Abstract
We identified the fatty acid synthesis (FAS) initiation enzyme in Pseudomonas aeruginosa as FabY, a β-ketoacyl synthase KASI/II domain-containing enzyme that condenses acetyl coenzyme A (acetyl-CoA) with malonyl-acyl carrier protein (ACP) to make the FAS primer β-acetoacetyl-ACP in the accompanying article (Y. Yuan, M. Sachdeva, J. A. Leeds, and T. C. Meredith, J. Bacteriol. 194:5171-5184, 2012). Herein, we show that growth defects stemming from deletion of fabY can be suppressed by supplementation of the growth media with exogenous decanoate fatty acid, suggesting a compensatory mechanism. Fatty acids eight carbons or longer rescue growth by generating acyl coenzyme A (acyl-CoA) thioester β-oxidation degradation intermediates that are shunted into FAS downstream of FabY. Using a set of perdeuterated fatty acid feeding experiments, we show that the open reading frame PA3286 in P. aeruginosa PAO1 intercepts C8-CoA by condensation with malonyl-ACP to make the FAS intermediate β-keto decanoyl-ACP. This key intermediate can then be extended to supply all of the cellular fatty acid needs, including both unsaturated and saturated fatty acids, along with the 3-hydroxyl fatty acid acyl groups of lipopolysaccharide. Heterologous PA3286 expression in Escherichia coli likewise established the fatty acid shunt, and characterization of recombinant β-keto acyl synthase enzyme activity confirmed in vitro substrate specificity for medium-chain-length acyl CoA thioester acceptors. The potential for the PA3286 shunt in P. aeruginosa to curtail the efficacy of inhibitors targeting FabY, an enzyme required for FAS initiation in the absence of exogenous fatty acids, is discussed.
INTRODUCTION
Pseudomonas aeruginosa is a versatile Gram-negative pathogen, being the causative agent of a wide range of both community-associated (folliculitis and otitis externa) and health care-associated (pneumonia, urinary tract, and bacteremia) bacterial infections (15, 34, 39, 55). While generally not considered a normal member of the human flora, the near ubiquitous environmental distribution of P. aeruginosa provides a ready reservoir for exposure and ensuing opportunistic infection. Infections due to P. aeruginosa are particularly prevalent among immunodeficient individuals in whom cutaneous or mucosal barriers have been breached by ventilators, catheters, or through trauma, as seen in burn units. In >28,000 cases of all health care-associated infections reported to the U.S. National Healthcare Safety Network during a 22-month period beginning in 2006, 7.9% were attributed to P. aeruginosa (24). Chronic Pseudomonas infections are especially problematic in the lungs of cystic fibrosis patients, in part due to a genetic defect that facilitates bacterial colonization through diminished mucociliary clearance (6). The breadth of difficult-to-treat P. aeruginosa-related infections, coupled with an impressive array of intrinsic and adaptive antibacterial resistance mechanisms (3, 38), makes developing new antipseudomonas drugs a challenging priority.
We initiated a target evaluation program focused on fatty acid synthesis (FAS) in P. aeruginosa. FAS plays a multifaceted role in both maintaining bacterial viability and virulence in P. aeruginosa, suggesting that inhibition of FAS in vivo may have added benefit beyond simply blocking division through depleting fatty acid pools available for phospholipid biosynthesis. Aside from being compulsory components of membrane phospholipids (12), fatty acids are utilized by multiple primary and secondary metabolic pathways in P. aeruginosa. Lipopolysaccharides (LPS), which are lipoglycans located in the external leaflet of the outer membrane, are dependent on 3-hydroxy (3-OH) acyl-ACP (acyl carrier protein) FAS intermediates for complete acylation and in turn the establishment of the permeability barrier (43). Lipoproteins, a large class of lipidated membrane proteins that includes components of the LPS transport machinery and of the resistance-nodulation cell division (RND)-type drug efflux pumps (44), depend on phospholipid donors for acylation (40). Even partial FAS inhibition could therefore induce LPS hypoacylation, decrease LPS transport, and/or cripple efflux pumps, raising the potential for synergistic combinations between FAS inhibitors and membrane-impermeable or efflux-susceptible antibiotics. Fatty acids are also used in the assembly of two important metabolic enzyme cofactors, lipoate and biotin. Lipoate is an essential cofactor of α-ketoacid dehydrogenases (49), including pyruvate dehydrogenase. Pyruvate dehydrogenase connects glycolytic flux to the tricarboxylic acid (TCA) cycle by forming acetyl coenzyme A (acetyl-CoA) from pyruvate. P. aeruginosa lacking pyruvate dehydrogenase does not express the type III secretion system, critical machinery involved in delivering cytotoxic effectors, due to the metabolic defect (45). More recently, the essential cofactor biotin has also been proposed to hijack FAS biosynthetic enzymes for its own synthesis (11, 37). Biotin is required by enzymes involved in amino acid metabolism, gluconeogenesis, and FAS itself. The assembly of the high-affinity ferric iron siderophore pyoverdine is dependent on the fatty acid myristate carrier in P. aeruginosa (22). Finally, orchestrating the complex intercellular signaling and hierarchal gene regulation that is a hallmark of the P. aeruginosa social lifestyle are the three acylated quorum-sensing signal molecules [Pseudomonas quinolone signal (PQS), N-(3-oxododecanoyl)-l-homoserine lactone, and N-butanoyl-l-homoserine lactone] (30) and the cis-2-decenoic fatty acid diffusible signal factor (DSF) (14). Collectively, these systems coordinate the expression of hundreds of genes (54), including pertinent virulence factors such as rhamnolipids, pyocyanin, and extracellular proteases, as well as the transition from planktonic to antibiotic-recalcitrant growth within biofilms. In addition to being an essential target in and of itself, the central roles of fatty acids in intrinsic antimicrobial resistance, in supporting diverse intermediary metabolism, in sensing environmental cues, and in mobilizing a suite of virulence factors all make FAS a promising antibacterial target in P. aeruginosa.
The initiating step of FAS in Escherichia coli is catalyzed by the enzyme β-acetoacetyl-ACP synthase encoded by fabH (52), which condenses malonyl-ACP with acetyl-CoA to form β-acetoacetyl-ACP. The enzyme is defined by the signature β-ketoacyl synthase III (KASIII) domain, which is present in multiple, highly similar genes within the genome of P. aeruginosa PAO1 (56, 57). Our initial attempts in the accompanying article (56) to assign the FabH-type activity of P. aeruginosa to a single KASIII ortholog were unsuccessful; rather, it was determined that the predominant β-acetoacetyl-ACP synthase is encoded by a new class of highly divergent KASI/II-type synthases named fabY (formerly PA5174). Deletion of fabY confirmed a pleiotropic phenotypic consistent with decreased FAS flux, as siderophore production, swarming motility, rhamnolipids, and fatty acid-dependent quorum-sensing signals (PQS and homoserine lactones) in tandem with the expression of their cognate regulatory targets were attenuated (56). Although the doubling time of the ΔfabY deletion mutant was three times longer in liquid media, the viability of the ΔfabY deletion mutant indicated that another P. aeruginosa gene(s) is capable of fatty acid initiation. In addition, we observed that the growth defect could be partially complemented by inclusion of exogenous free fatty acids in the growth media. We thus set out to address the fabY-independent route of FAS in P. aeruginosa and to understand how fatty acids are incorporated into de novo phospholipids. We herein show that the KASIII domain-containing enzyme from the previously unannotated open reading frame PA3286 shunts fatty acid degradation intermediates from the β-oxidation pathway by condensing octanoyl-CoA (C8-CoA) with malonyl-ACP to make β-keto-decanoyl-ACP, a key building block common to saturated fatty acids (SFA), unsaturated fatty acids (UFA), and LPS.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All P. aeruginosa strains were derived from the wild-type PAO1 strain (51), while Escherichia coli strains were derived from the reference strain BW25113 (CGSC7636). Strains were grown in LB-Miller medium at 37°C unless otherwise noted. Antibiotic markers were selected with gentamicin (Gm) (100 μg/ml in P. aeruginosa and 10 μg/ml in E. coli), carbenicillin (Carb) (150 μg/ml in P. aeruginosa and 100 μg/ml in E. coli), tetracycline (Tet) (125 μg/ml in P. aeruginosa and 15 μg/ml in E. coli), chloramphenicol (Cam) (20 μg/ml in E. coli), and kanamycin (Kan) (50 μg/ml in E. coli). Bacterial strains and plasmids used in this study are listed in Table 1.
Table 1.
Bacterial strains, plasmids, and primers used in this study
| Bacterial strain, plasmid, or primer | Relevant genotype or phenotypea or primer sequence | Source or reference |
|---|---|---|
| E. coli strains | ||
| BW25113 | E. coli Κ-12 Wt [Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514] | CGSCb |
| TMY32 | BW25113 fabH::camR(pET-PA5174); Kanr Camr | 56 |
| TMT47 | BW25113 fabH::camR(pET-PA3286); Kanr Camr | This study |
| P. aeruginosa strains | ||
| NB52019 | P. aeruginosa PAO1 prototroph K767 | K. Poole |
| TMT01 | NB52019 ΔPA3333 (fabH2) | 56 |
| TMT02 | NB52019 ΔPA0999 (pqsD) | 56 |
| TMT12 | NB52019 ΔPA3286 | 56 |
| TMT15 | NB52019 ΔPA0998 (pqsC) | 56 |
| TMT16 | NB52019 ΔPA0998 ΔPA0999 ΔPA3333 ΔPA3286 | 56 |
| TMT39 | NB52019 ΔfabY (PA5174)::aacC1; Gmr | 56 |
| TMT44 | TMT12 attB::PA3286+; Gmr | This study |
| Plasmids | ||
| pMini-CTX1 | Chromosomal integration vector; FRT oriT int+ori (pMB1) tetR-FRT attP+ MCS; Tetr | 26 |
| pRC9 | pMini-CTX-1 with araC-PBAD promoter cassette | C. Dean |
| pTMT131 | pRC9 with the PA3286 gene and 483 bp upstream | This study |
| pFLP2 | E. coli-P. aeruginosa shuttle vector with Flp recombinase; Carbr | 25 |
| pET24b(+) | IPTG-inducible T7 promoter for protein expression; Kanr | Novagen |
| pET-PA3286 | pET24b(+) with PA3286 and C-terminal His tag; Kanr | This study |
| pKD3 | oriRλ camR; Camr | 13 |
| pTMT123 | pEX18ApGW-PA5174::aacC1 fabHEc; Gmr | 56 |
| pTMT124 | pEX18ApGW-PA5174::aacC1 fabH′ fragment; Gmr | 56 |
| Primers | ||
| CTX-PA3286 EcoRVc | TGCGACGCTGGCGATCTTCCTTGTGAATAC | This study |
| CTX-PA3286 XhoIc | CGGGCCCCCCCTCGAAACCAGCTCGCGGAAG | This study |
| PA3286 for NdeId | GTAGCTCATATGCATAAAGCCGTCATC | This study |
| PA3286 rev HindIIId | GTAGCTAAGCTTGTGTTTACGCAGGATC | This study |
| fabH Ec KO P1 | CGCCACATTGCCGCGCCAAACGAAACCGTTTCAACCATGGGCCGCCTACCTGTGACGGAA | 56 |
| fabH Ec KO P2 | CCGCCCCAGATTTCACGTATTGATCGGCTACGCTTAATGCATGAACTTCATTTAAATGGCGCG | 56 |
Carbr, carbenicillin resistant; Camr, chloramphenicol resistant; Kanr, kanamycin resistant; Gmr, gentamicin resistant; Tetr, tetracycline resistant; FRT, Flp recombinase target; MCS, multicloning site.
Strain CGSC7636 at the Coli Genetic Stock Center (CGSC).
Cloned using the In-fusion system (Clontech).
Cloned into vectors by DNA digestion and T4 DNA-catalyzed ligation.
Growth analysis.
For P. aeruginosa and E. coli strains, starter cultures were made by scraping cells off LB agar plates with appropriate selection antibiotics. Cultures were typically resuspended to ∼2 × 105 CFU/ml in LB with or without fatty acid supplement (1 to 100 μg/ml) and inducer (1 mM isopropyl-β-d-1-thiogalactopyranoside [IPTG]). Cultures were inoculated into clear 96-well flat-bottom untreated microplates (catalog no. 3370; Costar) and incubated at 37°C. The optical density at 600 nm (OD600) was recorded every 10 min on a Spectramax plate reader with intermittent shaking.
Fatty acid composition.
For readily soluble fatty acids (C2:0 to C10:0), bacteria were streaked onto LB agar plates containing 100 μg/ml of the sodium salt of a given fatty acid (Sigma or CDN Isotopes for deuterated fatty acids) and incubated overnight at 37°C. For longer-chain fatty acids (C14:0 and C16:0), liquid cultures were inoculated (2 × 105 CFU/ml) into LB with 2 mg/ml of fatty acid-free bovine serum albumin (BSA; Sigma) carrier and grown overnight to stationary phase at 37°C. To induce the fatty acid transporters of E. coli, the medium was spiked with 10 μg/ml of unlabeled palmitate (C16:0) along with 100 μg/ml of perdeuterated decanoate. Biomass was scraped from the agar surface or collected by centrifugation, suspended in phosphate-buffered saline (PBS), and washed three times with PBS. Lipids were saponified, methylated, extracted, and washed according to the Sherlock microbial identification system (Microbial ID, Inc., Newark, DE). Fatty acid methyl ester (FAME) composition was determined by gas chromatography with flame ionization detection (GC-FID) or with mass spectrometry (GC-MS). An HP 6890 gas chromatograph with an HP-5MS 30 m column was connected to an HP 5973 mass selective detector. Results were analyzed on the HP MSD ChemStation (version D.01.02). Structural assignments were made by comparison of retention times to authentic FAME standards, as well as from the mass spectra.
Synthetic lethal scoring between fabY and KASIII domain orthologs.
The conjugation vector pEX18ApGW (9) carrying genes encoding carbenicillin resistance (Carbr), oriT transfer origin, and sucrose counterselection (sacB) was used to deliver an aacC1 (Gm) resistance cassette with either a transcriptionally coupled and functional E. coli fabH gene (pTMT123) or an inactive fabH fragment (pTMT124) (56) (Table 1). The cassettes were flanked by ∼1 kb of homologous DNA flanking the fabY gene in P. aeruginosa PAO1. Vectors were mobilized in trans using E. coli Stellar cells (Clontech) transformed with the helper plasmid pRK2013 (16) as the conjugation-proficient donor strain along with different recipient P. aeruginosa strains as has been described previously (56). Merodiploid colonies were confirmed by colony PCR and outgrown for 4 h in 1 ml of LB only at 37°C. Serially diluted aliquots were then spread on LB agar containing 7% sucrose (LB–7% sucrose agar) (without NaCl) to counterselect colonies harboring unresolved plasmid. In parallel, aliquots were plated on LB-sucrose agar with Gm or Gm plus decanoate (100 μg/ml) to select colonies from which fabY was replaced with fabH (pTMT123) or deleted (pTMT124). Colonies were counted after incubation at 37°C for 24 h (all pTMT123 constructs) or 48 h for slow-growing mutants (pTMT124 selected on Gm). In cases where less than 10 colonies arose and spontaneous sucrose tolerance was suspected, colonies were patched onto LB agar with Carb to confirm loss of plasmid backbone. The entire experiment was repeated three separate times in each P. aeruginosa recipient strain background.
Complementation of ΔPA3286.
The pBAD-containing pRC9 plasmid derivative of the site-specific integration vector pMini-CTX1 (26) was used to complement the ΔPA3286 deletion strain. The PA3286 gene, along with 483 bp upstream encompassing the putative native promoter region, was amplified from P. aeruginosa PAO1 genomic DNA template using the primer pair CTX-PA3286 EcoRV/CTX-PA3286 XhoI (Table 1). The fragment was introduced into pRC9 using the In-fusion system (Clontech), removing the pBAD regulatory element in the process. The resulting vector (pTMT131) was transferred into P. aeruginosa ΔPA3286 by conjugation, after which the integrated vector backbone encoding the tetracycline resistance (Tetr) gene was removed by FLP-catalyzed recombination using pFLP2 (25) to generate the complemented strain TMT44 (Table 1).
PA3286 complementation in E. coli.
The PA3286 open reading frame was cloned by restriction and ligation into the IPTG-inducible expression vector pET24b(+) (Novagen) using the PCR product of primers PA3286 for NdeI/PA3286 rev HindIII (rev stands for reverse) (Table 1). The expression plasmid pET-PA3286 was transformed into E. coli BW25113, in which the fabH gene was deleted using the fabHEc::camR (fabHEc is the fabH gene from E. coli) cassette and the Red recombinase system as described previously (56). The TMT47 strain was verified by flanking PCR, and maintained on 1 mM IPTG for PA3286 induction.
Recombinant PA3286 protein expression and purification.
Recombinant PA3286 was obtained according to the protocol described for PA5174 (56). Briefly, E. coli BL21(DE3) Rosetta 2 (Novagen) cells were transformed with the His tag expression vector pET-PA3286 (Table 1). Colonies were inoculated into 1 liter of LB medium and grown at 37°C with shaking until mid-exponential growth (OD600 of 0.6) before induction with 1 mM IPTG. After 3 h of expression at 37°C, the biomass was harvested by centrifugation (5,000 × g, 10 min, room temperature [RT]) and stored at −80°C. The pellet was subjected to 2 rounds of freeze-thaw cycles, incubated in lysis solution (1× Bugbuster [Novagen], 5 kU/ml recombinant lysozyme [Novagen], 25 U/ml benzonuclease [Novagen]), and then clarified by centrifugation. The supernatant was gently shaken with nickel-nitrilotriacetic acid (Ni-NTA) His bind resin (1 h on ice). The slurry was loaded into an empty column, washed with 50 ml of binding buffer (50 mM Tris, 500 mM NaCl, 10 mM imidazole [pH 7.5]), 20 ml of wash buffer (binding buffer with 50 mM imidazole [total concentration]), and eluted (binding buffer plus 200 mM imidazole [total concentration]). Fractions containing protein of the expected size (43 kDa, 373 amino acids without His tag) were pooled and concentrated (Amicon Ultra, 10-kDa molecular size cutoff; Millipore). The sample buffer was exchanged with storage buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM d/l-dithiothreitol, and 10% glycerol) by three rounds of dilution (10 ml each) and concentration, after which the aliquots were flash frozen at −80°C.
Recombinant PA3286 β-ketoacyl synthase enzyme activity assay.
Conformation-sensitive urea-PAGE was used to separate and analyze acyl-ACP condensation products (46). Malonyl-ACP was produced in situ using a previously described procedure (56). For each saturated straight-chain acyl coenzyme A (acyl-CoA) acceptor substrate tested (C2 to C16 in length; Sigma), 200 μM acyl-CoA along with 0.1 μg of PA3286 (final concentration of 70 nM) was added, and the reaction mixtures were incubated at room temperature for 1 h. The products were separated by 0.5 M urea–16% PAGE and stained with Coomassie blue dye.
RESULTS
Exogenous fatty acids C8 and longer rescue growth of P. aeruginosa ΔfabY.
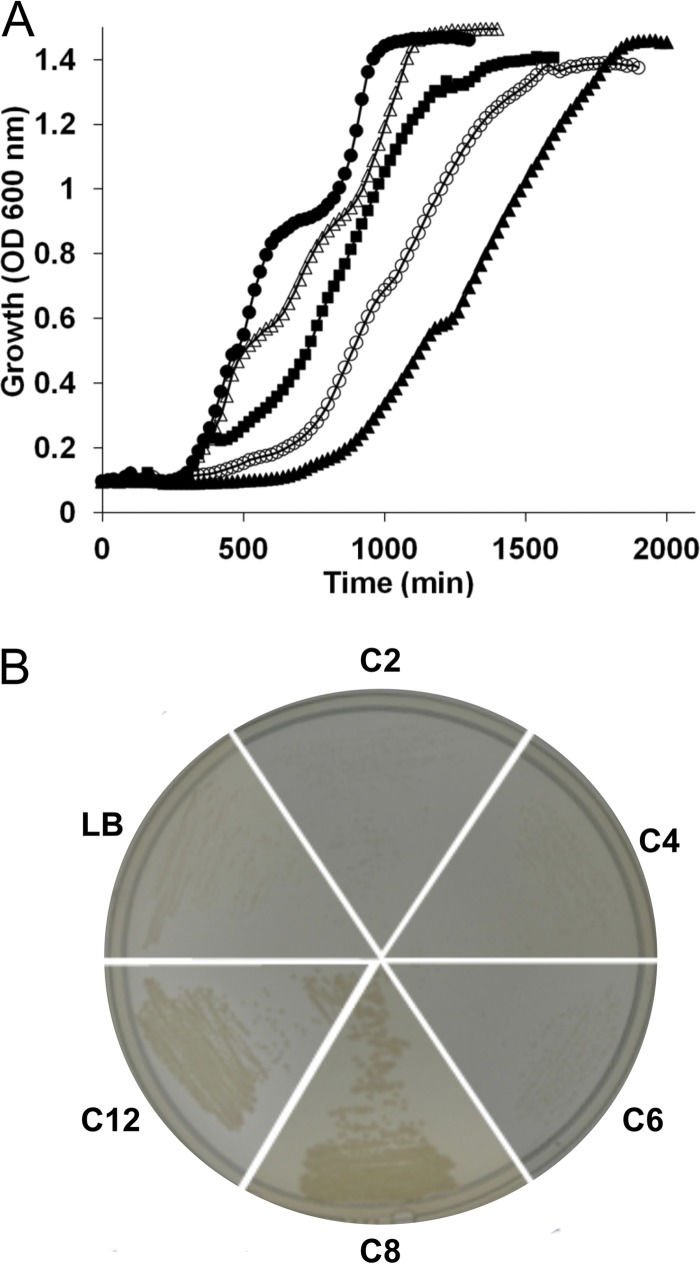
The P. aeruginosa ΔfabY strain has a pronounced growth defect both in LB liquid medium and on agar plates. The results of initial experiments suggested that growth on agar could at least partially be rescued by inclusion of the fatty acid decanoate (C10) in the media. In liquid media, inclusion of as low as 1 μg/ml of C10 in liquid LB culture also restored growth, with rates approaching those of the wild type at 100 μg/ml (Fig. 1A). To determine whether a specific fatty acid chain length or range is required to suppress the growth-defective phenotype, the ΔfabY strain was streaked onto LB agar plates supplemented with 100 μg/ml of straight-chain saturated fatty acids from C2 to C16 in length (Fig. 1B). The average colony size was noticeably larger for C8 and longer fatty acids, whereas C6 down to C2 had minimal effect in comparison to LB agar alone. P. aeruginosa PAO1 can thus utilize exogenous medium- and long-chain fatty acids to compensate for decreased de novo FAS flux in the absence of fabY.
Fig 1.
Fatty acid rescue of the P. aeruginosa ΔfabY mutant. (A) Growth curves in LB medium alone (P. aeruginosa PAO1 [●] and ΔfabY mutant [▲]) or in LB medium supplemented with decanoate (1 μg/ml [○], 10 μg/ml [■], or 100 μg/ml [△]) for the ΔfabY mutant. Growth was measured at 37°C by recording the optical density at 600 nm. (B) The ΔfabY strain was streaked onto LB agar supplemented with the indicated fatty acid (C2 to C12) at 100 μg/ml. The plates were incubated at 37°C for 16 h before imaging.
P. aeruginosa shunts exogenous medium-chain-length fatty acids.
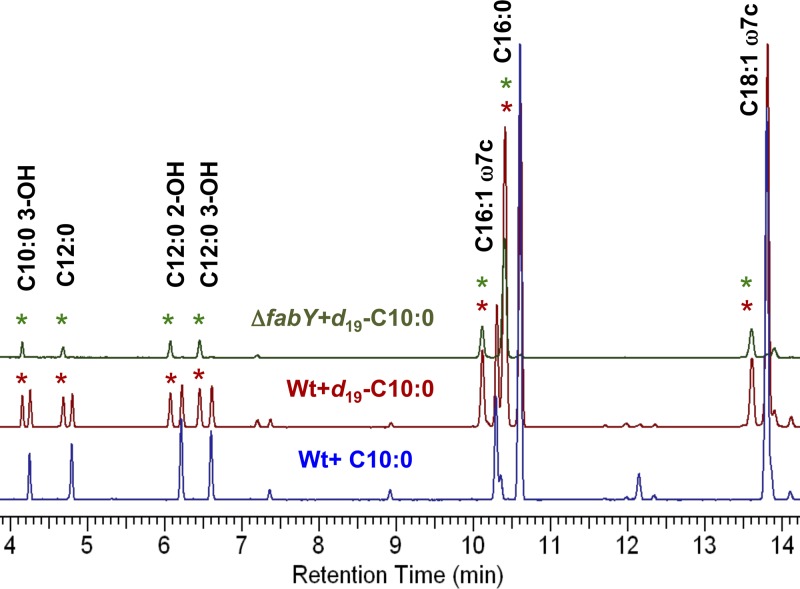
Exogenous fatty acids are taken up and degraded 2 carbons at a time by the fatty acid degradation (fad) β-oxidation cycle (2, 10). The range of substrates that can enter and be efficiently utilized by the β-oxidation cycle is defined by a combination of fatty acid inducer specificity and acyl-CoA synthetase (FadD) substrate preference. In E. coli for instance, only fatty acids longer than C12 induce transcription of fad genes, while fatty acids must be at least C8 to serve as FadD substrates (36, 53). The requirement for acyl chain lengths of C8 or longer for growth rescue in P. aeruginosa ΔfabY could therefore reflect two scenarios; either a longer-chain fatty acid is required for shunting into de novo FAS downstream of FabY or fatty acids must be efficiently utilized by the β-oxidation cycle in order to be degraded to acetyl-CoA. In the latter case, high concentrations of intracellular acetyl-CoA pools would then ultimately be responsible for rescuing growth by improving turnover rates of a putative low-affinity FabH/FabY-type enzyme. To differentiate between these two possibilities, we fed the wild-type and ΔfabY P. aeruginosa strains perdeuterated C10 and analyzed the fatty acid composition as fatty acid methyl ester (FAME) derivatives by gas chromatography with flame ionization detection (GC-FID) and mass spectrometry (Fig. 2 and 3). If the decanoate (C10) substrate is being completely degraded to acetyl-CoA, the label should become dispersed with random reincorporation into fatty acids, while a shunt would retain the label and generate unique deuterium-containing fatty acid peaks. Indeed, the FAME GC trace of the wild type revealed additional peaks specific to samples fed perdeuterated C10 (in comparison with unlabeled C10), eluting slightly ahead of every major constituent fatty acid peak (Fig. 2). Since the amount of deuterium incorporated into FAMEs is inversely correlated with retention time (42), the shortened retention time along with peak symmetry and relative abundance (∼40% of unlabeled FAME peak area) suggested a shunt mechanism. We then fed perdeuterated C10 to the P. aeruginosa ΔfabY strain, which has a rate of de novo FAS flux of less than 5% of the wild type as determined by phospholipid macromolecular labeling with radiolabeled acetate (56). In this case, only the labeled peak was observed, as would be expected in the ΔfabY genetic background, since the incorporation rate of the shunt pathway greatly exceeds de novo FAS initiation.
Fig 2.
Fatty acid composition analysis of P. aeruginosa. (A) Fatty acid methyl esters (FAMEs) prepared from strains grown overnight on LB agar supplemented with either 100 μg/ml of decanoate (C10:0) or perdeuterated decanoate (d19-C10:0) were analyzed by gas chromatography with flame ionization detection. FAME peaks that contain deuterated fatty acids analyzed by mass spectrometry in Fig. 3 are indicated with an asterisk. Wt, wild type.
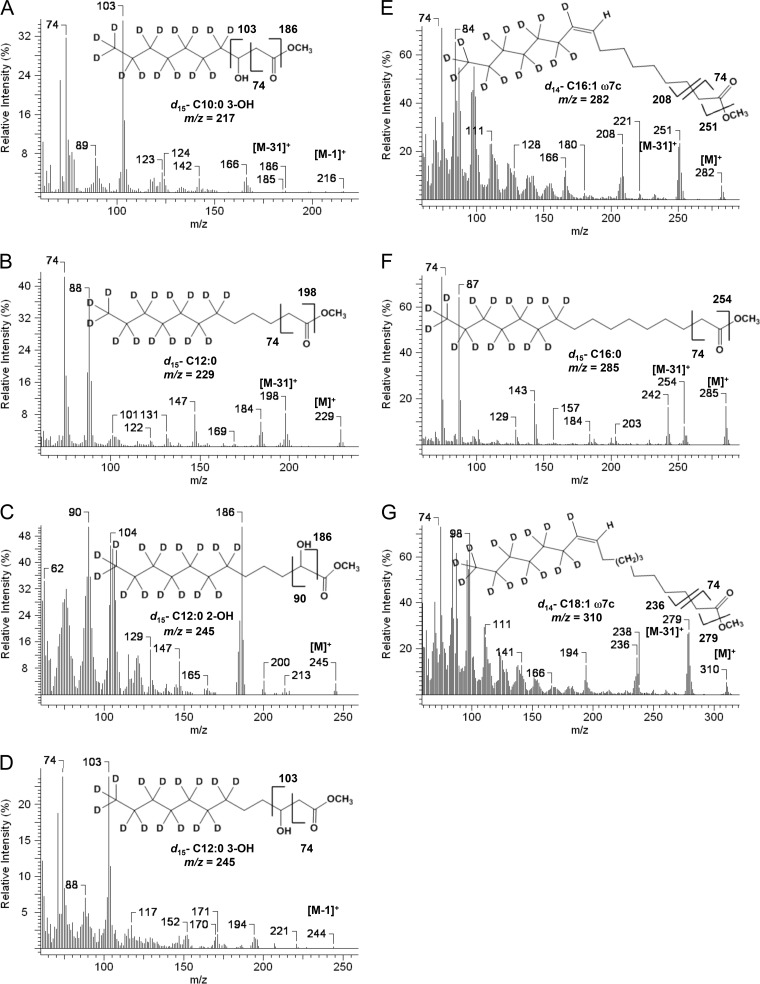
Fig 3.
Mass spectra of deuterated FAMEs. Unique FAME peaks appearing after supplementation with perdeuterated decanoate (indicated by asterisks in Fig. 2) were analyzed by mass spectrometry. The parent molecular ions ([M]+) and select fragment ions are indicated, including for the McLafferty ion (m/z = 74). The corresponding unlabeled FAME mass spectra are included for comparison in the supplemental information (see Fig. S1 in the supplemental material).
The structure of each deuterium-labeled FAME peak was assigned using mass spectrometry (Fig. 3). In comparison to the corresponding unlabeled FAME peak (see Fig. S1 in the supplemental material), the deuterated FAME molecular ions were 15 Da heavier and had odd mass numbers for all of the saturated and hydroxylated fatty acids. The odd masses suggested that the terminal ω-carbon was deuterated (-CD3). The McLafferty rearrangement ion, which is a characteristic fragmentation ion for a methyl ester with an α-methylene group, remained at 74 Da. The C-2 carbon was thus unsubstituted. The C10:0 3-OH (3-hydroxy decanoate) spectra contained the diagnostic carboxyl terminus ion resulting from fragmentation between C-3 and C-4 (Fig. 3A). The resulting mass likewise indicated that C-3 was unsubstituted (103 Da). By inference, the terminal 7 carbons [CD3(CD2)6-; 15 deuteriums total] were assigned as being fully deuterated. The unsaturated C16:1ω7c and C18:1ω7c FAMEs were only 14 Da heavier (Fig. 3E and G), as would result from extraction of a single deuterium at C-3 during FabA-catalyzed isomerization of trans-2-decenoyl-ACP to cis-3-decenoyl-ACP during anaerobic unsaturated fatty acid (UFA) biosynthesis (33). Similar analysis of the other spectra supports the conclusion that the 7 terminal carbon atoms of each fatty acid were fully substituted with deuterium. Since the cultures had been fed perdeuterated C10 fatty acid, deuterium labels were uniformly lost at C-2 and C-3. The data are consistent with a fatty acid metabolism model whereby P. aeruginosa degrades C10 via one round of the β-oxidation cycle to make C8-CoA. This intermediate is then shunted into FAS, at which point the C10 fatty acid is rebuilt by using unsubstituted malonyl-ACP as the substrate. The terminally labeled β-keto-decanoyl-ACP ester can then be used to supply all the different types of fatty acids needed by the cell, including saturated (SFA), unsaturated (UFA), and 3-hydroxyl fatty acids.
PA3286 is required for de novo FAS initiation in the absence of fabY and for fatty acid shunting.
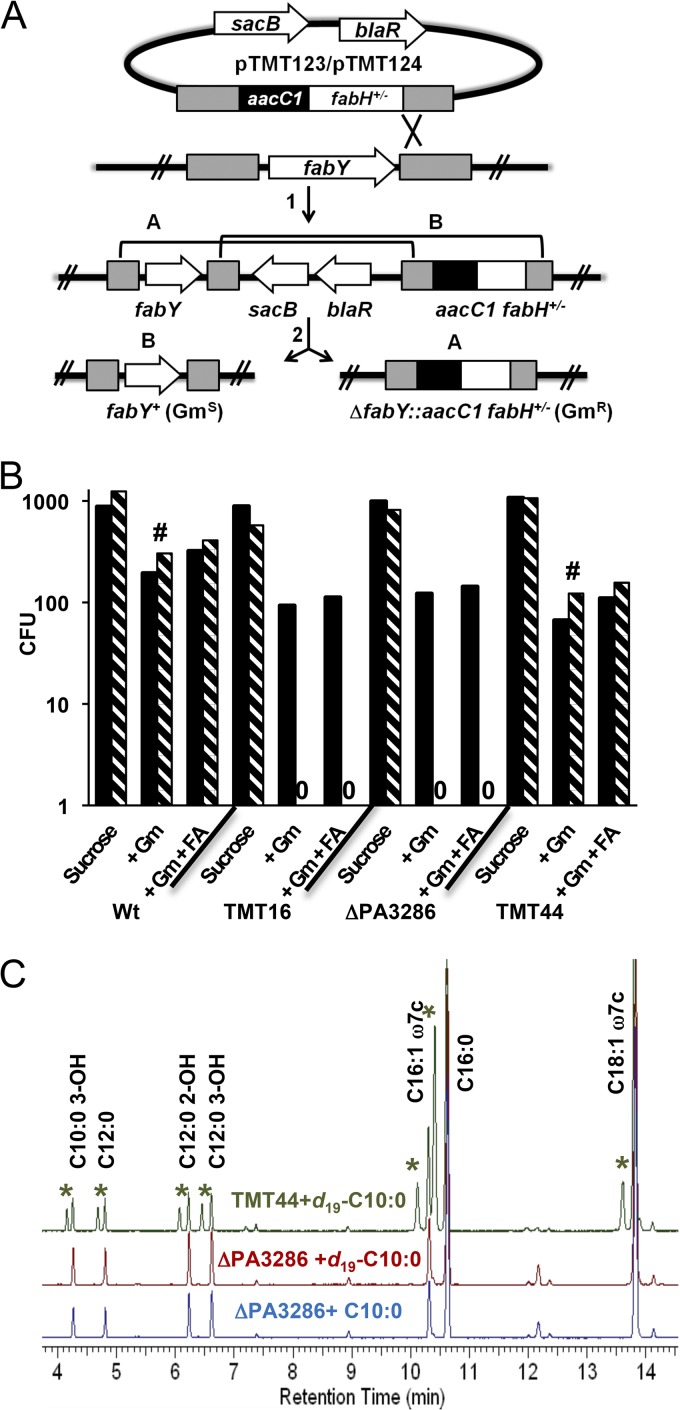
On the basis of the pattern of labeled fatty acid incorporation (Fig. 3), we hypothesized a fatty acid shunt consisting of a single enzyme that condenses C8-CoA with malonyl-ACP to make β-keto-decanoyl-ACP. This hypothetical catalytic activity is reminiscent of FabY/FabH, which condense acetyl-CoA with malonyl-ACP; the only difference being the acyl chain length of the CoA thioester acceptor. Since fatty acids were shunted in the absence of fabY and recombinant FabY displays an absolute substrate preference for short-chain acyl-CoA acceptors (56), we focused our search among the previously identified four KASIII domain-containing proteins sharing similarity to FabH of E. coli (56). If one of the KASIII domain-containing proteins were responsible for fatty acid shunting, then C10 supplementation should not rescue growth in a ΔfabY ΔKASIII genetic background. Allelic exchange plasmids were designed to replace FabY with either a gentamicin resistance (Gmr) marker alone or in tandem with the functional ortholog fabHEc from E. coli to serve as a positive control (Fig. 4A). The fabY targeting plasmids were integrated into the chromosomes in a panel of strains with single KASIII domain-containing protein deletions (PA3333, PA0998, PA0999, and PA3286) as well as in a strain harboring all four KASIII deletions (TMT16). Merodiploid P. aeruginosa intermediates were verified by flanking PCR analysis and passively outgrown before plating on LB-sucrose agar to counterselect unresolved clones. Aliquots were plated in parallel on plates with Gm to select for colonies in which the fabY allele had been replaced, as well as on plates with Gm and C10 fatty acid supplement. The fabY::Gmr deletion could be established with efficiencies comparable to the fabHEc exchange allele in the wild-type background (Fig. 4B), although as expected (56), the fabY deletion (Gm selection) strain grew slowly in comparison to the fabHEc exchange control without C10 supplementation (Gm plus fatty acid [FA] selection). Conversely, the fabY deletion could not be established in the TMT16 genetic background even with C10 supplementation, although the fabHEc allele could readily be established. The fabY deletion is thus synthetic lethal when introduced into the KASIII domain knockout strain TMT16. We then repeated this experiment in each of the single deletion strains to decipher whether a lone KASIII domain gene was the synthetic lethal partner of fabY or if the relationship was more complex. The merodiploid ΔPA3286 deletion strain resolved with efficiencies that closely mirrored that of TMT16 (Fig. 4B), whereas the other three single deletion strains (ΔPA3333, ΔPA0998, and ΔPA0999) behaved akin to the wild-type P. aeruginosa genetic background (see Fig. S2 in the supplemental material). Reintroduction of a functional copy of the PA3286 gene into the chromosome of the ΔPA3286 mutant at the phage CTX attB site (TMT44) allowed the fabY deletion to now be established, indicating no confounding polarity issues and confirming a true synthetic lethal pairing between fabY and PA3286.
Fig 4.
Synthetic lethal analysis for fabY with KASIII domain fabH orthologs and fatty acid rescue. (A) Vectors conferring aacC1-mediated gentamicin (Gm) resistance that targeted the P. aeruginosa fabY gene were designed to either exchange fabH of E. coli (pTMT123 fabH+) or to delete fabY (pTMT124). Merodiploid intermediates (after step 1) were confirmed by PCR analysis and passively resolved by outgrowth in LB medium (step 2). Aliquots were plated on LB-sucrose agar to counterselect unresolved clones and to enumerate the sum of A- and B-type recombination events. In parallel, aliquots were plated on LB-sucrose agar plus Gm and on LB-sucrose agar plus Gm with 100 μg/ml of the fatty acid supplement decanoate (FA) to select only the recombinants arising through A-type recombination. (B) The pTMT123/124 vectors were individually introduced into the wild-type (Wt) P. aeruginosa, TMT16 (ΔPA0998 ΔPA0999 ΔPA3333 ΔPA3286), ΔPA3286, and TMT44 (ΔPA3286 attB::PA3286+). CFU resulting from resolving either pTMT123 (black bars) or pTMT124 (hatched bars) were determined by counting colonies that appeared after 24 h or 48 h (#) of incubation at 37°C. The selection media and strain background are indicated below the x axis. Data are representative of 3 separate experiments. The results for the ΔPA0998, ΔPA0999, and ΔPA3333 mutant strains are shown in Fig. S2 in the supplemental material. (C) Gas chromatograms with FID detection of FAME extracts obtained from P. aeruginosa strains grown on LB agar with either 100 μg/ml of decanoate (C10:0) or perdeuterated decanoate (d19-C10:0). Asterisks indicate FAME peaks unique to d19-C10:0-fed samples with structures assigned as in Fig. 3.
The synthetic lethal relationship between PA3286 and fabY indicates shared acetyl-CoA:malonyl-ACP condensing activity. Unlike the P. aeruginosa ΔfabY strain, the ΔPA3286 deletion strain is aphenotypic with respect to growth (56). This suggests that the presumptive FAS initiating activity of PA3286 that allows ΔfabY to grow, albeit slowly, is secondary to another main cellular role utilizing similar substrates. To determine whether PA3286 was indeed the hypothesized C8-CoA:malonyl-ACP condensing enzyme responsible for fatty acid shunting, we fed perdeuterated C10 to the ΔPA3286 mutant. Whereas perdeuterated C10 fatty acids noticeably had a negative impact on the growth of the wild type and in particular the ΔfabY mutant on LB agar, no inhibitory isotope effect was observed for the ΔPA3286 strain (data not shown). Since the primary isotope effect arising through breaking of C-D bonds in shunted fatty acids to make UFA by the anaerobic FAS pathway was likely responsible, this hinted at a lack of shunting in the ΔPA3286 mutant. We next analyzed the FAME profile by GC-MS as had been done previously (Fig. 4C). The GC trace for the ΔPA3286 mutant fed perdeuterated C10 showed no en bloc incorporation of deuterated fatty acids and was nearly superimposable with traces derived from samples that had been grown with unlabeled C10. In the complemented PA3286 strain TMT44, deuterated FAME peaks having retention times and mass spectra identical to those of deuterated FAMEs assigned for the wild type reappeared (Fig. 2). The data support the identification of the PA3286 enzyme as the enzyme responsible for shunting fatty acids in vivo, as well as implicating PA3286 secondary enzymatic activity as the source of FAS initiation in the ΔfabY background.
PA3286 specifically shunts C8-CoA in vivo.
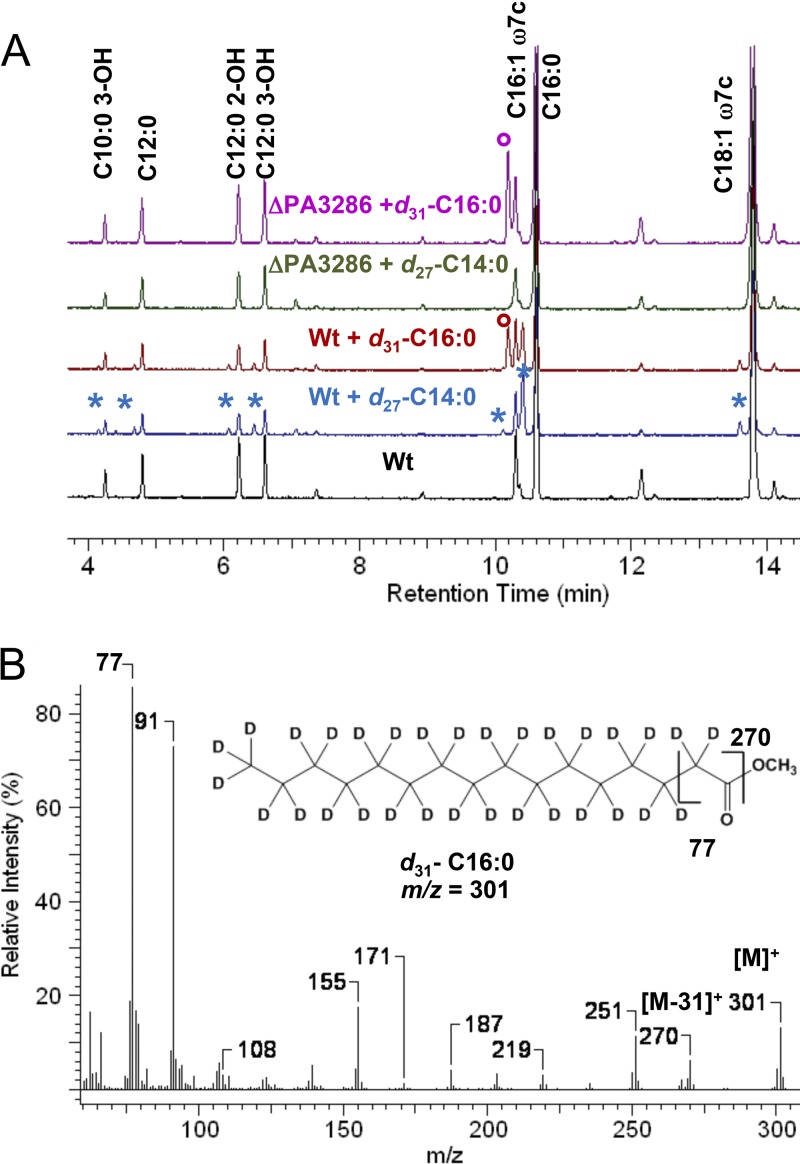
While decanoate is likely intercepted as a C8-CoA β-oxidation cycle intermediate by PA3286 (Fig. 3 and 4C), we wanted to determine whether this was specific to C10 or for all fatty acids in general. Perdeuterated long-chain fatty acids (C14 and C16) were therefore fed to the wild-type and ΔPA3286 P. aeruginosa strains and analyzed by GC-MS (Fig. 5A). FAME peaks with retention times and mass spectra consistent with the terminal 7 carbon atoms being fully substituted with deuterium atoms were once again observed only in the wild-type strain upon either C14 or C16 supplementation. An additional FAME peak was present in the C16-fed samples for both the wild-type and ΔPA3286 strains (marked by ○). Analysis of the mass spectrum identified the fatty acid as perdeuterated d31-C16 (Fig. 5B), based on the parent molecular ion mass (m/z = 301) and a fully deuterated McLafferty ion [CD2 = C(OD)+-OCH3; m/z = 77]. Long-chain fatty acid-CoA esters can be recycled en bloc and incorporated into de novo phospholipids by the glycerol-phosphate and acylglycerol-phosphate acyltransferases PlsB/PlsC in P. aeruginosa (60). Under these growth conditions, the PA3286 shunt clearly occurs in tandem with the direct en bloc PlsB/PlsC-catalyzed incorporation into phospholipids. Collectively, the data suggest that PA3286 has high in vivo substrate specificity for C8-CoA intermediates and that the shunt is relevant during the catabolism of both long- and medium-chain exogenous fatty acids.
Fig 5.
Fatty acid composition analysis of P. aeruginosa strains fed long-chain perdeuterated fatty acids. (A) Gas chromatograms with FID detection of FAME extracts obtained from stationary-phase cultures of P. aeruginosa grown in liquid LB-BSA medium supplemented with either 100 μg/ml of perdeuterated tetradecanoate (d27-C14:0) or hexadecanoate (d31-C16:0). Asterisks indicate deuterated FAMEs of identical structure to those already assigned in Fig. 3 based on time of elution and mass spectra analysis. FAME peaks unique to d31-C16:0-fed cultures are indicated (○). (B) Mass spectra analysis of the d31-C16:0-specific FAME with the parent molecular ion ([M]+) and diagnostic fragment ions labeled, including for the uniformly deuterium-labeled McLafferty ion (m/z = 77).
PA3286 cross complements fabH and confers a fatty acid shunt in E. coli.
Fatty acid metabolism in especially complex in pseudomonads, with an unusually large genetic allocation to both the catabolism and anabolism of fatty acids (32, 50, 51). To further confirm the two activities of PA3286 observed in P. aeruginosa PAO1, to probe the substrate specificity and to determine whether PA3286 requires partner proteins for intercepting β-oxidation cycle intermediates, we introduced the PA3286 gene into the model E. coli strain BW25113. In the presence of a high-copy-number plasmid harboring PA3286, we were able to delete the otherwise essential fabH gene that encodes the FAS initiating enzyme β-acetoacetyl synthase III in enterobacteria. Unlike the fabY gene that achieved full restoration of the growth rate (56), the PA3286 gene only partially complemented fabH and indicates weak intrinsic β-acetoacetyl-ACP synthase catalytic activity (Fig. 6A). This is consistent with our inability to identify PA3286 among a P. aeruginosa genomic cosmid library, which is introduced at low copy number without any strong endogenous E. coli promoters (56).
Fig 6.
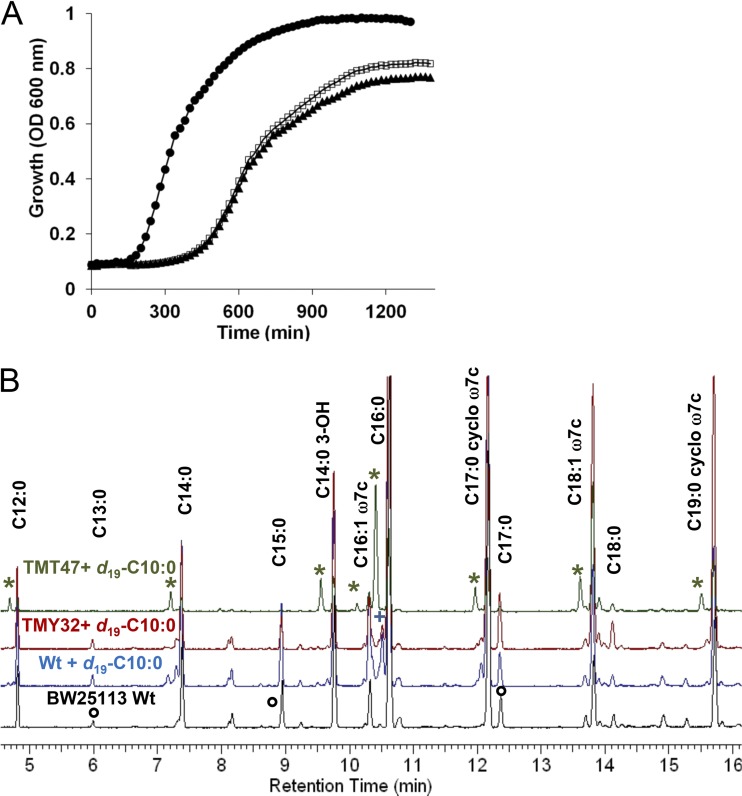
Complementation of E. coli fabH by the P. aeruginosa PA3286 gene. (A) The E. coli strain TMT47 [fabH::camR(pET-PA3286)] was grown at 37°C in LB medium alone (▲) or with 1 mM IPTG (□) and compared to the parent Wt strain (●). Growth was monitored by measuring the optical density at 600 nm. (B) The gas chromatogram-FID trace for FAME samples prepared from E. coli strains grown on LB agar with perdeuterated decanoate (d19-C10:0; 100 μg/ml) and palmitate (C16:0; 10 μg/ml) for induction of fatty acid degradation genes. The parent wild-type E. coli BW25113 and TMY32 [fabH::camR(pET-PA5174)] strains were included as controls. Asterisks indicate deuterated FAMEs of identical structures to those already assigned in Fig. 3 based on time of elution and mass spectra analysis. Mass spectra and assignment of deuterated FAME peaks unique to E. coli expressing PA3286 (C14:0, C14:0 3-OH, C17:0 cyclo ω7c, and C19:0 cyclo ω7c) along with the terminally labeled d7-C16:0 (+) peak present only in the absence of PA3286 are shown in Fig. S3 in the supplemental material. Odd-numbered acyl chain FAMEs with reduced abundance in strain TMT47 are indicated (○).
We next fed the recombinant E. coli strain TMT47 [fabH::catR(pET-PA3286)] a mixture of perdeuterated C10 and unlabeled C16 fatty acid (10:1 [wt/wt]) in order to induce the fad genes involved in β-oxidation (Fig. 6B). The wild-type E. coli strain did not incorporate the deuterium label for any of the peaks, indicating that a medium-chain fatty acid shunt in order to extend exogenous fatty acids is not native to E. coli, in agreement with previous reports (47). Likewise, replacement of fabH with fabY of P. aeruginosa did not result in fatty acid labeling. Only when fabH was replaced with PA3286 did deuterium-substituted FAME peaks become evident. Structure assignment based on the mass spectra of all fatty acids common to P. aeruginosa and E. coli (labeled with asterisks; spectra similar to those in Fig. 3), as well as those specific to E. coli (see Fig. S3 in the supplemental material), were entirely consistent with complete labeling of the terminal 7 carbons. Hence, PA3286 is intrinsically specific for C8-CoA and can intercept β-oxidation intermediates without the need for a partner protein. The second aspect specific to the FAME profile of TMT47 is the near absence of odd-numbered carbon fatty acids (indicated by ○ in Fig. 6B) and of terminally labeled d7-C16:0 (indicated by + in Fig. 6B; mass spectrum is included in Fig. S3E in the supplemental material). In E. coli, odd-numbered fatty acids arise though utilization of propionyl primers as alternative substrates to acetyl-CoA for FAS by FabH (23, 27). The low-abundance d7-C16:0 peak that is present only in perdeuterated C10-fed E. coli wild-type or TMY32 [fabH::catR(pET-FabY)] may arise through utilization of d7-butyryl-CoA produced by the β-oxidation cycle as a FAS primer by either FabH or FabY, respectively. In line with the previously observed in vivo substrate specificity in P. aeruginosa, PA3286 did not efficiently utilize either short-chain acyl-CoA primer in E. coli. These short-chain acyl-CoA substrates are present at much lower intracellular concentrations in comparison to the glycolytic product/tricarboxylic acid cycle (TCA) intermediate acetyl-CoA. Thus, while PA3286 can condense enough of the abundant acetyl-CoA substrate to maintain FAS initiation in the absence of fabH, other low-affinity short-chain acyl-CoA substrates present in trace amounts are not appreciably utilized.
Characterization of recombinant PA3286 β-ketoacyl ACP synthase activity.
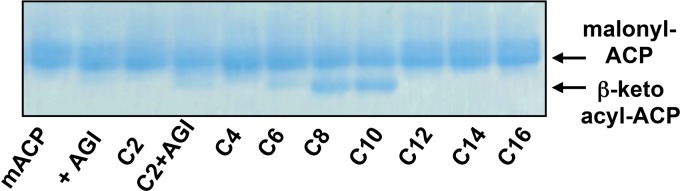
The PA3286 open reading frame is predicted to encode a 350-amino-acid protein (http://www.ncbi.nlm.nih.gov/). However, repeated attempts to express recombinant PA3286 failed to produce soluble protein. We thus chose an upstream start codon and cloned the gene (encoding a 373-amino-acid protein) into a pET-24b(+) expression vector so as to append a C-terminal His tag for purification. The longer protein was readily soluble and amenable to characterization. The substrate specificity of the PA3286 enzyme was explored using various straight-chain saturated acyl-CoA (C2 to C16) as acceptors along with malonyl-ACP, as has previously been described for FabY (56). Reaction products were separated by conformation-sensitive urea-PAGE (46), and the gels were stained with Coomassie blue. PA3286 only weakly accepted short-chain acyl-CoA substrates, including acetyl-CoA, butyryl-CoA, and hexanoyl-CoA (Fig. 7). The substrate specificity suggests why shorter exogenous fatty acids did not rescue growth in the P. aeruginosa ΔfabY mutant (Fig. 1B), even though they can be utilized as the sole carbon source (31). The two best substrates were clearly C8-CoA and C10-CoA. When the experiment was repeated with C8-CoA and C10-CoA acceptors using radiolabeled [2-14C]malonyl-ACP, radioactivity was incorporated into the faster-migrating bands (data not shown). This confirmed PA3286 is a malonyl-ACP condensing KAS as opposed to an acyl-CoA:ACP transacylase. CoA thioester acceptors with longer acyl chains (C12, C14, and C16) were all excluded as substrates. The in vitro substrate specificity is consistent with the in vivo characteristics of PA3286, and together, these results support our assignment of PA3286 as a β-keto-decanoyl-ACP synthase that condenses malonyl-ACP with C8-CoA originating from the fatty acid β-oxidation degradation cycle.
Fig 7.
Acyl-CoA substrate specificity of recombinant PA3286. Reaction products using malonyl-ACP (mACP) and saturated straight-chain acyl-CoAs (C2 to C16) as potential PA3286 substrates were separated with conformation-sensitive urea-PAGE. The gels were stained with Coomassie blue dye. The position of the malonyl-ACP-only control (malonyl-ACP) is shown to the right of the gel. + AGI, plus E. coli FabAGI coupling enzymes.
DISCUSSION
While it is clear that FAS in P. aeruginosa is a central metabolic pathway upon which the expression of multiple virulence and regulatory factors depend, exogenous C10 fatty acid noticeably muted the growth-defective phenotype in the ΔfabY strain (Fig. 1). It might be expected, therefore, that the efficacy of a small-molecule inhibitor of FabY would likewise be subject to the fatty acid content of the environment where it is administered. This is a particularly pressing concern with any intended anti-pseudomonal agent, as an important segment of the intended patient population has cystic fibrosis. P. aeruginosa can be found in the respiratory tracts of 80% of all cystic fibrosis patients by the time they are 18 years old (20), an environment where there is an already abundant lipid nutrient source in the form of pulmonary surfactant. Pulmonary surfactant, being composed of 90% lipids (mostly dipalmitoyl phophatidylcholine) along with 10% surfactant proteins (1, 19), is an important in vivo nutrient source for P. aeruginosa (31, 48). Transcription of fad β-oxidation cycle genes is induced in vivo, and mutants deficient in fatty acid degradation exhibited decreased fitness in a mouse lung infection model (32). Our goal in the present work was to address how the ΔfabY strain is rescued by exogenous fatty acids and whether the mechanism(s) might potentially compromise an inhibitor targeting FabY in P. aeruginosa.
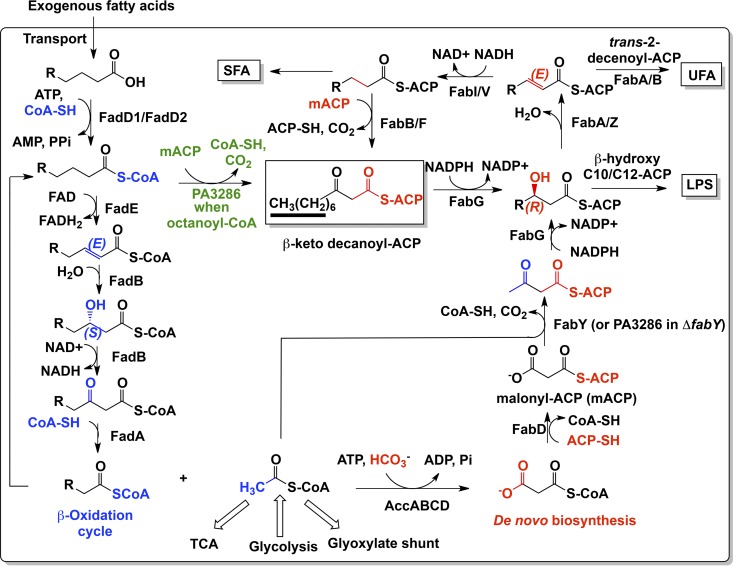
Based on the perdeuterated feeding experiments with P. aeruginosa and the defined isogenic mutants (Fig. 2 to 5), the synthetic lethal relationship between PA3286 and fabY (Fig. 4B), the PA3286 cross complementation of fabH activity and fatty acid shunting in E. coli (Fig. 6), and in vitro characterization of recombinant PA3286 (Fig. 7), we propose that fatty acids rescue the P. aeruginosa ΔfabY mutant through incorporation via the PA3286-mediated β-oxidation cycle to the FAS shunt (Fig. 8). In this pathway, C8-CoA intermediates originating from the oxidation of exogenous fatty acids C8 and longer are intercepted by PA3286 and condensed with malonyl-ACP to make the FAS intermediate β-keto-decanoyl-ACP. PA3286 belongs to the β-ketoacyl-acyl carrier protein synthase III family (KASIII), with E. coli FabH being the prototypical member (52). As in E. coli FabH and most other characterized orthologs (18), PA3286 contains the class-defining Cys-His-Asn catalytic triad common to KASIII enzymes (56), and so the catalytic function of condensing malonyl-ACP with an acyl-CoA is not altogether unexpected. However, KASIII domain-containing enzymes generally exhibit high preference for short-chain acyl-CoA substrates (18). The use of longer-chain acyl-CoA primers by KASIII domain synthases does have precedent, as the Mycobacterium tuberculosis FabH prefers long-chain acyl-CoA substrates in vitro and in vivo during the synthesis of mycolic acids (5, 8). With an overall sequence identity between E. coli FabH and PA3286 of 27%, there is significant difference between the two proteins that may contribute to its substrate specificity. Structural information of PA3286 is needed to shed more light on how the KASIII fold has been co-opted by PA3286 into accommodating longer acyl chains and whether there are common themes with FabH from M. tuberculosis.
Fig 8.
The proposed PA3286-mediated fatty acid β-oxidation to synthesis shunt of P. aeruginosa PAO1. After facilitated diffusion across the outer membrane through FadL (not shown), fatty acids are trapped in the cytoplasm by FadD1/FadD2-catalyzed vectorial esterification with CoA (32). Further metabolism depends on acyl-CoA ester chain length. Long-chain fatty acid CoA esters (C16/C18-CoA) can either be recycled en bloc and incorporated into de novo phospholipids by the glycerol-phosphate and acylglycerol-phosphate acyltransferases PlsB/PlsC (not shown) (60), or as with medium-chain acyl CoA esters (C14 to C10-CoA), be further degraded by the β-oxidation pathway (in blue). Once the preferred C8-CoA substrate chain length is reached, the CoA thioester is intercepted by PA3286 and condensed with malonyl-ACP to form the key uncommitted fatty acid intermediate β-keto-decanoyl-ACP (green). The β-keto-decanoyl-ACP metabolite can be utilized by the anaerobic unsaturated fatty acid (UFA) pathway, the saturated fatty acid (SFA) pathway, and in lipopolysaccharide (LPS) biosynthesis (C10:0/C12:0 3-OH). The terminal 7 carbons in β-keto-decanoyl-ACP that remain labeled with deuterium when fed perdeuterated fatty acids are underlined. PA3286 does not significantly contribute to de novo FAS biosynthesis (in red) (56) but becomes essential in the P. aeruginosa ΔfabY background (Fig. 4B) due to a cellular requirement for basal FabH-type acetyl-CoA:malonyl-ACP condensation activity. For clarity, not all putative FAS and β-oxidation isozymes are shown. TCA, tricarboxylic acid cycle.
The β-keto-decanoyl ACP thioester shunt product generated by PA3286 is a versatile precursor that is uncommitted to the biosynthesis of any specific essential fatty acid (Fig. 8). Pathways for the biosynthesis of LPS (3-OH decanoyl-ACP/3-OH dodecanoyl-ACP), UFA (via trans-2-decenoyl-ACP), and SFA can all utilize nutrient-acquired fatty acids for extension without having to degrade them down to C2-CoA before reincorporation via de novo FAS. This is critical, as FAS is bioenergetically the most costly synthetic process for any membrane component (59); for instance, building an n-acyl-ACP thioester (where n ≥ C10) from C2-CoA consumes [(n/2) − 1] ATP molecules along with [(n/2) − 1] × 2 reducing equivalents. In comparison, degradation of n-acyl-CoA β-oxidation intermediates down to C2-CoA before reassembly into n-acyl-ACP thioesters by FAS consumes a net [(n/2) − 1] ATP molecules. However, the PA3286 C8 shunt uses only [(n/2) − 4] ATP molecules to accomplish the same feat, yielding a further net savings of 3 ATP molecules per metabolized n-acyl fatty acid molecule (Fig. 8). The use of a C8 fatty acid chain length for shunting by P. aeruginosa is a metabolically savvy choice, maximizing energy conservation while retaining fatty acid anabolic versatility. P. aeruginosa now joins certain Vibrio species which are also capable of extending exogenous fatty acids en bloc using the FAS pathway (7, 21, 28). In the case of Vibrio, exogenous medium-chain fatty acids are directly ligated to free ACP to form acyl-ACP thioesters. While mechanistically distinct and independent of the β-oxidation pathway, the end result is equivalent to the PA3286 shunt in that the acyl-ACP produced can be utilized for incorporation into LPS, SFA, and UFA (29).
It has been reported that the entire FAS can be bypassed by exogenous fatty acid uptake from serum in certain Gram-positive pathogens, including Streptococcus (4). The essentiality of the FAS pathway has not been questioned in Gram-negative bacteria, in part due to the dependence of LPS biogenesis on FAS supplied 3-hydroxyacyl-ACP precursors (41). The LPS of P. aeruginosa contains both 3-hydroxydecanoyl and 3-hydroxydodecanoyl acyl chains (35). Although the PA3286 shunt can supply 3-hydroxydecanoyl-ACP (Fig. 8), it should be noted that FabG is still needed to introduce the β-hydroxyl group. Further, even if a C16/C18 fatty acid source were available for direct incorporation into UFA/SFA membrane phospholipids by the combined activities of the acylglycerol-phosphate acyltransferases PlsB/PlsC and the aerobic desaturases DesB/DesC/DesA (60), at least one more turn from the FAS cycle enzymes would still be necessary to form 3-hydroxydodecanoyl-ACP. Hence, it seems unlikely that PA3286 would impact the efficacy of FAS inhibitors beyond FabY. In the absence of having access to a FabY-specific small-molecule inhibitor, whether FabY can be entirely bypassed by uptake of exogenous fatty acids still must remain an open question. However, if one assumes that treatment with a FabY inhibitor will phenocopy the ΔfabY deletion strain, then it certainly seems likely that a FabY inhibitor could be compromised by exogenous fatty acids. More than 95% of the total extracted fatty acids were terminally labeled with deuterium in the ΔfabY sample (Fig. 2). This suggests that the shunt can satisfy the majority of the cellular needs for acyl-ACP primers, in effect making β-acetoacetyl synthase (FabY/FabH) activity superfluous. Indeed, there may even be intrinsic cellular regulatory mechanisms to repress de novo FAS initiation by FabY, as synthesis and degradation in most bacteria are tightly regulated in order to maintain homeostasis and to avoid a futile metabolic cycle (17, 58). We are currently studying these putative regulatory mechanisms in order to understand how the PA3286 shunt is integrated into the global fatty acid metabolism network of P. aeruginosa.
Supplementary Material
ACKNOWLEDGMENTS
We thank Herbert P. Schweizer (Colorado State University) for the pMini-CTX1 plasmid and Charles Dean (Novartis) for the pRC9 plasmid. We also thank Karen Dohrman and Gary Jackoway of Microbial ID, Inc. (Newark, DE) for expert technical assistance in FAME analysis.
Footnotes
Published ahead of print 29 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Bernhard W, et al. 1997. Lung surfactant in a cystic fibrosis animal model: increased alveolar phospholipid pool size without altered composition and surface tension function in cftrm1HGU/m1HGU mice. Thorax 52:723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Black PN, DiRusso CC. 1994. Molecular and biochemical analyses of fatty acid transport, metabolism, and gene regulation in Escherichia coli. Biochim. Biophys. Acta 1210:123–145 [DOI] [PubMed] [Google Scholar]
- 3. Breidenstein EB, Fuente-Nunez C, Hancock RE. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19:419–426 [DOI] [PubMed] [Google Scholar]
- 4. Brinster S, et al. 2009. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 458:83–86 [DOI] [PubMed] [Google Scholar]
- 5. Brown AK, et al. 2005. Probing the mechanism of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III mtFabH: factors influencing catalysis and substrate specificity. J. Biol. Chem. 280:32539–32547 [DOI] [PubMed] [Google Scholar]
- 6. Brugha RE, Davies JC. 2011. Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and new treatments. Br. J. Hosp. Med. (Lond.) 72:614–619 [DOI] [PubMed] [Google Scholar]
- 7. Byers DM. 1989. Elongation of exogenous fatty acids by the bioluminescent bacterium Vibrio harveyi. J. Bacteriol. 171:59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi KH, Kremer L, Besra GS, Rock CO. 2000. Identification and substrate specificity of beta-ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis. J. Biol. Chem. 275:28201–28207 [DOI] [PubMed] [Google Scholar]
- 9. Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30 doi:10.1186/1471-2180-5-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clark DP, Cronan JE. 1996. Two-carbon compounds and fatty acids as carbon sources, p 343–358 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 1 American Society for Microbiology, Washington, DC [Google Scholar]
- 11. Cronan JE, Lin S. 2011. Synthesis of the alpha, omega-dicarboxylic acid precursor of biotin by the canonical fatty acid biosynthetic pathway. Curr. Opin. Chem. Biol. 15:407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cronan JE, Rock CO. 1996. Biosynthesis of membrane lipids, p 612–636 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 1 American Society for Microbiology, Washington, DC [Google Scholar]
- 13. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davies DG, Marques CN. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191:1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Driscoll JA, Brody SL, Kollef MH. 2007. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67:351–368 [DOI] [PubMed] [Google Scholar]
- 16. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujita Y, Matsuoka H, Hirooka K. 2007. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 66:829–839 [DOI] [PubMed] [Google Scholar]
- 18. Gajiwala KS, et al. 2009. Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme. FEBS Lett. 583:2939–2946 [DOI] [PubMed] [Google Scholar]
- 19. Galabert C, Jacquot J, Zahm JM, Puchelle E. 1987. Relationships between the lipid content and the rheological properties of airway secretions in cystic fibrosis. Clin. Chim. Acta 164:139–149 [DOI] [PubMed] [Google Scholar]
- 20. Geller DE. 2009. Aerosol antibiotics in cystic fibrosis. Respir. Care 54:658–670 [DOI] [PubMed] [Google Scholar]
- 21. Giles DK, Hankins JV, Guan Z, Trent MS. 2011. Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol. Microbiol. 79:716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hannauer M, et al. 2012. Biosynthesis of the pyoverdine siderophore of Pseudomonas aeruginosa involves precursors with a myristic or a myristoleic acid chain. FEBS Lett. 586:96–101 [DOI] [PubMed] [Google Scholar]
- 23. Heath RJ, Rock CO. 1996. Inhibition of beta-ketoacyl-acyl carrier protein synthase III (FabH) by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 271:10996–11000 [DOI] [PubMed] [Google Scholar]
- 24. Hidron AI, et al. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 29:996–1011 [DOI] [PubMed] [Google Scholar]
- 25. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 26. Hoang TT, Kutchma AJ, Becher A, Schweizer HP. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–72 [DOI] [PubMed] [Google Scholar]
- 27. Ingram LO, Chevalier LS, Gabba EJ, Ley KD, Winters K. 1977. Propionate-induced synthesis of odd-chain-length fatty acids by Escherichia coli. J. Bacteriol. 131:1023–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang Y, Chan CH, Cronan JE. 2006. The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry 45:10008–10019 [DOI] [PubMed] [Google Scholar]
- 29. Jiang Y, Morgan-Kiss RM, Campbell JW, Chan CH, Cronan JE. 2010. Expression of Vibrio harveyi acyl-ACP synthetase allows efficient entry of exogenous fatty acids into the Escherichia coli fatty acid and lipid A synthetic pathways. Biochemistry 49:718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jimenez PN, et al. 2012. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76:46–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kang Y, Nguyen DT, Son MS, Hoang TT. 2008. The Pseudomonas aeruginosa PsrA responds to long-chain fatty acid signals to regulate the fadBA5 beta-oxidation operon. Microbiology 154:1584–1598 [DOI] [PubMed] [Google Scholar]
- 32. Kang Y, Zarzycki-Siek J, Walton CB, Norris MH, Hoang TT. 2010. Multiple FadD acyl-CoA synthetases contribute to differential fatty acid degradation and virulence in Pseudomonas aeruginosa. PLoS One 5:e13557 doi:10.1371/journal.pone.0013557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kass LR, Brock DJ, Bloch K. 1967. Beta-hydroxydecanoyl thioester dehydrase. I. Purification and properties. J. Biol. Chem. 242:4418–4431 [PubMed] [Google Scholar]
- 34. Kerr KG, Snelling AM. 2009. Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp. Infect. 73:338–344 [DOI] [PubMed] [Google Scholar]
- 35. King JD, Kocincova D, Westman EL, Lam JS. 2009. Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 15:261–312 [DOI] [PubMed] [Google Scholar]
- 36. Klein K, Steinberg R, Fiethen B, Overath P. 1971. Fatty acid degradation in Escherichia coli. An inducible system for the uptake of fatty acids and further characterization of old mutants. Eur. J. Biochem. 19:442–450 [DOI] [PubMed] [Google Scholar]
- 37. Lin S, Hanson RE, Cronan JE. 2010. Biotin synthesis begins by hijacking the fatty acid synthetic pathway. Nat. Chem. Biol. 6:682–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22:582–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moore NM, Flaws ML. 2011. Epidemiology and pathogenesis of Pseudomonas aeruginosa infections. Clin. Lab Sci. 24:43–46 [PubMed] [Google Scholar]
- 40. Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65:239–259 [DOI] [PubMed] [Google Scholar]
- 41. Parsons JB, Rock CO. 2011. Is bacterial fatty acid synthesis a valid target for antibacterial drug discovery? Curr. Opin. Microbiol. 14:544–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Patton GM, Lowenstein JM. 1979. Measurements of fatty acid synthesis by incorporation of deuterium from deuterated water. Biochemistry 18:3186–3188 [DOI] [PubMed] [Google Scholar]
- 43. Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Remans K, Vercammen K, Bodilis J, Cornelis P. 2010. Genome-wide analysis and literature-based survey of lipoproteins in Pseudomonas aeruginosa. Microbiology 156:2597–2607 [DOI] [PubMed] [Google Scholar]
- 45. Rietsch A, Mekalanos JJ. 2006. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 59:807–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rock CO, Cronan JE. 1981. Acyl carrier protein from Escherichia coli. Methods Enzymol. 7(Part C):341–351 [DOI] [PubMed] [Google Scholar]
- 47. Silbert DF, Ruch F, Vagelos PR. 1968. Fatty acid replacements in a fatty acid auxotroph of Escherichia coli. J. Bacteriol. 95:1658–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Son MS, Matthews WJ, Jr, Kang Y, Nguyen DT, Hoang TT. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 75:5313–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Spalding MD, Prigge ST. 2010. Lipoic acid metabolism in microbial pathogens. Microbiol. Mol. Biol. Rev. 74:200–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stanier RY, Palleroni NJ, Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159–271 [DOI] [PubMed] [Google Scholar]
- 51. Stover CK, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 52. Tsay JT, Oh W, Larson TJ, Jackowski S, Rock CO. 1992. Isolation and characterization of the beta-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J. Biol. Chem. 267:6807–6814 [PubMed] [Google Scholar]
- 53. Weeks G, Shapiro M, Burns RO, Wakil SJ. 1969. Control of fatty acid metabolism. I. Induction of the enzymes of fatty acid oxidation in Escherichia coli. J. Bacteriol. 97:827–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Williams P, Camara M. 2009. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 12:182–191 [DOI] [PubMed] [Google Scholar]
- 55. Wu DC, Chan WW, Metelitsa AI, Fiorillo L, Lin AN. 2011. Pseudomonas skin infection: clinical features, epidemiology, and management. Am. J. Clin. Dermatol. 12:157–169 [DOI] [PubMed] [Google Scholar]
- 56. Yuan Y, Sachdeva M, Leeds JA, Meredith TC. 2012. Fatty acid biosynthesis in Pseudomonas aeruginosa is initiated by the FabY class of β-ketoacyl acyl carrier protein synthases. J. Bacteriol. 194:5171–5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang YM, Frank MW, Zhu K, Mayasundari A, Rock CO. 2008. PqsD is responsible for the synthesis of 2,4-dihydroxyquinoline, an extracellular metabolite produced by Pseudomonas aeruginosa. J. Biol. Chem. 283:28788–28794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang YM, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222–233 [DOI] [PubMed] [Google Scholar]
- 59. Zhang YM, Rock CO. 2009. Transcriptional regulation in bacterial membrane lipid synthesis. J. Lipid Res. 50(Suppl):S115–S119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhu K, Choi KH, Schweizer HP, Rock CO, Zhang YM. 2006. Two aerobic pathways for the formation of unsaturated fatty acids in Pseudomonas aeruginosa. Mol. Microbiol. 60:260–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.