Abstract
We have thoroughly investigated the abrB2 gene (sll0822) encoding an AbrB-like regulator in the wild-type strain of the model cyanobacterium Synechocystis strain PCC6803. We report that abrB2 is expressed from an active but atypical promoter that possesses an extended −10 element (TGTAATAT) that compensates for the absence of a −35 box. Strengthening the biological significance of these data, we found that the occurrence of an extended −10 promoter box and the absence of a −35 element are two well-conserved features in abrB2 genes from other cyanobacteria. We also show that AbrB2 is an autorepressor that is dispensable to cell growth under standard laboratory conditions. Furthermore, we demonstrate that AbrB2 also represses the hox operon, which encodes the Ni-Fe hydrogenase of biotechnological interest, and that the hox operon is weakly expressed even though it possesses the two sequences resembling canonical −10 and −35 promoter boxes. In both the AbrB2-repressed promoters of the abrB2 gene and the hox operon, we found a repeated DNA motif [TT-(N5)-AAC], which could be involved in AbrB2 repression. Supporting this hypothesis, we found that a TT-to-GG mutation of one of these elements increased the activity of the abrB2 promoter. We think that our abrB2-deleted mutant with increased expression of the hox operon and hydrogenase activity, together with the reporter plasmids we constructed to analyze the abrB2 gene and the hox operon, will serve as useful tools to decipher the function and the regulation of hydrogen production in Synechocystis.
INTRODUCTION
Cyanobacteria are ancient photoautotrophic prokaryotes that are regarded as the progenitors of oxygenic photosynthesis (33, 39) and the plant chloroplast (8). Over time, cyanobacteria have evolved as the largest and most diverse groups of bacteria (44) and have colonized most waters and soils of our planet. The hardiness of cyanobacteria is due to their efficient photosynthesis, which uses nature's most abundant resources, solar energy, water, CO2, and mineral nutrients, to produce a large part of the atmospheric oxygen and organic assimilates for the food chain (52). On a global scale, cyanobacteria fix an estimated 25 gigatons of carbon from CO2 per year into energy-dense biomass (37, 49). To perform this huge CO2 fixation, cyanobacteria use 0.2 to 0.3% (49) of the total solar energy, 178,000 TW, reaching the Earth's surface (22). Thus, the amount of energy passing through cyanobacteria exceeds by more than 25 times the energy demand of human society (about 15 TW), roughly 1,000 times the total nuclear energy produced on Earth.
Furthermore, the availability of molecular tools for gene manipulation make cyanobacteria promising “low-cost” microbial cell factories for the carbon-neutral sustainable production of alkanes (10, 41), bioplastics (1), hydrogen (2, 12), and lipids (25, 43), while saving arable soils for crops (50). In light of their tremendous importance, deeper investigation into the mechanisms by which cyanobacteria convey solar energy to the environment is justified. In this frame, investigating the photobiological production of hydrogen by cyanobacteria has both fundamental and applied research values. As the basic research interest, it addresses the paradox of the antagonistic production of oxygen and hydrogen (O2 inhibits H2 production). As a biotechnological interest, it may lead to the sustainable production of a high-energy fuel (26), which burns cleanly in producing only water as its by-product.
The pentameric hydrogenase enzyme (HoxEFUYH; Hox for hydrogen oxidation) of cyanobacteria produces H2 through the reversible reaction 2 H+ + 2 e− ↔ H2, which uses NAD(P)H as the source of electrons originating from photosynthesis and/or sugar catabolism, as well as a nickel-iron cluster and several iron-sulfur centers as redox cofactors (5). The Hox enzyme has been studied mostly in the best-characterized unicellular cyanobacterium, Synechocystis strain PCC6803 (here designated Synechocystis), which harbors a small genome (less than 4 Mb [see CyanoBase, http://genome.kazusa.or.jp/cyanobase/]) that is easily manipulable (14, 29, 38). The active Hox enzyme, matured by the HoxW protease (47) and assembled using the six-subunit HypABCDEF complex (5), has been recently characterized as a truly bidirectional enzyme with a bias toward H2 production (30). The five genes hoxEFUYH are clustered in a octacistronic operon that also contains three open reading frames of unknown function. This operon is weakly expressed as a polycistronic transcript, which initiates 168 bp upstream of the start codon of the proximal hoxE gene (15, 35). The transcriptional regulation of the hox operon is complex in responding to various environmental conditions (21), and it involves at least three proteins (36). The LexA-related protein Sll1626, which appears to regulate carbon assimilation rather than DNA repair (9), was found to activate the transcription of the hox genes through binding to the promoter of the hox operon (15, 35). In addition, two AbrB-like regulators, which have their putative DNA-binding domain in the C-terminal region instead of in the usual N-terminal region, as occurs in other prokaryotes (19), were found to operate in hox regulation. In Bacillus subtilis, the AbrB regulator is implicated in the regulation of about 100 genes involved in sporulation, biofilm formation, antibiotic production, and development of competence for DNA uptake, but its promoter recognition consensus sequence and mode of interaction with DNA remain unclear (6). In Synechocystis, the AbrB1 (Sll0359) protein was found to be indispensable to cell life in the wild-type (WT) strain (34) and the glucose-tolerant mutant (19). Furthermore, AbrB1 was shown to bind to the promoter region of its own gene and to activate transcription of the hox operon through binding to the hox operon promoter (34). In contrast, AbrB2 (Sll0822) was studied only in the glucose-tolerant strain, which possesses several specific mutations that may interfere, at least indirectly, with the studied process (20). In the glucose-tolerant mutant, AbrB2 appeared to (i) be dispensable to cell life, (ii) bind to the promoter regions of its own gene and of other genes involved in nitrogen and carbon assimilations (19, 24, 51), and (iii) negatively influence expression of the hox operon via an unknown mechanism (19).
In this study, we thoroughly investigated the function and regulation of the abrB2 gene in the wild-type strain of Synechocystis, because it is the organism that actually occurs in nature. We demonstrate that abrB2 is expressed from an atypical promoter that possesses an extended −10 element to compensate for the absence of a −35 box. Furthermore, we demonstrate through gene deletion and overexpression that AbrB2 represses its own gene, as well as the hox operon, which is of biotechnological interest. We think that our abrB2-deleted mutant with an improved hydrogenase activity and healthy growth, and also the reporter plasmids we constructed to analyze the promoters of the abrB2 gene and the hox operon, will serve as useful tools to decipher the regulation and the function of the hydrogen production machine in cyanobacteria.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Synechocystis PCC6803 was grown at 30°C or 39°C (depending on the strain) under continuous white light (2,500 lx, 31.25 μE m−2 s−1) on BG11 medium (40) enriched with 3.78 mM Na2CO3 (9).
E. coli strains used for gene manipulations (TOP10; Invitrogen), production of recombinant proteins [BL21(DE3); Novagen], or conjugative transfer to Synechocystis (CM404; [31]) of replicative plasmids (Table 1) derived from our temperature-controlled expression vector pFC1 (32) were grown on LB medium at 30°C (CM404 and TOP10 harboring pFC1 derivatives) or 37°C [TOP10 and BL21(DE3)]. Antibiotic selection was as follows: for E. coli, ampicillin (Ap) at 100 μg ml−1 or 50 μg ml−1, kanamycin (Km) at 50 μg ml−1, and spectinomycin (Sp) at 100 μg ml−1 for E. coli; for Synechocystis, Km at 50 to 300 μg ml−1, Sp at 5 μg ml−1, and streptomycine (Sm) at 5 μg ml−1.
TABLE 1.
Characteristics of plasmids used in this study
| Plasmid use and name | Relevant feature(s) | Reference |
|---|---|---|
| Targeted deletion of abrB2 in Synechocystis | ||
| pGEMT | AT overhang Ampr cloning vector | Promega |
| pUC4K | Source of Kmr marker gene | Pharmacia |
| pΔsll0822 | pGEMT with Synechocystis sll0822 flanking sequences, with sll0822 coding sequence (from bp 7 to 384) replaced by SmaI site | This study |
| pΔsll0822::Kmr | pΔsll0822 with Kmr marker inserted into unique SmaI site | This study |
| High-level expression of abrB2 in Synechocystis | ||
| pFC1 | Replicating plasmid for heat-inducible gene expression in Synechocystis | 32 |
| pSll0822 | pFC1 with sll0822 CS cloned between NdeI-EcoRI sites | This study |
| Analysis of hox and abrB2 promoters in Synechocystis | ||
| pSB2A | Kmr Smr/Spr replicative promoter probe plasmid harboring unique SnaBI site in front of its promoterless cat reporter gene | 27 |
| pPS1 | Kms derivative of pSB2A generated after cleavage with NsiI and religation | This study |
| pJD1 | hox PRa (bp −794 to +173 relative to TSS) cloned at SnaBI site of pPS1 | This study |
| pJD2 | hox PR (bp −522 to +173 relative to TSS) cloned at SnaBI site of pPS1 | This study |
| pJD3 | hox PR (bp −403 to +173 relative to TSS) cloned at SnaBI site of pPS1 | This study |
| pJD4 | hox PR (bp −170 to +173, relative to TSS) cloned at SnaBI site of pPS1 | This study |
| pPS2 | abrB2 PR (bp −75 to +88 relative to TSS) cloned at SnaBI site of pSB2A | This study |
| pPS2 m-8 | pPS2 harboring T→G mutation at −8 position relative to TSS | This study |
| pPS2 m-13 | pPS2 harboring T→G mutation at −13 position relative to TSS | This study |
| pPS2 m-15 | pPS2 harboring T→G mutation at −15 position relative to TSS | This study |
| pPS2 m-4746 | pPS2 harboring TT→GG mutation at positions −47 to −46 relative to TSS | This study |
| Production of AbrB2 in E. coli | ||
| pET14b | Ampr E. coli plasmid for production of 6×His-tagged proteins | Novagen |
| pSS1 | pET14b plasmid with sll0822 CS cloned between NdeI and BamHI sites | This study |
PR, promoter region.
Construction of the DNA cassette for targeted deletion of the Synechocystis abrB2 gene.
The two regions of Synechocystis DNA (each about 300 bp in length) flanking the abrB2 (sll0822) protein coding sequence (CS) were independently amplified by PCR, using the primers sll0822M-Fw and sll0822R4 for the upstream region and sll0822M-Rv and sll0822A4 for the downstream region (see Table S1 in the supplemental material). These two DNA regions were fused through standard PCR-driven overlap extension (17) in a single DNA segment harboring a SmaI restriction site in place of the abrB2 CS. After cloning in pGEMT (Table 1), the resulting plasmid was opened at the unique SmaI site, where we cloned the Kmr resistance cassette (a HincII fragment of pUC4K) in the same orientation as the abrB2 CS it replaced. The resulting Δsll0822::Kmr deletion cassette was verified by PCR and nucleotide sequencing (BigDye kit; ABI, Perkin-Elmer).
Construction of the vector for high-level expression of the abrB2 gene in Synechocystis.
Our temperature-controlled expression vector pFC1, which replicates autonomously in E. coli and several cyanobacteria (32), was used for high-level expression of the Synechocystis abrB2 gene. pFC1 harbors the λcI857 temperature-sensitive repressor-encoding gene that tightly controls the activity of the otherwise-strong λpR promoter located downstream, which is followed by the λcro ribosome-binding site (5′-AGGA-3′) and ATG start codon (in bold) embedded within the unique NdeI restriction site (5′-CATATG-3′) for in-frame fusion of protein coding regions. The abrB2 CS was PCR amplified from Synechocystis DNA, using specific primers (sll0822FL1 and sll0822FL2 [see Table S1 in the supplemental material]), which flanked the abrB2 CS between unique NdeI and EcoRI restriction sites, for cloning into pFC1, opened with the same enzymes. The Smr/Spr resulting plasmid, pSll0822, for high-level expression of abrB2 at 39°C was verified by PCR and nucleotide sequencing (BigDye kit; ABI Perking Elmer).
RNA isolation.
Aliquots of 200 ml of mid-log-phase cultures (2.5 × 107cells ml−1) were rapidly harvested by vacuum filtration (less than 1 min), resuspended in 4 ml of 50 mM Tris 50 (pH 8), 5 mM EDTA, immediately frozen in an Eaton press chamber cooled in a dry ice and ethanol bath, and disrupted (250 MPa). RNA was extracted and purified with the Qiagen RNeasy kit as we have previously described (18) and then treated with 20 U of DNase I, RNase-free (Applied Biosystems) for 30 min at 37°C. RNA concentration and purity (A260/A280 ratio, >1.9) were determined with a Nanodrop apparatus (Thermo Scientific) and migration on an agarose gel to verify the absence of RNA degradation. The absence of contaminant DNA was verified with the Taq DNA-dependent DNA polymerase (Invitrogen) using primers specific to the control gene rnpB (see Table S1 in the supplemental material).
RT-PCR and quantitative PCR.
cDNAs were synthesized from 5 μg of total RNA by using Moloney murine leukemia virus reverse transcriptase (RT; Invitrogen). Samples were then incubated for 20 min at 37°C with 4 U RNase H (Applied Biosystems) to eliminate RNA templates, and the cDNA concentration was measured with Nanodrop apparatus adjusted to 1 μg μl−1 by dilution in H2O. Quantitative PCR (qPCR) assays of the expression levels of the studied genes and the well-known constitutive control gene rnpB (see Table S1 in the supplemental material) were performed with Mesa Green qPCR MasterMix Plus for SYBR assay (RT-SYS2X-03+WOU; Eurogentec) according to the manufacturer's instructions. The gene-specific primers were chosen so as to generate DNA fragments of similar length, between 199 bp and 234 bp. Each reaction was performed in a 50-μl reaction mixture containing 5 ng cDNA, 0.04 μM specific primer, 1× Mesa Green qPCR MasterMix Plus buffer, and 2 mM MgCl2 in an iCycler iQ 96-well reaction plate covered with adhesive film (Bio-Rad). Samples were incubated in an iQ5 multicolor real-time PCR detection system (Bio-Rad) for 2 min at 50°C, 2 min at 95°C, and 45 cycles of 15 s at 95°C and 1 min at 60°C. Each assay was performed in triplicate, allowing the mean threshold cycle value (CT) to be calculated from standard curve by using the iQ5 optical system software (Bio-Rad). Each gene-specific standard curve was made by 4-fold serial dilution of wild-type strain cDNA (ranging from 312.5 to 0.3 ng) compared to the log input cDNA concentration for each primer (data not shown). For each primer tested, the regression value (ΔCT versus cDNA concentration) was less than 0.1, indicating approximately equal amplification efficiencies. Then, for each studied gene, the CT value was converted to the gene copy number per ng of template cDNA.
Determination of the TSS of sll0822 by 5′-random amplification of cDNA ends (5′-RACE).
Aliquots of 25 μg of total RNAs of the Synechocystis WT strain were treated with shrimp alkaline phosphatase (SAP), which does not affect the 5′-triphosphate extremity of full-length mRNAs but dephosphorylates the 5′-monophosphate end of degraded RNA. Then, RNAs were treated with tobacco acid pyrophosphatase (TAP; Epicentre) to convert the 5′-triphosphate extremity of full-length mRNA into the 5′-monophosphate without modifying the 5′-OH end of degraded RNA. The 5′-monophosphate extremity of the full-length mRNAs was ligated to an RNA anchor (see Table S1 in the supplemental material) with the T4 RNA ligase (Invitrogen). The resulting chimeric RNAs were reverse transcribed with SuperScript II (Invitrogen) and the sll0822 gene-specific primer sll0822P1 (see Table S1). This first strand of cDNA was amplified by PCR using both the DNA anchor at the 5′ extremity and the sll0822P1 primer at the 3′ side. The resulting DNA was sequenced (BigDye kit; ABI Perkin-Elmer) to identify the first sll0822 nucleotide located immediately downstream of the DNA anchor, which is the sll0822 transcription start site (TSS) (see Fig. S1 in the supplemental material).
Construction of transcriptional fusions to the cat reporter gene and the CAT assay.
The studied promoter regions were amplified by PCR with specific oligonucleotides designed to introduce blunt-ended restriction sites in a way that allowed the elimination of all nucleotide substitutions upon restriction cleavage (see Table S1 in the supplemental material). These promoters were cloned in the unique SnaBI site preceding the promoterless cat (chloramphenicol acetyltransferase) reporter gene of our promoter probe vector pSB2A or of its Kms derivative, pPS1 (Table 1), which replicate in Synechocystis with the same copy number as the chromosome (27). Site-directed mutagenesis of the sll0822 promoter was done through standard PCR-driven overlap extension (17) using specific mutagenic primers (see Table S1) and the forward and reverse primers SnaBIFW22 and SnaBIRV22. The sequence of each promoter insert was verified, before and after propagation in Synechocystis. Then, 1 × 109 to 2 × 109 cells grown on standard plates to mid-log phase were rapidly harvested and disrupted with a chilled Eaton press prior to CAT assay (28).
AbrB2 production and purification from E. coli.
The abrB2 coding sequence was PCR amplified from Synechocystis DNA with the primers abrB2-NdeI-Fw and abrB2-BamHI-Rv (see Table S1 in the supplemental material), digested with both NdeI and BamHI, and cloned into the pET14b plasmid opened with the same enzymes for in-frame fusion to the 6×His tag. The resulting plasmid, pSS1 (Table 1), was introduced into E. coli BL21(λDE3), which was grown to mid-log phase prior to the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h to produce the 6×His-AbrB2 protein. Cells were harvested by centrifugation at 7,000 rpm for 5 min at 4°C, suspended in buffer A (30 mM Tris-HCl [pH 7.5], 400 mM NaCl, 20 mM imidazole) containing 2 mg/ml lysozyme (Sigma-Aldrich), and incubated for 1 h on ice. After sonication on ice (6 pulses, 20 s each) and centrifugation (13,000 rpm for 30 min at 4°C), cell lysates were loaded onto a 1 ml Ni-nitrilotriacetic acid–agarose column (Qiagen) preequilibrated with buffer A and washed with 10 bed volumes of 30 mM imidazole in buffer A. The proteins eluted with 300 mM imidazole in buffer A were analyzed by SDS-PAGE. AbrB2-containing fractions were pooled, concentrated with Amicon Ultra centrifugal filter devices (Millipore), diluted in buffer B (30 mM Tris-HCl [pH 7.5], 100 mM NaCl), loaded onto a heparin column (GE Healthcare) preequilibrated with buffer B, and washed with 10 bed volumes of buffer B. AbrB2 was eluted with a linear 0-to-500 mM NaCl gradient in 30 mM Tris (pH 7.5), and the AbrB2-enriched fractions were pooled, concentrated, and stored in 30 mM Tris (pH 7.5), 400 mM NaCl. AbrB2 protein purity, estimated by SDS-PAGE, was greater than 95%.
Electromobility shift assays (EMSAs).
The studied promoter regions were PCR amplified from Synechocystis DNA with specific primers (see Table S1 in the supplemental material) purified, 3′-end-labeled with digoxigenin (DIG gel shift kit, 2nd generation; Roche), incubated with AbrB2, migrated on a Novex 6% DNA retardation gel (Invitrogen), blotted on a positively charged nylon membrane (Roche) with a semidry transfer apparatus (Apelex), and cross-linked onto the membrane with a 2-min UV-C (254 nm) exposure (Stratalinker). The DNA bands were revealed with anti-DIG antibodies by chemiluminescence, using Hyperfilm ECL (Amersham Pharmacia) and Kodak developer.
Hydrogenase activity measurements.
Hydrogenase activities were measured by two standard methods. First, hydrogen evolution was measured with a modified Clark-type electrode (Hansatech, United Kingdom) (48) as described previously by some of us (46). A total of 2.5 × 107 cells (1-ml culture at an optical density at 580 nm of 1) were harvested by a 5-min centrifugation at 5,000 rpm, resuspended in 25 μl of 50 mM Tris-HCl (pH 7.5) buffer, and introduced into 500 μl of a solution containing 50 mM Tris-HCl (pH 7.5), 20 mM Na-dithionite, and 5 mM methylviologen as the electron donor to hydrogenase. Calibration was performed using the amplitude of the electrical signal from the electrode in the presence of an aliquot of H2-saturated water as a concentration reference. Second, hydrogenase activity (the H/D exchange rate of labeled dihydrogen) was determined by membrane-inlet mass spectrometry (MIMS) on cell suspensions, as we have described previously (3, 7).
RESULTS
The abrB2 gene is dispensable to the viability of the wild-type strain of Synechocystis.
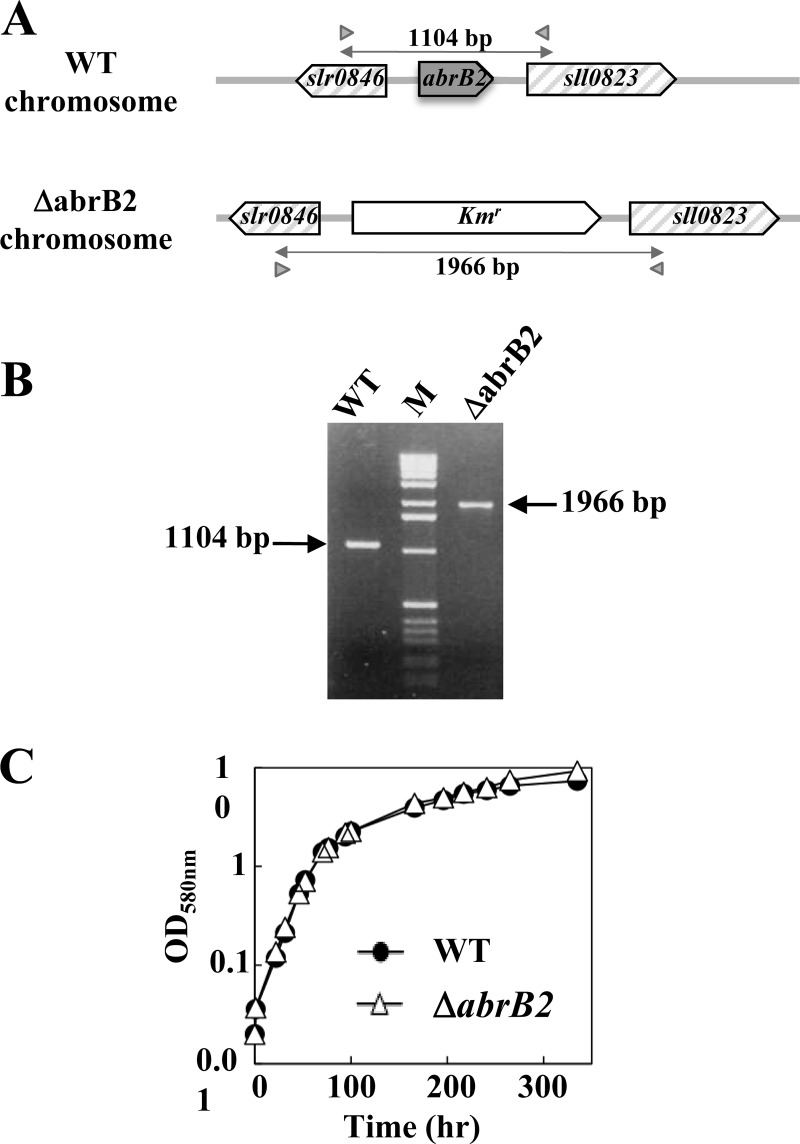
In the framework of our long-term interest in gene regulation, we have investigated the AbrB2 regulator (Sll0822 in CyanoBase) of the model cyanobacterium Synechocystis. Therefore, we constructed the ΔabrB2::Kmr deletion cassette (Table 1), in which the complete coding sequence for the 129-amino-acid AbrB2 protein has been deleted and replaced by a transcription terminatorless Kmr marker for selection (Fig. 1A). After transformation in the WT strain of Synechocystis, we verified through PCR (Fig. 1B) and DNA sequencing (data not shown) that the Kmr marker had properly replaced the abrB2 gene in all copies of the polyploid chromosome (13, 23). We also verified through RT-PCR and quantitative RT-PCR that the mutant completely lacked abrB2 mRNA (data not shown). These data, together with the fact that the ΔabrB2::Kmr mutant grows as healthy as the WT strain (Fig. 1C), showed that the AbrB2 protein is dispensable to the growth of Synechocystis. This finding is consistent with, but not identical to, the previous observation that the insertion of the Kmr marker at 177 bp downstream of the ATG start codon of abrB2 (possibly leading to the synthesis of an aberrant protein) did not impair the viability but strongly reduced the growth rate of the glucose-tolerant mutant of Synechocystis (19), which has several specific mutations compared to the WT strain (20), i.e., the organism actually occurring in nature.
Fig 1.
Influence of the AbrB2 regulator on growth of Synechocystis under standard conditions. (A) Schematic representation of the abrB2 chromosome locus in the WT strain (see CyanoBase) and the abrB2-deleted mutant (ΔabrB2::Kmr) constructed in this study. The genes are represented by boxes pointing in the direction of their transcription. The PCR primers specific to the slr0846 and sll0823 genes are represented by the small gray triangles, and the sizes of the products they generated upon amplification of the abrB2 chromosome locus of the WT (1,104 bp) and abrB2-deleted (ΔabrB2::Kmr; 1,966 bp) chromosomes are indicated by double arrows. (B) UV light image of the agarose gel, showing the 1,104-bp and 1,966-bp PCR product typical of the WT and abrB2-deleted chromosomes, respectively; the results show that the abrB2-deleted mutant harbors no WT copies of the chromosome. (C) Growth of the WT strain and the ΔabrB2 mutant (this experiment was repeated three times and yielded statistical deviations too small to be represented).
AbrB2 negatively regulates expression of its own gene through binding to its own promoter.
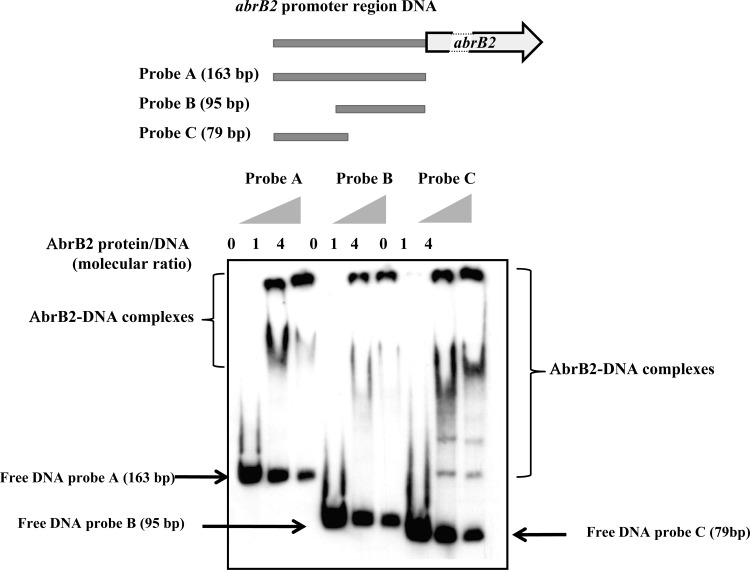
The Synechocystis AbrB2 protein was translationally fused to the 6×His tag for facile purification from recombinant E. coli cells to near homogeneity through nickel affinity chromatography. Then, this 6×His-AbrB2 protein was used for an EMSA to determine its ability to bind the DIG-labeled, 163-bp-long promoter region (Fig. 2) shared by the divergently transcribed genes abrB2 and slr0846 (Fig. 1). We found that AbrB2 binds on this 163-bp full promoter region, in agreement with a previous observation (19), and on the two subfragments tested here (Fig. 2); thus, the promoter region contains several AbrB2-binding motifs (see below). As negative-control experiments, we verified the absence of binding of (i) AbrB2 on a heterologous (noncyanobacterial) DNA (see Fig. S2 in the supplemental material) and (ii) the bovine serum albumin (BSA) protein on the abrB2 promoter DNA (see Fig. 5B, below; see also Fig. S2 in the supplemental material). The obvious interpretation of the binding of AbrB2 on its own promoter is that AbrB2 is an autoregulator. To test this hypothesis, we performed a thorough in vivo analysis of the abrB2 promoter by using our promoter probe vector, which harbors the promoterless cat reporter gene for promoter analysis and replicates autonomously in Synechocystis with a similar copy number as the chromosome (27). We cloned the whole intergenic region (163 bp) between the divergently transcribed genes abrB2 and slr0846 in front of the cat reporter gene, and we selected the pabrB2-cat reporter plasmid (pPS2 [Table 1]) expressing the cat gene from the abrB2 promoter. This pabrB2-cat plasmid (Table 1) was introduced in both the WT strain and our abrB2 deletion mutant (ΔabrB2::Kmr), where it replicated stably, as expected (data not shown). The level of cat expression driven by the abrB2 promoter in the WT strain (44 CAT units [Fig. 3A]) was similar to that directed by the other promoters we previously analyzed (references 11 and 45 and references therein). This finding indicates that the abrB2 promoter has an average strength. As the usual control, we verified that the empty promoter probe vector with no promoter insert produced no CAT activity. Very interestingly, the abrB2 promoter was found to be more active (about 3-fold) in the abrB2 deletion mutant (131 CAT units), i.e., in the absence of the AbrB2 protein, thereby demonstrating that AbrB2 negatively regulates the expression of its own gene. Collectively, our data unambiguously demonstrate, for the first time, that AbrB2 is an autorepressor.
Fig 2.
Electrophoretic migration shift assay of binding of the AbrB2 regulator to the promoter region of its own gene. (Top) Schematic representation of the abrB2 gene, showing its 163-bp promoter region (gray line) and its protein-coding sequence (boxed arrows), which is interrupted (dashed lines) for the sake of size limitation. The positions and sizes of the three DNA fragments of the abrB2 promoter region used as targets for AbrB2 binding are indicated as probes A, B, and C. (Bottom) Analysis of the electrophoretic mobility of the DIG-labeled segments of the abrB2 promoter region, following incubation with increasing amounts of purified 6×His-AbrB2 protein. Arrows and regions delineated by braces indicate the positions of the free DNA probes and the retarded DNA-protein complexes, respectively.
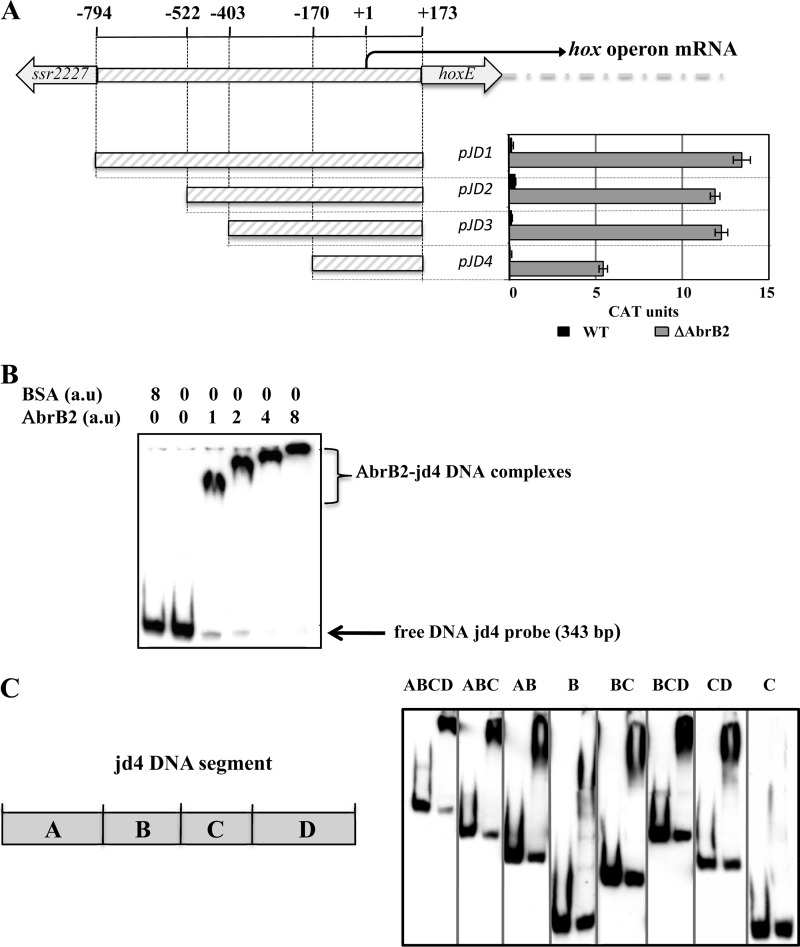
Fig 5.
Analysis of the hoxEFUYH operon promoter through transcriptional fusion to the cat reporter gene and electrophoretic migration shift assay results for binding of the AbrB2 regulator. (A, top) Schematic representation of the hoxEFUYH operon promoter region (thick gray lines) located between the hoxE protein-coding sequence and the opposite (putative) transposase gene, ssr2227. The nucleotide positions within the hoxEFUYH operon promoter region are indicated relative to the transcription start site (bent arrow), taken as the origin of distance (noted as +1). For the sake of clarity, the operonic genes downstream of hoxE are indicated by the large dashed gray line. (A, bottom) Deletion analysis of the hox operon promoter region transcriptionally fused to the cat reporter gene of our replicative promoter probe vector. CAT specific activities driven by the resulting reporter plasmids replicating in Synechocystis WT or ΔabrB2 (hatched gray bars) are expressed in nmol of chloramphenicol acetylated/min/mg of protein. They are the mean values ± standard deviations calculated from three independent experiments. (B and C) Analysis of the electrophoretic mobility of the DIG-labeled JD4 segment, or subsegments thereof, noted as A, B, C, and D, of the hox operon promoter, following incubation with increasing amounts of purified 6×His-AbrB2 regulator or the BSA negative-control protein. Arrows and braces indicate the positions of the free DNA probes and the retarded DNA-protein complexes, respectively.
Fig 3.
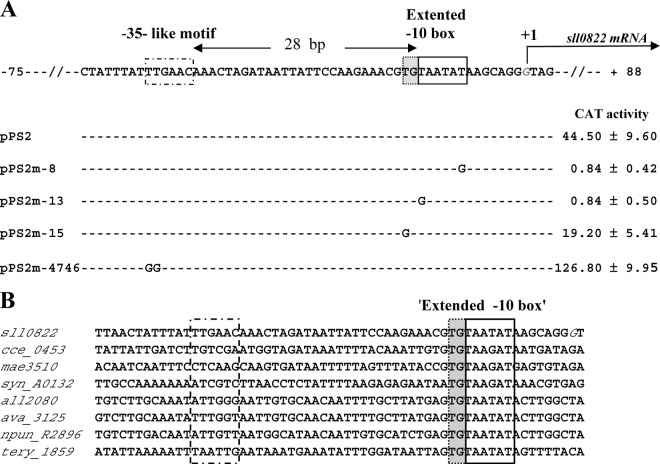
Mutational analysis in Synechocystis of the abrB2 promoter transcriptionally fused to the cat reporter gene of our replicative promoter probe vector. (A) Sequence alignment of the nontranscribed DNA strand of the WT (pPS2 reporter plasmid) and mutant (pPS2m reporter plasmids) abrB2 promoters. Sequence resembling the −35 and extended −10 promoter elements are boxed, and the origin of transcription (+1; G nucleotide in italics) that we mapped in this study (see Fig. S1 in the supplemental material) is indicated by the bent arrow. Nucleotide substitutions in the mutant promoters (m-8, m-13, m-15, and m-4746) are shown in uppercase letters, while the conserved nucleotides are indicated by dashed lines. For each reporter strain, the CAT activity is the mean value of three measurements performed on two independent cellular extracts; 1 CAT unit = 1 nmol of chloramphenicol acetylated/min/mg of protein. (B) Sequence alignments of the abrB2 promoter regions from various cyanobacteria resembling Synechocystis in that their abrB2 gene is divergently transcribed with an opposite gene. Besides Synechocystis PCC6803 (sll0822), these cyanobacteria are as follows: Cyanothece ATCC 51142 (cce_0453), Microcystis aeruginosa NIES-843 (mae_35130), Nostoc PCC 7120 (all2080), Anabaena variabilis ATCC 29413 (ava_3125), Nostoc punctiforme PCC 73102 (npun_R2896), and Trichodesmium erythraeum ISM101 (tery_1859). The positions of the conserved extended −10 promoter element and of the nonconserved sequence resembling a −35 element in Synechocystis are indicated by boxes.
The core promoter of the abrB2 gene possesses an “extended −10 element” which compensates for the absence of a −35 box.
Using the classical 5′-RACE technique (42), which has performed well in our hands (11, 45), we mapped the TSS of abrB2. It is the G nucleotide located 87 bp upstream of the ATG start codon (see Fig. S1 in the supplemental material). Then, we performed a mutational analysis of the abrB2 promoter to identify its cis-acting elements. We studied the 5′-TAatAT-3′ hexamer (lowercase letters indicate nucleotides that are not widely conserved) matching the canonical −10 box of σ70-type Escherichia coli promoters (16) with regard to both sequence (5′-TAatAT-3′) and its position (−13 to −8) from the TSS, the above-mentioned G nucleotide we used as the origin of distance (noted as +1 [see Fig. S1 in the supplemental material]). As anticipated, this 5′-TAatAT-3′ abrB2 element behaves as a −10 promoter box, as it is crucial to transcription. Indeed, transversion mutagenesis of either its presumably important T nucleotides (5′-TAatAT-3′ to 5′-GAatAT-3′ and 5′-TAatAT-3′ to 5′-TAatAG-3′) completely abolished abrB2 promoter activity (Fig. 3A; reporter plasmids pPS2m-13 and pPS2m-8, respectively), as we previously observed with the −10 box of several Synechocystis promoters (references 11 and 45 and references therein). In the abrB2 promoter, we also noticed the presence of a 5′-TTGAac-3′ motif, resembling a canonical −35 box (16) in sequence (5′-TTGACA-3′) but not position (28 bp instead of 17 ± 1 bp, the usual spacing distance from the −10 box). We mutagenized this 5′-TTGAac-3′ abrB2 element and found that it was not crucial to promoter activity (Fig. 3A, pPS2m-4746 plasmid), unlike a −35 promoter box. Instead, this abrB2 5′-TTGAac-3′ motif appeared to negatively influence the activity of the abrB2 promoter (Fig. 3A). Then, having in mind that the absence of a −35 promoter box can be compensated by the presence of an “extended −10 box” mediating all contacts with the RNA polymerase σ70 factor (4), we mutagenized the upstream T of the presumptive extended −10 box, 5′-TGTAATAT-3′ of the abrB2 promoter. As anticipated, this mutation (5′-TGTAATAT-3′ to 5′-GGTAATAT-3′ decreased the abrB2 promoter activity (Fig. 3A, pPS2m-15 plasmid), similarly to what we found in the case of the “extended −10 box” of the secA promoter (28). In agreement with its crucial importance for abrB2 transcription, we found the “extended −10 box,” 5′-TGTAATAT-3′, to be highly conserved in the promoter region of the abrB2 genes from other cyanobacteria (Fig. 3B). Collectively, our data indicate that the abrB2 promoter possesses an extended −10 element to compensate for the absence of a −35 box and that these features are conserved in other cyanobacteria.
AbrB2 negatively regulates both the expression and the hydrogenase activity of the hoxEFUYH operon.
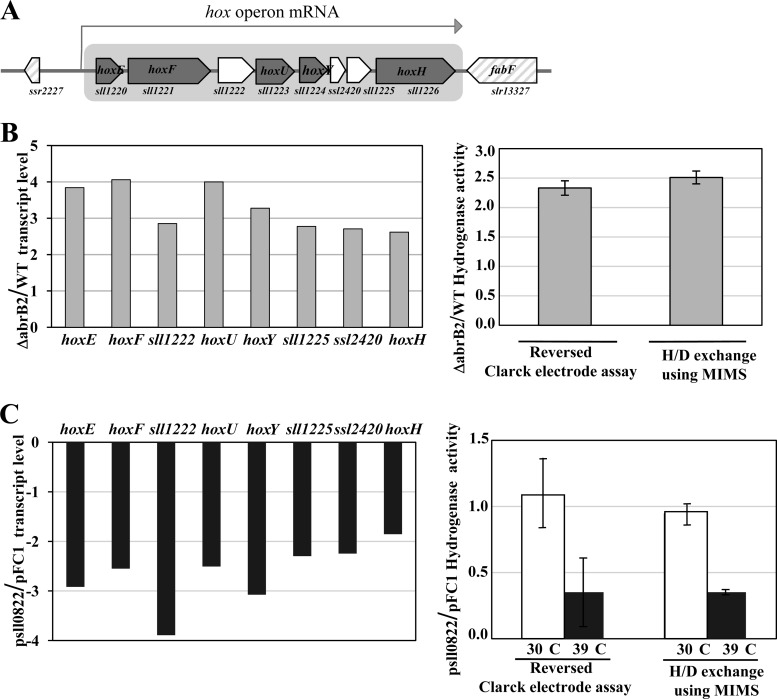
We used quantitative RT-PCR to analyze the influence of the AbrB2 regulator on the transcript abundance of the eight-gene hox operon, namely, sll1220 (hoxE), sll1221 (hoxF), sll1222, sll1223 (hoxU), sll1224 (hoxY), ssl2420, sll1225, and sll1226 (hoxH). Therefore, total RNAs were isolated from WT and abrB2-deleted (ΔabrB2::Kmr) cells growing under standard conditions and were subsequently hybridized with the gene-specific RT-PCR primers (see Table S1 in the supplemental material) designed to amplify a segment of each of the eight protein-coding sequences of the hox operon (Fig. 4A). All eight transcripts were found to be at least 2.5-fold more abundant in the abrB2-deleted mutant than in the WT strain (Fig. 4B). As a negative control, we verified the absence of abrB2 transcripts in the abrB2-deleted mutant (data not shown). Together, these data show that AbrB2 negatively regulates the hox operon in WT cells of Synechocystis, in agreement with what was observed in the glucose-tolerant mutant (19). From these results one can anticipate that the hydrogenase activity should increase in response to the absence of the AbrB2 regulator. Therefore, we used the two classical methods to measure the level of hydrogenase activity, which appeared to be at least 2-fold higher in our abrB2-deleted mutant than in the WT strain (Fig. 4B), as expected. To confirm that AbrB2 negatively regulates the expression of the hox operon and the activity of the hydrogenase in Synechocystis, we constructed an abrB2 overexpression mutant, as follows. We cloned the abrB2 protein-coding sequence into our temperature-controlled expression vector, pFC1, which replicates autonomously in cyanobacteria and tightly controls the production of the studied protein according to the growth temperatures, i.e., no production at 30°C and strong production after 24 h of induction at 39°C (32, 38). The resulting pSll0822 plasmid was introduced by conjugative transfer in Synechocystis cells, which were transferred for 24 h at 39°C to verify the strong induction of abrB2 expression (27-fold [data not shown]) and the concomitant downregulation (by at least 2-fold) of hox expression and hydrogenase activity (Fig. 4C). Collectively, these results demonstrate that AbrB2 negatively regulates expression of the hox operon and activity of the hydrogenase in Synechocystis.
Fig 4.
Analysis of expression of the hydrogenase-encoding genes and of hydrogenase activity in various strains of Synechocystis harboring a wild-type abrB2 gene (WT), a deletion of abrB2 (Δsll0822::Kmr), a plasmid (psll0822) for high-level expression of abrB2 inducible by 24 h of growth at 39°C, or the empty expression vector (pFC1). All results are expressed as means ± standard deviations of the data obtained after 3 to 6 biological repetitions of each assay. (A) Schematic representation of the locus of the octacistronic hox operon. The genes are represented by boxes pointing in the direction of their transcription; boxes are gray in the case of the hox genes. (B, left) Histogram plots of the ratios of transcript abundance (measured by quantitative real-time PCR) of each gene of the octacistronic hox operon in the abrB2-deleted mutant over the WT strain. (Right) Histogram representation of the hydrogenase activity ratios in ΔabrB2 over WT cells measured with the MIMS assay and a reversed Clark-type electrode, as indicated. (C, left) Histograms showing the transcript level ratios of each gene of the octacistronic hox operon, in cells propagating psll0822 over those propagating pFC1, which were all grown for 24 h at 39°C prior to qRT-PCR analysis. (Right) Histogram representation of the hydrogenase activity ratios of cells propagating psll0822 over those propagating pFC1, before (white rectangles) or after (black rectangles) 24 h of growth at 39°C, prior to MIMS and Clark-type electrode assays, as indicated.
AbrB2 negatively regulates the activity of the hox operon promoter.
We used our promoter probe vector to test whether AbrB2 negatively controls the promoter of the hoxEFUYH operon. As Synechocystis promoters can be complex in harboring several cis-acting regulatory elements (references 9 and 45 and references therein), we cloned in our vector the whole hox promoter region occurring between the opposite genes hoxE and ssr2227, along with the first 5 bp of their protein-coding sequences (Fig. 5A). This 967-bp hox promoter region extends from −794 to +173 nucleotides relative to the transcription start site (the A nucleotide mapped by other workers [15, 35], which we used as the origin of distance). Our hoxprom-cat reporter plasmid (pJD1 [Fig. 5A]) weakly expressed the cat gene in Synechocystis (less than 1 CAT unit), thereby indicating that the hox promoter was weakly active, in agreement with the low abundance of hox transcripts (34). However, the low activity of the hox promoter seemed at variance with the occurrence in this promoter of the two sequences resembling canonical promoter boxes, −35 (TTGctc) and −10 (TAacAa), which are located at correct distances from each other (18 bp) and from the transcription start site (7 bp). This apparent discrepancy prompted us to speculate that the AbrB2 regulator, which negatively regulates the expression of the hox operon (Fig. 4), operates at the level of hox promoter activity. To verify this hypothesis, we introduced the hox-cat reporter plasmid pJD1 in the abrB2-deleted mutant (ΔabrB2::Kmr), and cat expression was found to be much higher (about 13 CAT units [Fig. 5A]) than in WT cells, as expected.
Then, to meaningfully examine the EMSA analysis of the binding of AbrB2 on the hox promoter, we subcloned the long hox regulatory region (967 bp) in our promoter probe vector. Then, we serially deleted the 5′ end of the hox regulatory region while keeping its 3′ end intact, because such promoter downstream regions can contain negative cis-acting elements (9, 28, 45). As expected, we identified a shorter hox regulatory region (343 bp; plasmid pJD4) that retained a promoter activity that was negatively regulated by the AbrB2 protein (i.e., higher in ΔabrB2::Kmr cells than WT cells) (Fig. 5A). Together, our data show that AbrB2 negatively regulates the expression of the hox operon at the level of its promoter activity.
AbrB2 represses expression of the hox operon through binding to its promoter and flanking regions.
Using the suitably sized hox operon promoter region (343 bp; JD4 DNA) (Fig. 5A), we performed EMSAs to show that AbrB2 binds on the hox promoter region (Fig. 5B), as expected. The apparent size of the AbrB2-hox retardation complex appeared to increase in parallel with the increase in the molecular ratio of AbrB2 over hox DNA, indicating that several AbrB2 molecules can bind to each hox DNA molecule. As the usual controls for specific affinity of AbrB2 for hox DNA, we verified the absence of interaction between the hox DNA and the BSA control protein and that the abundance of the DIG-labeled hox-AbrB2 complex was decreased by the presence of an excess of unlabeled hox DNA (see Fig. S2 in the supplemental material).
We also tested the binding of AbrB2 on various segments of the 343-bp JD4 hox promoter region in an attempt to better localize the AbrB2-binding sites, and we named the segments A, B, C, and D for the sake of clarity. Fragment A (100 bp) is located upstream of fragment B (74 bp), which harbors the core promoter with the −35-like and −10-like boxes, followed by the downstream fragments C (69 bp) and D (100 bp), in that order (Fig. 5C). We found that AbrB2 bound to all tested subregions, with the noticeable exception of fragment C alone (Fig. 5C). The occurrence of distant AbrB2-binding regions in the hox promoter regions suggests the possible involvement of a DNA looping mechanism in the AbrB2-mediated regulation of hox transcription.
DISCUSSION
It is important to thoroughly study the mechanisms controlling the expression of the cyanobacterial genes encoding the Ni-Fe bidirectional hydrogenase in order to better understand its role in the global metabolism of the cell and possibly generate new cell factories for better production of H2. The pentameric hydrogenase Hox enzyme (HoxEFUYH) is mostly studied in the best-characterized cyanobacterium Synechocystis PCC6803. In Synechocystis, the five hox genes are grouped together with three genes of unknown function in an octacistronic “hox” operon (Fig. 4), which generates rare transcripts (5, 36). In the context of our long-term interest in gene expression, we decided to test whether this was due to the weak activity of the hox operon promoter. Therefore, we cloned the hox promoter region upstream of the promoterless cat reporter gene of our promoter probe plasmid vector, which replicates autonomously in Synechocystis at the same copy number as the chromosome (27). Our hoxprom-cat reporter plasmid (pJD1 [Fig. 5]) directed a very low level of cat expression in Synechocystis (less than 1 CAT unit), showing, for the first time, that the hox promoter is weakly active. This finding is at variance with the occurrence in the hox promoter of sequences resembling the canonical −35 (TTGctc) and −10 (TAacAa) promoter boxes (15, 35). Consequently, we speculated that the hox promoter might be controlled by a negative regulator. Hence, we became interested in the AbrB2 protein, which resembles the pleiotropic AbrB repressor of B. subtilis (6, 19). Working with the glucose-tolerant mutant of Synechocystis, which harbors several mutations with unknown physiological consequences (20), it was shown that the disruption of the sll0822 gene (here, abrB2) increased by about 2-fold the abundance of the hox transcripts monitored with DNA microarrays (19). Consequently, we decided to test the influence of AbrB2 on the expression and promoter activity of the hox operon. For this purpose, we used the wild-type strain of Synechocystis, because it is actually this strain that was originally isolated from nature. First, we deleted the abrB2 gene from all copies of the Synechocystis chromosome, which is polyploid (13, 23), and we found that the corresponding mutant grew as well as the wild-type strain (Fig. 1). This result was similar, but not identical, to what was observed in the glucose-tolerant mutant, in which the inactivation of the abrB2 gene strongly reduced the growth rate (19). This discrepancy might result from the differences in the strains (see above), growth conditions, and/or gene manipulation protocols employed by the two laboratories. While the previous workers introduced the Kmr marker inside the abrB2-coding sequence (at 177 bp downstream of the ATG start codon), which might thereby encode an aberrant protein impeding cell growth, we replaced the full abrB2-coding sequence with the Kmr marker to preclude the synthesis of AbrB2 (we have verified the absence of the abrB2 gene [Fig. 1] and its transcripts [data not shown]).
Using our abrB2-deleted mutant, we verified through quantitative RT-PCR that the absence of AbrB2 increased (at least 2.5-fold) the transcript abundance of all eight genes of the hox operon (Fig. 4). In agreement, we showed, for the first time, that the amount of active hydrogenase was also increased (about 2-fold) in the absence of AbrB2 (Fig. 4). To confirm these findings, we constructed an abrB2 overexpression mutant, by cloning the abrB2 protein-coding sequence into our temperature-controlled expression vector, pFC1, which replicates autonomously in Synechocystis and strongly produces the studied protein after 24 h of induction at 39°C (32, 38). As expected, the resulting cells strongly expressed abrB2 (27-fold more than noninduced cells) and concomitantly downregulated (at least 2-fold) hox expression and hydrogenase activity (Fig. 4). Furthermore, we showed that AbrB2 negatively regulated the activity of the hox promoter through binding to it (Fig. 5). Together, these novel findings demonstrate unambiguously that AbrB2 represses the hox operon. Furthermore, we found distant AbrB2-binding regions in the hox promoter region (Fig. 5), thereby suggesting the possible involvement of a DNA looping mechanism in the AbrB2-mediated repression of hox transcription. A similar hypothesis has been proposed for LexA-mediated regulation of the hox operon (36).
We also studied the expression and the regulation of the abrB2 gene. First, we mapped its transcription start site (see Fig. S1 in the supplemental material), and we analyzed its promoter through mutations and transcriptional fusions to the promoterless cat reporter gene of our promoter probe plasmid vector (27). We report that abrB2 is expressed from an atypical promoter harboring an extended −10 element (5′-TGTATAAT-3′) that compensates the absence of a −35 box (5′-TTGACA-3′) (Fig. 3), similarly to what we found previously for the secA gene (28). Confirming the biological significance of our results, we found the occurrence of an extended −10 promoter element and the absence of a −35 box to be two well-conserved features in the abrB2 genes from various cyanobacteria (Fig. 3). Also, interestingly, we found the abrB2 promoter to be about 3-fold more active in the abrB2 deletion mutant (131 CAT units) than in the WT strain (44 units), thereby demonstrating that AbrB2 negatively regulates expression of its own gene. Furthermore, we verified through EMSA analysis (Fig. 2) that AbrB2 binds on its own promoter, in agreement with a previous observation (19). Collectively, our data demonstrate unambiguously that AbrB2 is an autorepressor that also represses the hox operon.
Finally, when looking for a DNA motif that occurs in the abrB2 promoter and the hox promoter subfragments A, B, and D, which all bind AbrB2, but not in the C segment of the hox promoter, which does not bind AbrB2, we identified a consensus motif, TT(N5)AAC, as being possibly involved in AbrB2 binding (see Fig. S2 in the supplemental material). In agreement with this hypothesis, the mutation of the TT(N5)AAC motif (TTGAACAAAC to GGGAACAAAC), which overlaps the presumptive −35 box of the abrB2 promoter, appeared to increase (not decrease) the activity of the abrB2 promoter (Fig. 3).
We believe that our abrB2-deleted mutant with an improved hydrogenase activity and also our reporter plasmids for the analysis of the abrB2 gene and hox operon in various host strains with relevant genetic backgrounds will help in deciphering the regulation and the function of the hydrogen production machine, so as to improve it.
Supplementary Material
ACKNOWLEDGMENTS
J.D. and P.S. were, respectively, recipients of Ph.D. and postdoctoral fellowships from the CEA (France). This work was supported by the Agence Nationale de la Recherche Grants ANR-09-BIOE-002-01 (EngineeringH2cyano) and the CNRS (Centre National recherche scientifique) Programme Interdisciplinaire Energie PIE2 (Reprogramhydrogen), as well as the HélioBiotec platform, funded by the European Union, Région PACA, French Ministry of Research, and the CEA.
We thank Patrick Carrier and Pierre Richaud for their help in the MIMS analysis of the hydrogenase activity.
Footnotes
Published ahead of print 3 August 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abed RM, Dobretsov S, Sudesh K. 2009. Applications of cyanobacteria in biotechnology. J. Appl. Microbiol. 106:1–12 [DOI] [PubMed] [Google Scholar]
- 2. Angermayr SA, Hellingwerf KJ, Lindblad P, de Mattos MJ. 2009. Energy biotechnology with cyanobacteria. Curr. Opin. Biotechnol. 20:257–263 [DOI] [PubMed] [Google Scholar]
- 3. Aubert-Jousset E, Cano M, Guedeney G, Richaud P, Cournac L. 2011. Role of HoxE subunit in Synechocystis PCC6803 hydrogenase. FEBS J. 278:4035–4043 [DOI] [PubMed] [Google Scholar]
- 4. Barne KA, Bown JA, Busby SJ, Minchin SD. 1997. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the extended-10′ motif at promoters. EMBO J. 16:4034–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carrieri D, Wawrousek K, Eckert C, Yu J, Maness PC. 2011. The role of the bidirectional hydrogenase in cyanobacteria. Bioresour. Technol. 102:8368–8377 [DOI] [PubMed] [Google Scholar]
- 6. Coles M, et al. 2005. AbrB-like transcription factors assume a swapped hairpin fold that is evolutionarily related to double-psi beta barrels. Structure 13:919–928 [DOI] [PubMed] [Google Scholar]
- 7. Cournac L, Guedeney G, Peltier G, Vignais PM. 2004. Sustained photoevolution of molecular hydrogen in a mutant of Synechocystis sp. strain PCC 6803 deficient in the type I NADPH-dehydrogenase complex. J. Bacteriol. 186:1737–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deusch O, et al. 2008. Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol. Biol. Evol. 25:748–761 [DOI] [PubMed] [Google Scholar]
- 9. Domain F, Houot L, Chauvat F, Cassier-Chauvat C. 2004. Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 53:65–80 [DOI] [PubMed] [Google Scholar]
- 10. Ducat DC, Way JC, Silver PA. 2011. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 29:95–103 [DOI] [PubMed] [Google Scholar]
- 11. Garcin P, et al. 2012. A transcriptional-switch model for Slr1738-controlled gene expression in the cyanobacterium Synechocystis. BMC Struct. Biol. 12:1 doi:10.1186/1472-6807-12-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghirardi ML, Dubini A, Yu J, Maness PC. 2009. Photobiological hydrogen-producing systems. Chem. Soc. Rev. 38:52–61 [DOI] [PubMed] [Google Scholar]
- 13. Griese M, Lange C, Soppa J. 2011. Ploidy in cyanobacteria. FEMS Microbiol. Lett. 323:124–131 [DOI] [PubMed] [Google Scholar]
- 14. Grigorieva G, Shestakov S. 1982. Transformation in the cyanobacterium Synechocystis sp. 6803. FEMS Microbiol. Lett. 13:367–370 [Google Scholar]
- 15. Gutekunst K, et al. 2005. LexA regulates the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 as a transcription activator. Mol. Microbiol. 58:810–823 [DOI] [PubMed] [Google Scholar]
- 16. Hawley DK, McClure WR. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924–932 [DOI] [PubMed] [Google Scholar]
- 18. Houot L, et al. 2007. Cadmium triggers an integrated reprogramming of the metabolism of Synechocystis PCC6803, under the control of the Slr1738 regulator. BMC Genomics 8:350 doi:10.1186/1471-2164-8-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishii A, Hihara Y. 2008. An AbrB-like transcriptional regulator, Sll0822, is essential for the activation of nitrogen-regulated genes in Synechocystis sp. PCC 6803. Plant Physiol. 148:660–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanesaki Y, et al. 2012. Identification of substrain-specific Mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res. 19:67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kiss E, Kos PB, Vass I. 2009. Transcriptional regulation of the bidirectional hydrogenase in the cyanobacterium Synechocystis 6803. J. Biotechnol. 142:31–37 [DOI] [PubMed] [Google Scholar]
- 22. Kruse O, Rupprecht J, Mussgnug JH, Dismukes GC, Hankamer B. 2005. Photosynthesis: a blueprint for solar energy capture and biohydrogen production technologies. Photochem. Photobiol. Sci. 4:957–970 [DOI] [PubMed] [Google Scholar]
- 23. Labarre J, Chauvat F, Thuriaux P. 1989. Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the cyanobacterium Synechocystis strain PCC 6803. J. Bacteriol. 171:3449–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lieman-Hurwitz J, et al. 2009. A cyanobacterial AbrB-like protein affects the apparent photosynthetic affinity for CO2 by modulating low-CO2-induced gene expression. Environ. Microbiol. 11:927–936 [DOI] [PubMed] [Google Scholar]
- 25. Liu X, Fallon S, Sheng J, Curtiss R., III 2011. CO2-limitation-inducible green recovery of fatty acids from cyanobacterial biomass. Proc. Natl. Acad. Sci. U. S. A. 108:6905–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maeda T, Sanchez-Torres V, Wood TK. 2008. Metabolic engineering to enhance bacterial hydrogen production. Microb. Biotechnol. 1:30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marraccini P, Bulteau S, Cassier-Chauvat C, Mermet-Bouvier P, Chauvat F. 1993. A conjugative plasmid vector for promoter analysis in several cyanobacteria of the genera Synechococcus and Synechocystis. Plant Mol. Biol. 23:905–909 [DOI] [PubMed] [Google Scholar]
- 28. Mazouni K, Bulteau S, Cassier-Chauvat C, Chauvat F. 1998. Promoter element spacing controls basal expression and light inducibility of the cyanobacterial secA gene. Mol. Microbiol. 30:1113–1122 [DOI] [PubMed] [Google Scholar]
- 29. Mazouni K, Domain F, Cassier-Chauvat C, Chauvat F. 2004. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol. Microbiol. 52:1145–1158 [DOI] [PubMed] [Google Scholar]
- 30. McIntosh CL, Germer F, Schulz R, Appel J, Jones AK. 2011. The [NiFe]-hydrogenase of the cyanobacterium Synechocystis sp. PCC 6803 works bidirectionally with a bias to H2 production. J. Am. Chem. Soc. 133:11308–11319 [DOI] [PubMed] [Google Scholar]
- 31. Mermet-Bouvier P, Cassier-Chauvat C, Marraccini P, Chauvat F. 1993. Transfer and replication of RSF1010-derived plasmids in several cyanobacteria of the genera Synechocystis and Synechococcus. Curr. Microbiol. 27:323–327 [Google Scholar]
- 32. Mermet-Bouvier P, Chauvat F. 1994. A conditional expression vector for the cyanobacteria Synechocystis sp. strains PCC6803 and PCC6714 or Synechococcus sp. strains PCC7942 and PCC6301. Curr. Microbiol. 28:145–148 [DOI] [PubMed] [Google Scholar]
- 33. Mulkidjanian AY, et al. 2006. The cyanobacterial genome core and the origin of photosynthesis. Proc. Natl. Acad. Sci. U. S. A. 103:13126–13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oliveira P, Lindblad P. 2008. An AbrB-Like protein regulates the expression of the bidirectional hydrogenase in Synechocystis sp. strain PCC 6803. J. Bacteriol. 190:1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oliveira P, Lindblad P. 2005. LexA, a transcription regulator binding in the promoter region of the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 251:59–66 [DOI] [PubMed] [Google Scholar]
- 36. Oliveira P, Lindblad P. 2009. Transcriptional regulation of the cyanobacterial bidirectional Hox-hydrogenase. Dalton Trans. 2009:9990–9996 [DOI] [PubMed] [Google Scholar]
- 37. Paumann M, Regelsberger G, Obinger C, Peschek GA. 2005. The bioenergetic role of dioxygen and the terminal oxidase(s) in cyanobacteria. Biochim. Biophys. Acta 1707:231–253 [DOI] [PubMed] [Google Scholar]
- 38. Poncelet M, Cassier-Chauvat C, Leschelle X, Bottin H, Chauvat F. 1998. Targeted deletion and mutational analysis of the essential (2Fe-2S) plant-like ferredoxin in Synechocystis PCC6803 by plasmid shuffling. Mol. Microbiol. 28:813–821 [DOI] [PubMed] [Google Scholar]
- 39. Rasmussen B, Fletcher IR, Brocks JJ, Kilburn MR. 2008. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature 455:1101–1104 [DOI] [PubMed] [Google Scholar]
- 40. Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strains histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1–61 [Google Scholar]
- 41. Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. 2010. Microbial biosynthesis of alkanes. Science 329:559–562 [DOI] [PubMed] [Google Scholar]
- 42. Scotto-Lavino E, Du G, Frohman MA. 2006. Amplification of 5′ end cDNA with ‘new RACE.’ Nat. Protoc. 1:3056–3061 [DOI] [PubMed] [Google Scholar]
- 43. Sheng J, Vannela R, Rittmann BE. 2011. Evaluation of methods to extract and quantify lipids from Synechocystis PCC 6803. Bioresour. Technol. 102:1697–1703 [DOI] [PubMed] [Google Scholar]
- 44. Shi T, Falkowski PG. 2008. Genome evolution in cyanobacteria: the stable core and the variable shell. Proc. Natl. Acad. Sci. U. S. A. 105:2510–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soni B, Houot L, Cassier-Chauvat C, Chauvat F. 2012. Prominent role of the three Synechocystis PchR-like regulators in the defense against metal and oxidative stresses. Open J. Biochem. 1:1 http://www.rossscience.org/ojbb/articles/2227-7021-1-1.htm [Google Scholar]
- 46. Sybirna K, et al. 2008. Shewanella oneidensis: a new and efficient system for expression and maturation of heterologous [Fe-Fe] hydrogenase from Chlamydomonas reinhardtii. BMC Biotechnol. 8:73 doi:10.1186/1472-6750-8-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tamagnini P, et al. 2007. Cyanobacterial hydrogenases: diversity, regulation and applications. FEMS Microbiol. Rev. 31:692–720 [DOI] [PubMed] [Google Scholar]
- 48. Wang R, Healey FP, Myers J. 1971. Amperometric measurement of hydrogen evolution in chlamydomonas. Plant Physiol. 48:108–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Waterbury JB, Warson SW, Guillard RRL, Brand JE. 1979. Widerspread occurrence of a unicellular, marine, planktonic, cyanobacterium. Nature 277:293–294 [Google Scholar]
- 50. Williams PG. 2009. Panning for chemical gold: marine bacteria as a source of new therapeutics. Trends Biotechnol. 27:45–52 [DOI] [PubMed] [Google Scholar]
- 51. Yamauchi Y, Kaniya Y, Kaneko Y, Hihara Y. 2011. Physiological roles of the cyAbrB transcriptional regulator pair Sll0822 and Sll0359 in Synechocystis sp. strain PCC 6803. J. Bacteriol. 193:3702–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zehr JP. 2011. Nitrogen fixation by marine cyanobacteria. Trends Microbiol. 19:162–173 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.