed] [Google Scholar]
- 57. Strom MS, Lory S. 1987. Mapping of export signals of Pseudomonas aeruginosa pilin with alkaline phosphatase fusions. J. Bacteriol. 169:3181–3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Strom MS, Nunn DN, Lory S. 1993. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc. Natl. Acad. Sci. U. S. A. 90:2404–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Toutain CM, Zegans ME, O'Toole GA. 2005. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 187:771–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Whitchurch CB, Hobbs M, Livingston SP, Krishnapillai V, Mattick JS. 1991. Characterization of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in eubacteria. Gene 101:33–44 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.