Abstract
Nipah virus (NiV) is a highly pathogenic, negative-strand RNA paramyxovirus that has recently emerged from flying foxes to cause serious human disease. We have analyzed the role of the nonstructural NiV C protein in viral immunopathogenesis using recombinant virus lacking the expression of NiV C (NiVΔC). While wild-type NiV was highly pathogenic in the hamster animal model, NiVΔC was strongly attenuated. Replication of NiVΔC was followed by the production of NiV-specific antibodies and associated with higher recruitment of inflammatory cells and less intensive histopathological lesions in different organs than in wild-type-NiV-infected animals. To analyze the molecular basis of NiVΔC attenuation, we studied early changes in gene expression in infected primary human endothelial cells, a major cellular target of NiV infection. The transcriptomic approach revealed the striking difference between wild-type and mutant NiV in the expression of genes involved in immunity, with the particularly interesting differential patterns of proinflammatory cytokines. Compared to wild-type virus, NiVΔC induced increased expression of interleukin 1 beta (IL-1β), IL-8, CXCL2, CXCL3, CXCL6, CCL20, and beta interferon. Furthermore, the expression of NiV C in stably transfected cells decreased the production of the same panel of cytokines, revealing a role of the C protein in the regulation of cytokine balance. Together, these results suggest that NiV C regulates expression of proinflammatory cytokines, therefore providing a signal responsible for the coordination of leukocyte recruitment and the chemokine-induced immune response and controlling the lethal outcome of the infection.
INTRODUCTION
Nipah virus (NiV) is a recently emergent, highly pathogenic, zoonotic paramyxovirus, first recognized following a 1998-1999 outbreak of severe febrile encephalitis in Malaysia and Singapore (9, 61). Subsequent outbreaks of NiV occurred in India and almost annually in Bangladesh (6, 32, 53). The large Malaysian outbreak was marked by severe, fatal encephalitis, with about 40% mortality (11), whereas the smaller, more recent Bangladeshi and Indian outbreaks displayed higher mortality rates (75 to 92%), potential human-to-human transmission, and an increased occurrence of severe respiratory disease (6, 23, 31). In addition to its high lethality, NiV exhibits a broad host range and is able to infect bats, pigs, humans, cats, dogs, and other species (9, 10, 46), probably due to the use of highly conserved ephrin B2 and B3 receptors (3). NiV is closely related to Hendra virus, another zoonotic paramyxovirus, which had emerged in Australia in 1994 (9, 25, 26). These two viruses constitute the recently recognized genus Henipavirus (41).
The NiV genome is composed of a negative-sense, nonsegmented RNA strand 18,246 nucleotides (nt) in length for the Malaysian isolate and 18,252 nt for the Bangladeshi isolate (24–26). The genome contains six transcription units encoding six viral structural proteins and three predicted nonstructural proteins, C, V, and W. As in other paramyxoviruses, the C protein of NiV (166 amino acids [aa]) is expressed from an alternative open reading frame within the phosphoprotein (P) gene, whereas the V and W proteins are expressed by transcriptional RNA editing (15). At a unique, highly conserved RNA-editing site in the P gene, the viral polymerase inserts a single, nontemplated G residue that results in a frameshift and the expression of the V protein. Insertion of two nontemplated G residues results in expression of the W protein (9, 25, 26). While the C proteins of NiV and other paramyxoviruses are unique and share no sequence similarity with the P protein (15), the V and W proteins share an amino-terminal 407-aa domain with P, and each possesses a unique carboxyl-terminal domain consisting of 52 aa for V and 47 aa for W (9, 25, 26).
The majority of these alternative P-gene products have been shown to function as inhibitors of the host innate immune response (12, 20, 21, 30). The NiV V and W proteins counteract interferon signaling by interacting with STAT1 and preventing its activation in transfected cells (2, 48, 51). NiV V, like other paramyxovirus V proteins, binds to mda-5 and thereby inhibits the downstream signaling events leading to beta interferon (IFN-β) synthesis (8). NiV W can also prevent IFN production through an unidentified mechanism that requires its nuclear localization and inhibits the Toll-like receptor 3 (TLR3) signaling pathway (50). The NiV C protein has also been shown to inhibit the activation of an antiviral state, but the mechanism is unknown (43). Together those studies, as well as experiments showing that the C, V, and W proteins can inhibit replication of a minigenome (52), have been performed using plasmid-based expression analysis. Finally, using the recombinant NiV, lacking the expression of accessory proteins, NiV V and C were shown to be important in viral virulence, but in contrast to the other paramyxoviruses, none of the three NiV nonstructural proteins seemed to affect interferon signaling in infected cells (63), leaving the mechanism of their action unclear.
To assess the role of the C protein in NiV pathogenesis, we generated recombinant viruses lacking the expression of C and analyzed the molecular basis of decreased virus virulence. Study of the differences in the transcriptome signature between the wild-type (wt) and mutant viruses revealed modulation of proinflammatory cytokine expression in cells infected by the mutant virus. Furthermore, cells stably expressing NiV C have a significantly decreased capacity to produce proinflammatory cytokines, and hamsters infected with recombinant virus lacking the expression of NiV C (NiVΔC) developed successful inflammatory reactions and anti-NiV immune responses, thus limiting viral infection. The obtained results suggest an important role of the C protein in NiV pathogenesis by modulating the production of early-induced proinflammatory chemokines, leading consequently to an inappropriate immune response and lethal outcome of the infection.
MATERIALS AND METHODS
Cell culture.
Primary macrovascular human umbilical vein endothelial cells (HUVEC) were isolated from human umbilical cords by treating the umbilical vein with 0.1% collagenase for 30 min at 37°C as described previously (33, 39). Umbilical cords were obtained between 2006 and 2009 from healthy full-term newborns with written parental informed consent, according to the guidelines of the medical and ethical committees of Hospices Civils de Lyon and of Agence de Biomedecine, Paris, France. Cells from at least three donors were pooled for experiments. Cells were cultured in flasks coated with 0.2% gelatin in complete medium containing M199 medium (Gibco), 20% serum (Gibco), 100 μg/ml bovine brain extract (5), 14 mM HEPES (Gibco), 10 IU/ml heparin (Pfizer), and an antibiotic/antimycotic cocktail (Gibco). At passage 4, cells were seeded at 30,000 cells/cm2 for 8 h and then serum deprived for 16 h prior to NiV infection in complete medium. VeroE6 and HEK293 cells (derived from human embryonic kidneys, with assumed neuronal origin [49]) were maintained in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin, 0.1 mg streptomycin, 10 mM HEPES and 2 mM l-glutamine at 37°C in 5% CO2. BSR-T7/5 cells, a BHK-21 cell line stably expressing T7 RNA polymerase, were grown in Glasgow medium (Invitrogen, Carlsbad, CA) supplemented with 10% FCS, 29.5 g/liter tryptose phosphate broth (Eurobio, Paris, France), 2× nonessential amino acid mixture (Invitrogen), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, and 0.5 mg/ml Geneticin (Gibco). Activation of HEK293 cells was performed by transfection of poly(I·C) (Sigma) (10 ng/well of 6-well plates) using an oligofectamine reagent kit (Invitrogen) according to the manufacturer's recommendations.
Viruses, cell infection and virus titration.
Nipah virus (isolate UMMC1; GenBank AY029767) (7) was prepared on Vero-E6 cells as described previously (22) and used at a multiplicity of infection (MOI) of 1 to infect cells in vitro. To generate recombinant NiV, all recombinant plasmids containing full-length cDNA copies of Nipah virus were generated as described elsewhere (12). Specifically, the 18,246-nucleotide (nt) genome (corresponding to GenBank accession no. AY029767) was cloned into the pSL1180 cloning vector, resulting in a full-length cDNA clone, pFL-NiV-WT. T7 promoter and terminator sequences and hepatitis delta virus ribozyme sequences were appended via PCR. To knock out the C protein, two alternative constructs were generated. Plasmid pFL-NiV-ΔC1 was generated by mutating the two initiating methionine codons in the C open reading frame (ORF) to ACG. To further ensure the knockout of this ORF, a stop codon was also introduced into the C ORF, replacing the 4th codon coding for a serine residue. All nucleotide changes in the C ORF did not affect the P, V, or W ORF. In the pFL-NiV-ΔC2 construct, the 6th codon in the C ORF was replaced by a stop codon. To recover the recombinant viruses, BSR-T7/5 cells were cotransfected with plasmids encoding full-length NiV antigenomes (pFL-NiV-WT, pFL-NiV-ΔC1, and pFL-NiV-ΔC2) and pTM1 N, P, and L. At 72 h posttransfection, virus-containing medium was harvested from transfected cells and passaged onto Vero E6 cells. To generate virus stocks, Vero E6 cells were inoculated with virus at an MOI of 0.01. The virus-containing medium was harvested at 48 h postinfection (p.i.), clarified by low-speed centrifugation, aliquoted, and stored at −70°C. To confirm the presence of the introduced mutations, viral RNA was isolated from culture medium of Vero E6 cells infected with either virus, cDNA was generated and amplified by reverse transcription-PCR (RT-PCR), and the complete viral genome was sequenced.
HUVECs were infected with either wild-type (wt) or recombinant NiV, and at indicated times p.i., 150 μl of cell culture supernatant was collected and frozen and viral titration was performed as detailed elsewhere (39).
Infection of hamsters.
Eight-week-old golden hamsters (Mesocricetus auratus, Janvier, France), 5 to 6 animals/group, were anesthetized and infected intraperitoneally with 0.4 ml of wt NiV (NiVwt), NiVΔC1, and NiVΔC2 in the biosafety level 4 (BSL-4) laboratory. Some experiments were performed using UV-irradiated NiVΔC, where inactivation was achieved by a 30-min exposure at 4°C to UV irradiation from s 254-nm UV lamp. Surviving animals were bled on 0 and 21 days p.i., and the sera were frozen at −80°C until tested by enzyme-linked immunosorbent assay (ELISA). To study the pathology induced by NiV, tissue specimens (brain, lung, bladder, spleen, kidney, and serum) were collected daily over 6 days p.i. Tissues were frozen at −80°C for RT-PCR analysis for virus isolation or ELISA. For histopathological studies, tissues were fixed in 4% paraformaldehyde. All animals were handled in strict accordance with good practices as defined by the French national charter on the ethics of animal experimentation. Animal work was approved by the regional ethical committee (CREEA), and experiments were performed in the INSERM Jean Mérieux BSL-4 laboratory in Lyon, France.
Generation of stably transfected HEK293C (293C) cells.
An expression plasmid carrying NiV C was created using the Invitrogen Gateway cloning system. Primers were designed using the Vector NTI software program (Invitrogen) to encode attB1.1 and attB2.1 recombination sequences. The stop codon of the NiV C gene was removed to allow the addition of tags: NiV C for, GGGACAACTTTGTACAAAAAAGTTGGCATGATGGCCTCAATATTATTG; NiV C rev, GGGACAACTTTGTACAAAAAAGTTGGGATCAGTTTCCCGATCTTCTTT. PCR products, obtained from cDNA, coming from NiVwt-infected Vero cells, were migrated in a 1.5% agarose gel and purified by using a NucleoSpinExtract II kit (Macherey-Nagel). The correct sequence was then cloned into the entry vector pDONOR223 using GatewayBP Clonase II enzyme mix (Invitrogen). The corresponding pDONOR 223-Niv C was used to recombine with pcDNA-DEST40 using GatewayLR Clonase II enzyme mix (Invitrogen). The pcDNA-DEST40 expression vector provides the opportunity to clone a sequence as a fusion protein with a C-terminal V5 epitope and a poly-His tag to allow purification and detection. The obtained plasmid was sequenced (Eurofins MWG Operon) and analyzed using the Vector NTI software program (Invitrogen). Subconfluent HEK293 cells were then transfected with 1 μg of pcDNA-DEST40-NiVC using Lipofectamine (Invitrogen), and transfectants were selected in a medium containing 600 μg/ml of G418 for 21 days. Several stable clones were then obtained by limiting dilution in the presence of G418.
Western blot analysis.
Vero E6 cells were inoculated with recombinant and wild-type viruses at an MOI of 0.05. Culture supernatants and cell lysates were collected at 48 h postinfection and analyzed by Western blot analysis using rabbit polyclonal antibodies specific for the NiV proteins M, G, and C and to cellular actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abcam). Expression of the NiV C protein fused with the V5 epitope and a poly(His) tag in HEK293 cells was analyzed by Western blotting using either rabbit polyclonal serum or anti-V5 monoclonal antibody (MAb) (Invitrogen).
Immunofluorescence.
Cells were attached on 6-well plate glass coverslips (400,000 cells/well), and left at 37°C, 5% CO2, for 24 h; medium was then removed, and cells were washed with phosphate-buffered saline (PBS). Fixation was performed using 4% paraformaldehyde (PFA) in PBS (1 ml/well, 10 min) at room temperature, followed by a wash with PBS. Cells were then permeabilized using Triton X-100 (0.1% in PBS, 3 to 5 min, at room temperature) and washed again. Blocking was performed using 3% bovine serum albumin (BSA) in PBS for 30 min at room temperature. Cells were then incubated with primary rabbit polyclonal anti-NiV C antibody (Covalab) for 1 to 2 h at 37°C in humid chambers. After 3 washes in PBS (5 min each), cells were incubated with secondary anti-rabbit IgG fluorescein isothiocyanate (FITC) conjugate (Sigma) and 4′,6-diamidino-2-phenylindole (DAPI) (0.1 to 1 μg/ml) for 30 min at 37°C in wet chambers. After an additional 3 washes in PBS and 1 wash in water, mounting was performed using Fluoromount-G medium (Southern Biotech). All slides were analyzed using an Axioscope microscope equipped with Zeiss Axiovision software (Zeiss Germany).
Immunohistochemistry.
Paraformaldehyde-fixed tissues were trimmed and routinely processed. Paraffin sections, 5 μm thick, were placed on silanized slides and deparaffinized by xylene and serial ethanol washes. The sections were routinely stained with hematoxylin and eosin and examined by light microscopy. For immunohistochemistry, antigens were retrieved by microwaving the sections in citrate buffer, pH 6.0, at 99°C for 20 min. Endogenous peroxidase was blocked with 0.6% H2O2 in methanol for 20 min. The primary antibody was a mouse polyclonal, cross-reacting, anti-Hendra virus antibody used at a 1:2,000 dilution (38, 61). Positive signals were revealed by linking secondary antimouse antibodies and diaminobenzidine as the substrate. The slides were then stained with hematoxylin and coverslipped.
Microarray analysis.
HUVECs were prepared from two different pools, each containing 3 donors, cultured under the same conditions and infected with either NiVwt or NiVΔC (MOI of 1) or mock treated. Total RNA was extracted 8 h p.i. using an RNeasy kit (Qiagen) according to the manufacturer's protocol. Hybridization was performed on Codelink Uniset Human Whole Genome bioarrays (http://www.codelinkbioarrays.com), as described previously (39). Data extraction and raw data normalization were performed using the CodeLink Gene Expression Analysis v4.0 software program. Obtained data sets were deposited in the GEO database in accord with MIAME guidelines (accession numbers GSM813064, GSM813065, GSM813066, GSM813067, GSM814496, and GSM812297). Analysis of differentially expressed genes was performed by comparisons of mock versus NiVwt-infected and mock versus NiVΔC-infected groups for each of 2 independent experiments. Finally, data were converted to the Excel format, and data analysis was performed by using the Gene Spring v7.0 software program from Agilent.
Quantitative reverse transcription-PCR (RT-qPCR).
Total RNA was extracted from mock- and NiV-infected HUVECs (MOI = 1) 8 and 24 h postinfection using the RNeasy minikit according to the manufacturer's instructions, including additional step with RNase-free DNase (Qiagen). RNA was isolated from hamster organs (10 to 30 mg) using a tissue lyser (Qiagen), and reverse transcriptions and quantitative PCR were performed as described previously (39, 40). All samples were run in duplicate, and results were analyzed using the software program ABI Prism 7000 SDS. The GAPDH gene was used as a housekeeping gene for mRNA quantification and normalization. GAPDH and standard references for the corresponding genes were included in each run to standardize results in respect to RNA integrity, loaded quantity, and inter-PCR variations. Primers were designed using the Beacon 7.0 software program and validated for their efficacy close to 100%: NiV N forward (for), GGCAGGATTCTTCGCAACCATC; NiV N reverse (rev), GGCTCTTGGGCCAATTTCTCTG; IL-1β for, GCCTCTCTCACCTCTCCTACTC; IL-1β rev, ATCAGAATGTGGGAGCGAATGAC; CXCL2 for, GAAGGTTTGCAGATATTCTCTAGTC; CXCL2 rev, GCCGTCACATTGATCTTACTGG; CXCL3 for, AGCACCAACTGACAGGAGAG; CXCL3 rev, ACACATTAAGTCCCTTTCCAGC; CXCL6 for, AACGGGAAGCAAGTTTGTCTGG; CXCL6 rev, TCTTACTGGGTCCAGGGATCTCC; CXCL8 for, TGCACGGGAGAATATACAAATAGC; CXCL8 rev, TCTAGCAAACCCATTCAATTCCTG; CXCL10 for, GGAAGGTTAATGTTCATCATCCTAAGC; CXCL10 rev, TAGTACCCTTGGAAGATGGGAAAG; CXCL11 for, GGATGAAAGGTGGGTGAAAGGAC; CXCL11 rev, AACGTGAAAGCACTTTGTAAACTCC;CCL20 for, CCAAAGAACTGGGTACTCAACACTG; CCL20 rev, AGTTGCTTGCTGCTTCTGATTCG; IFN-β for, CTCCTAGCCTGTGCCTCTGG; IFN-β rev, TGCAGTACATTAGCCATCAGTCAC; human GAPDH for, CACCCACTCCTCCACCTTTGAC; human GAPDH rev, GTCCACCACCCTGTTGCTGTAG. Primers used for hamster study were the following: murine GAPDH for, GCATGGCCTTCCGTGTCC; murine GAPDH rev, TGTCATCATACTTGGCAGGTTTCT. Calculations were done using the 2ΔΔCT model, according to the MIQE guidelines, as described previously (39).
ELISA analysis.
Supernatants of mock- and NiV-infected HUVECs were collected 8, 24, 48 h p.i. and used to quantify the induction of interleukin 8 (IL-8) using the IL-8 Human Direct ELISA kit (Invitrogen) according to the manufacturer's instructions. All tests were performed with 2 sets of 3 donors, in duplicate.
To determine NiV-specific antibodies in serum, microtiter plates (Dominic Dutscher) were coated with 100 μl/well of NiV antigen, prepared as described previously (22), and incubated overnight at 4°C. Wells were blocked with PBS (1×)-milk (5%) for 30 min at 37°C and washed with PBS containing 0.05% Tween 20 between each step. Twofold dilutions of sera were incubated at 4°C overnight, and the reaction was revealed with the appropriate anti-hamster immunoglobulin G (IgG) (gamma specific) (Sigma-Aldrich, France) conjugated to peroxidase for 2 h at room temperature, followed by incubation with the Sigmafast OPD substrate system (Sigma-Aldrich, France) and stopped with sulfuric acid (12%). The optical density was read at 492 nm, and the ELISA titer was determined as the reciprocal value of the last serum dilutions giving the optical density reading above the background.
Statistical analysis.
Data were expressed as means ± standard deviations (SD) or as a percentage of survival. Statistical analyses were performed using the Mann-Whitney U test, chi-square test, and two-way analysis of variance (ANOVA) with Bonferroni's correction test, using the GraphPad Prism4 software program.
RESULTS
Generation and characterization of recombinant NiV deficient for C protein expression.
To investigate the role of the NiV C protein, we generated two recombinant viruses, carrying mutations that do not allow C expression, using our previously established reverse genetics system for constructed full-length NiV cDNA (12).
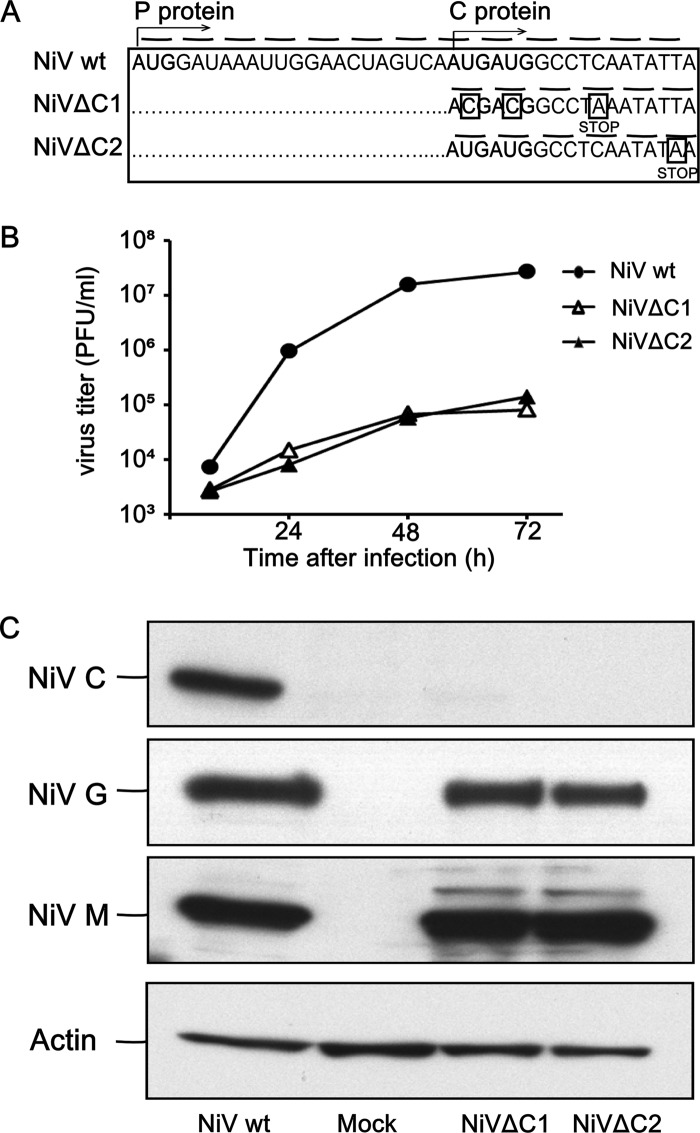
The expression of the C protein was abrogated in two alternative ways (Fig. 1A). In NiVΔC1, synthesis of C was blocked due to a replacement of two translation-initiating tandem AUG codon by ACG and by introduction of a translational stop codon which replaced the 4th codon. Alternatively, in NiVΔC2, synthesis of C was abrogated by a translational stop codon replacing the 6th codon in the C ORF. In both constructions, the introduced mutations did not affect the ORF of the P protein, which is in a +1 frame in comparison to the C ORF (Fig. 1A). The substitutions introduced in the P gene were further confirmed by sequencing of the RT-PCR products. In accord with previously published data (12, 63), both NiVΔC viruses showed a reduced replication capacity in primary human endothelial cells (HUVEC) (Fig. 1B). Western blot analysis of Vero E6 cells infected at an MOI of 0.05 with either NiVwt or mutated NiVs revealed that the synthesis of the C protein was eliminated in the case of both mutants (Fig. 1C).
Fig 1.
Generation of NiVΔC recombinant viruses. (A) Schematic illustration of mutation introduced into the viral genome in order to eliminate expression of the C protein. Nucleotide changes designed to knock out the expression of the C ORF are put in squares, and introduced stop codons are indicated. (B) Kinetic analysis of the production of infectious particles in the supernatant fluids of HUVEC infected with NiVwt, NiVΔC1, or NiVΔC2 (MOI = 0.01). (C) Vero E6 cells infected with NiVwt, NiVΔC1, or NiVΔC2 were harvested at 48 h p.i. and analyzed by Western blotting, using antibodies specific for the NiV proteins G, M, and C and actin.
We then evaluated the pathogenicity of recombinant viruses in the hamster model, which closely reproduces human NiV disease (60). Animals were inoculated intraperitoneally (i.p.) with either recombinant wild-type NiV (NiVwt) or one of the NiVΔC viruses (Fig. 2). Hamsters infected with either 100 or 1,000 PFU of NiVwt died within 6 days p.i. Nevertheless, an abrogation of C protein expression in both recombinant viruses dramatically reduced virus virulence, and some animals survived the infection even with the highest infectious dose of 1,000 PFU (Fig. 2B and C). These results demonstrated the decreased virulence of both rescued NiVs lacking C protein. NiVΔC2 was selected for additional analysis and is referred to in the text below as NiVΔC.
Fig 2.
Survival curves of hamsters infected with different doses of recombinant NiV. Hamsters (5 or 6 per group) were inoculated i.p. with indicated doses of NiVwt (A), NiVΔC1 (B), or NiVΔC2 (C). Animals were followed for signs of infection during 3 weeks. The results are expressed as percentages of surviving animals in each group. Differences in the rate of survival are statistically significant between NiVwt and NiVΔC for all groups injected with the same dose of virus (P < 0.01, chi-square test), except for the group injected with 1,000 PFU of NiVΔC1, which was not statistically different from the NiVwt group. Differences in virulence between NiVΔC1 and NiVΔC2 were not statistically significant.
NiVΔC infection induces an efficient proinflammatory and adaptive immune response.
We next analyzed kinetics of virus replication by RT-qPCR and immunohistology in different tissues of NiVwt- and NiVΔC-infected hamsters (Fig. 3). While NiV N expression was found regularly in all tissues from NiVwt-infected hamsters at each time point before the death of all infected animals on day 6 p.i., in NiVΔC-infected hamsters it was detected occasionally and at a lower level (Fig. 3A). Both viruses induced production of equivalent levels of NiV N transcripts in the brains of hamsters infected on day 1 p.i. (between 220 and 390 copies/μg of total RNA), although NiV replication in lungs and spleen was lower in NiVΔC-infected animals. Low levels of NiV N were still detected on day 5 p.i. in all analyzed organs from NiVΔC-infected hamsters, suggesting the existence of the residual virus without significant pathological effect, possibly due to the control of the adaptive immune response developed in the meantime. Histological analysis showed mild to moderate inflammation in some tissues from both NiVwt- and NiVΔC-infected animals, with the exception of the brain, where inflammation was not detected in NiVΔC-infected hamsters. More-detailed histological analysis of the type of inflammation suggested differences between NiVwt- and NiVΔC-infected organs. In the lung tissues, NiVwt induced moderate inflammation with the presence of necrosis and edema (Fig. 3C), while NiVΔC induced stronger inflammation without necrosis or edema (Fig. 3D). In the other organs, vasculitis and adjacent extravascular inflammation were more prominent in NiVwt infection. Viral antigens were found in organs from NiVwt-infected animals in most cases, starting from 3 days p.i., in a pattern similar to what has been shown in previous publications (60), but these were not found with NiVΔC.
Fig 3.
Immunopathogenesis of NiV replication in hamsters. (A) Kinetics of NiV replication in the organs of infected hamsters determined by RT-qPCR. Hamsters were infected i.p. with either 105 PFU of NiVwt (white histograms) or NiVΔC (black histograms), and every day p.i., 1 to 3 animals were euthanized and organs were analyzed by RT-qPCR, as described in Materials and Methods. The average NiV N RNA expression in indicated organs is shown, and bars present standard deviations. (B to D) Generation of inflammatory reaction in infected animals. Lung tissues from hamsters were infected with either mock (B), 105 PFU of NiVwt (C), or 105 PFU of NiVΔC and taken 5 days p.i. (D). Occasional nodular inflammation, edema, and necrosis (arrow) are observed (C), and more intense inflammation and cell infiltration with no edema or necrosis are seen (D). (E) Production of NiV-specific antibodies in the serum of hamsters infected with different doses of NiVΔC or UV-irradiated NiVΔC. Twenty-nine days after infection, serum was taken and analyzed by ELISA for the presence of virus-specific antibodies. Sera taken from animals before infection were always negative by ELISA. Histograms correspond to the average titer obtained from 3 to 6 animals, and bars present standard deviations.
Finally, we analyzed the generation of NiV-specific antibodies in infected animals by specific ELISA on 29 days p.i. (Fig. 3E). Because NiVwt induces rapid lethality in hamsters by day 6, specific antibodies were hardly detectable by that time point. However, in hamsters infected with different doses of NiVΔC, virus-specific antibodies were readily detected in the serum of surviving animals 29 days p.i. When NiVΔC was UV inactivated and the equivalent number of virus particles was injected into hamsters, production of NiV-specific antibodies was scarcely detectible, confirming the requirement for NiVΔC replication in animals for the generation of an adaptive immune response and production of virus-specific antibodies. Together, these results strongly suggest that in contrast to NiVwt, NiVΔC induces an efficient proinflammatory reaction and an adequate adaptive immune response that likely controls the infection.
Analysis of gene expression in endothelial cells infected with NiV and NiVΔC.
To better understand the function of the accessory NiV C protein, we have used a microarray approach to study early changes in the gene expression pattern in infected primary endothelial cells, HUVEC. Cells were infected by either NiVwt or NiVΔC, and RNA was extracted 8 h after infection and subjected to microarray analysis. The modulatory effect of NiV on gene expression was determined by permutation analysis, and we considered significant a minimal fold change of 1.3. Among the 55,000 targeted genes, 806 genes were found to be differentially expressed in NiVwt-infected cells in comparison to expression in noninfected cells, including 537 upregulated genes (fold change [FC] of ≥1.3 and ≤46) and 269 downregulated genes (FC of ≥−43 and ≤−1.3). Infection with NiVΔC modulated the expression of 1,343 genes, with 701 upregulated genes (FC of ≥1.3 and ≤46) and 642 downregulated (FC of ≥−43 and ≤−1.3) genes. Genes modulated after NiVwt and NiVΔC infection were then classified according to their gene ontology (GO) biological processes and their GO molecular functions and the biological functions of these genes were analyzed using the Ingenuity Pathway Analysis software program. The most importantly modulated cellular functions were those belonging to “immune response,” “organism injury,” and “cells signaling” [−log(P value) between 6.5 and 13]. We then concentrated our attention on those genes within the “immune response” and “cell signaling” groups which were differentially expressed after NiVwt and NiVΔC infection. Permutational analysis revealed particularly interesting differential expression patterns with inflammatory cytokines and chemokines. Although both viruses upregulated CCL8, CXCL10 and -11, and lambda 1 IFN (IFN-λ1), NiVΔC specifically increased production of interleukin 1 beta (IL-1β), IL-8, CXCL2, CXCL3, CXCL6, and CCL20 compared to results for NiVwt (Table 1). Only 2 cytokines were downregulated by NiVΔC, CCL7 and IL-16, although the differential effect was quite low.
Table 1.
Modulation of chemokine expression in HUVEC 8 h after either NiVwt or NiVΔC infection
| Profile and chemokine/cytokinea | GenBank accession no. | FC in expressionb |
Known receptor(s) | Recruited cellsc | |
|---|---|---|---|---|---|
| NiVwt | NiVΔC | ||||
| A | |||||
| CXCL10 (IP-10) | NM_001565 | 10.93 | 14.7 | CXCR3 | TH1, NK, Ms |
| CXCL11 (I-TAC) | NM_005409 | 3.54 | 3.7 | CXCR3 | TH1, NK, Ms |
| CCL8 (MCP-2) | NM_005623 | 4.88 | 5.07 | CCR3 | Bs, Mo, T, NK, DC |
| IL-29 (interferon λ1) | NM_172140 | 1.38 | 1.49 | Interferon type III receptor | Antiviral activity |
| B | |||||
| CCL3 (MIP1a) | BI837082 | −1.27 | 1.38 | CCR1, CCR5 | Eo, Bs, Mo, T, NK, DC |
| CCL5 (RANTES) | NM_002985 | 1.3 | 2.09 | CCR1, CCR3, CCR5 | Eo, Bs, Mo, T, NK, DC, Ms |
| CCL20 (MIP3a) | NM_004591 | −1.02 | 2.73 | CCR6 | Tm, B, DC |
| CXCL2 (MIP2a) | NM_002089 | 1.2 | 2.73 | CXCR2 | Neu, Mo |
| CXCL3 (MIP2β) | NM_002090 | 1.19 | 2.82 | CXCR2 | Neu, Mo |
| CXCL5 | NM_002994 | −1.2 | 1.43 | CXCR2 | Neu, Mo |
| CXCL6 | NM_002993 | 1.11 | 2.41 | CXCR1, CXCR2 | Neu, Mo |
| CXCL8 (IL-8) | NM_000584 | 1.01 | 1.51 | CXCR1, CXCR2 | Neu, Mo |
| IL-1 alpha | NM_000575 | 1.2 | 1.79 | IL1R | Inflammation, acute-phase response, fever |
| IL-1 beta | NM_000576 | 1.03 | 1.52 | IL1R | Inflammation, acute-phase response, fever |
| IL-6 | NM_000600 | 1.18 | 1.83 | IL6R | Fever and acute-phase response |
| C | |||||
| CCL7 (MCP3) | NM_006273 | −1.15 | −1.46 | CCR1, CCR2, CCR3 | Eo, Bs, Mo, T, NK, DC, CD4+ T and DC |
| IL-16 | NM_004513 | −1.25 | −1.53 | IL16R | Eo, Bs, Mo, T, NK, DC, CD4+ T, and DC |
A, cytokines upregulated during infection with both NiVwt and NiVΔC; B, cytokines upregulated only during infection with NiVΔC; C, cytokines downregulated during infection with NiVΔC.
FC, fold change: level of gene expression in NiV-infected cells in comparison to that in mock-infected cells as determined by microarray analysis. Analysis was carried out for each condition with two pools of cells including 3 donors.
Type of cells recruited by indicated chemokines (45): Neu, neutrophils; Eo, eosinophils; Bs, basophils; Mo, monocytes; T, T lymphocytes; TH1, TH1 lymphocytes; Tm, memory T cells; B, B lymphocytes; NK, NK cells; DC, dendritic cells; Ms, mast cells.
We then analyzed the expression of cytokines differentially upregulated after NiVΔC infection by using RT-qPCR (Fig. 4). HUVEC were infected with either NiVwt or NiVΔC, and RNAs were extracted 8 h and 24 h later and analyzed for the expression of the same panel of cytokines, determined to be upregulated in the microarray. In accord with the transcriptome results, all analyzed cytokines were significantly increased 8 h after infection with NiVΔC compared to findings for noninfected cells. In contrast, NiVwt induced only CXCL10 but not any other cytokines. In accord with the results obtained early after infection, significantly higher levels of CXCL2, -3, -8, -10, -11 and -20 mRNAs were detected 24 h post-infection with NiVΔC (Fig. 4B). However, in this later period, production of IL-1β and CXCL6 was not significantly increased further. Furthermore, at 24 h postinfection, NiVwt upregulated the production of CXCL10, CXCL11, and CCL20 mRNA but not other cytokines, suggesting that these chemokines are produced in the second wave after infection, requiring a longer period for their induction in endothelial cells. A high cytopathic effect of both NiVwt and NiVΔC infection at later time points prevented further analysis. Since several paramyxovirus accessory C proteins have been shown to be involved in the regulation of type 1 interferon (IFN) production and signaling (19), we finally analyzed production of IFN-β as well. Indeed, IFN-β mRNA was significantly increased in HUVEC cells infected with NiVΔC compared to that with NiVwt infection (Fig. 4C), suggesting the role of NiV C in the inhibition of IFN-β production.
Fig 4.
Analysis of cytokine expression in NiVwt- and NiVΔC-infected HUVEC. HUVEC were infected with either NiVwt or NiVΔC (MOI = 1) or mock infected; RNA was isolated 8 h (A) or 24 h (B) later and analyzed for cytokine expression by RT-qPCR or for the IFN-β mRNA expression 24 h and 48 h p.i. (C). (D) Induction of IL-8 production after NiVwt and NiVΔC infection of HUVEC in cell supernatants taken 8, 24, and 48 H after infection and analyzed by using an IL-8 ELISA kit. Results are expressed as a fold change in induction by virus infection, compared to results for mock infection, and correspond to the average for 6 individual experiments, and bars represent standard deviations. Statistically significant differences either between infected and noninfected cells or between NiVwt and NiVΔC infection are indicated (*, P < 0.05, * *, P < 0.01, Mann-Whitney U test).
We then tested whether NiVΔC modulates production of cytokines at the protein level. Indeed, production of CXCL8 (IL-8) was higher in the supernatant of NiVΔC-infected cells both 24 and 48 h postinfection (Fig. 4D). Together, these results suggested that the NiV C protein inhibits the production of proinflammatory cytokines in primary endothelial cells.
Stable expression of the transfected NiV C protein decreases cytokine production.
To analyze the role of NiV C outside the context of infection, we generated HEK293 cells stably expressing the NiV C protein (293C cells). Immunostaining of the NiV C protein revealed a predominant punctuated pattern of cytosolic expression (Fig. 5B). NiV C protein expression in 293C cells was further confirmed by Western blot analysis (data not shown).
Fig 5.
NiV C inhibits production of proinflammatory cytokines. (A and B) 293 cells (A) or 293C cells (B), stably expressing NiV C, were stained with anti-NiV C antibodies (green) and DAPI (blue) and observed by immunofluorescence microscopy (×100). The insert in each figure corresponds to higher magnification (×200). (C and D) Both 293 cells (black histograms) and 293C cells (white histograms) were infected with either NiVwt or NiVΔC (C) or stimulated with poly(I·C) (D) and analyzed 24 h later for the production of CXCL2, CXCL3, IL-8, and IFN-β by RT-qPCR. Representative results of 3 individual experiments are shown, and bars present standard deviations. Statistically significant differences between analyzed samples are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001; calculated using two-way ANOVA with Bonferroni's correction test.
We next analyzed whether the C protein could modulate cytokine production when stably expressed as in HEK293 cells (293C cells). We compared the production of several cytokines in 293 and 293C cells by RT-qPCR after infection with either NiVwt or NiVΔC (Fig. 5C). NiVwt induced significantly higher production of CXCL2 and -3, IL-8, and IFN-β in 293 cells than in 293C cells. This difference was slightly lower when NiVΔC was used for infection, although this virus induced higher expression of all analyzed cytokines in both cell lines than that with NiVwt. Moreover, expression of some analyzed cytokines in mock-infected cells was also lower in 293C cells than in 293 cells, suggesting that NiV C may regulate constitutive in addition to induced expression of proinflammatory cytokines. To further test whether the expression of C requires infectious conditions in cells to accomplish the cytokine inhibition, we stimulated both 293 and 293C cells with poly(I·C) and then analyzed production of cytokine RNA (Fig. 5D). Again, expression of C strongly decreased production of all tested cytokine RNAs, demonstrating that expression of NiV C alone, in the absence of an infectious context, is sufficient to inhibit the induction of cytokines, thus confirming its role in control of generation of the early proinflammatory response.
DISCUSSION
Understanding the mechanisms of immune evasion by emergent henipaviruses and identifying of the virus-encoded factors that contribute to their ability to rapidly disseminate in a variety of host species are essential for comprehension of their pathogenesis. This study aimed to elucidate the role of the nonstructural NiV C protein in virus pathogenicity. Most of the previous studies of the function of paramyxovirus C accessory proteins addressed the analysis of the modulation of type I IFN induction. Sendai virus (SeV) mutants unable to express either the C gene or the V gene induced elevated levels of host type I IFN production, and the product of the SeV C open reading frame can directly limit type I IFN induction (35, 54, 55). Measles virus that cannot express the C protein also induces more type I IFN than the wild-type virus (42). Furthermore, C knockout measles and rinderpest virus show reduced growth on cells that produce and respond to IFN and in this regard are more impaired than their equivalent V-knockout mutants (1, 13, 16). NiV C has been proposed previously to have interferon antagonist activity (43), although the other study showed the lack of an inhibitory effect of NiV C on IFN signaling in infected cells (63). Our results demonstrate that similarly to the other paramyxovirus C proteins, NiV C inhibits the production of IFN-β. Therefore, the C protein could play a role in NiV-induced inhibition of the production of type I interferon, observed recently in human cells (59), although the underlying mechanism of action and potential synergistic effect with NiV V and W activity remain to be determined. The effect of NiV C, however, was limited to IFN-β and did not include IFN-λ1, since NiVwt and NiVΔC induced production of IFN-λ similarly (Table 1).
Our transcriptomic analysis of infected primary human endothelial cells revealed a remarkable complexity of chemokine expression in early stages postinfection. Both NiVwt and NiVΔC induced the production of CCL8, CXCL10 and -11 and IFN-λ1. Recent studies showed that NiVwt could induce the secretion of IL-8, MCP-1, IFN-β, and THP1 monocyte chemotaxic factor in human microvascular endothelial cells (37) and IL-1-β, IL-6, tumor necrosis factor alpha (TNF-α), and gamma IFN in hamsters (47). In contrast to findings of a previously published study (37), we could detect the production of IFN-β and IL-8 mRNA early after NiV infection in human macrovascular endothelial cells, probably because our primary cultures did not contain cortisone, known to block the expression of proinflammatory cytokines (28). Moreover, our studies revealed that NiVΔC induces the expression of the whole new panel of other proinflammatory cytokines not observed for NiVwt, particularly those involved in chemotaxis and leukocyte recruitment, capable of enhancing cell adhesion to the vascular endothelium, including CCL3, CXCL2, -3, and -6 (4, 18). Most importantly, in accord with the pattern of chemokine induction, we have observed much higher accumulation of inflammatory cells in lungs of hamsters infected with NiVΔC than in those infected with NiVwt, followed with less-intensive pathological lesions and generation of the adaptive immune response (Fig. 3). IL-1-β, IL-6, and IL-8 have been shown to be major triggers and regulators of inflammatory responses to microbial pathogens, inducing further cytokine release, endothelial cell activation, acute-phase proteins synthesis, and fever. These cytokines could further stimulate an immune response, including antigen presentation, cytokine production, inflammation, phagocytosis, and cytotoxic activity (4, 18). Accordingly, asymptomatic Ebola virus infection was shown to be associated with the generation of a strong inflammatory response, not observed in patients with clinical symptoms (36), revealing the importance of the secretion of inflammatory cytokines for the virus clearance. Since chemokine balance is critical for the generation of an adequate immune response, it is possible that C plays a role in the modulating this balance and inhibiting the early proinflammatory response at the site of infection, thus helping NiV invasion. Similar to what has been shown with dendritic cells (45), endothelial cells may secrete distinct chemokines in several waves after virus infection, and the early production may have a particularly important effect on shaping the immune response. The other paramyxovirus C proteins have been shown to influence cytokine production: the measles virus C protein could control generation of the proinflammatory cytokines IL-6 and TNF-α, as well as interferon, in rhesus monkeys (14). However, to our knowledge this is the first report of broad regulation of proinflammatory chemokines by the paramyxovirus C protein. Most intriguingly, our study demonstrates the effect of NiV C on cytokine production in both virus-infected cells and cells stimulated with poly(I·C), demonstrating that it does not depend on the infectious content. Since transcription factors, such as STAT or NF-κB, are essential for chemokine production, NiV C, similar to what was observed with NiV V and W (12, 50), could interact with some of these factors, leading to an altered chemokine balance and consecutively to an inappropriate immune response.
NiVΔC viruses generated in this study were shown to be partially attenuated in a hamster model, suggesting the role of the accessory NiV C protein in viral pathogenesis. Infection resulted in the substantial delay of death and final survival of a significant number of animals even after the highest infectious dose (1,000 PFU). These data differ from those of a recent study that postulated that NiV lacking expression of the C protein is completely attenuated even at very high infectious doses (63). Although the same mutation was introduced in the NiV C gene in one of our mutants (NiVΔC2), the rest of the recombinant NiV construct was not identical in the virus used in this study and the NiV virus published by Yoneda et al. (62), particularly in intergenic regions. Multiple nucleotide changes introduced in order to facilitate the cloning procedure are likely to have an additional attenuation effect and could explain partial attenuation of recombinant wt NiV in comparison to unmodified wt NiV, shown in that study for low doses of the virus (62), and elimination of C protein expression may further reduce the virulence of already-attenuated recombinant virus. However, we could not exclude some other, unknown factors, which could account for those differences.
Accessory C proteins from the other paramyxoviruses have been shown to play an important role in viral pathogenesis. The Sendai virus C protein was shown to act as a virulence factor (34). It inhibits virus transcription and consecutively replication by binding the L polymerase (29), as well as signaling JAK/STAT in response to interferon (17), and plays a role in protein assembly and virus budding (27). Similar functions were proposed for the measles virus C protein, and its important role in virus virulence has been shown in vivo with different animal models, such as macaques, CD46-transgenic mice, cotton rats, or SCID mice engrafted with human thymus implants (16, 44, 56, 57). It is thus possible that NiV C, in addition to its rather unique role in the regulation of cytokine balance, has other functions important for virus pathogenicity and responsible for its lower replication capacity. Furthermore, NiV C may not function in the same way in all species susceptible to infection, particularly in fruit bats, a natural reservoir of henipavirus, where infection does not induce clinical disease. It has been demonstrated recently that NiV antagonizes interferon production and signaling pathways in cell lines obtained from fruit bats, as it does in human cells (58). Based on results from this study, we could hypothesize that production of some proinflammatory chemokines in bats may escape inhibition by NiV C, resulting in highly attenuated infection in this species.
In conclusion, differential expression of chemokines regulated by NiV C may provide a molecular code responsible for precise coordination of leukocyte trafficking under proinflammatory conditions, such as in infection. The molecular mechanism implicated in the regulation of cytokine production and activation of particular transcription factors during the first hours after infection remains to be elucidated. A search for the cellular partners of the NiV C protein appears essential to better understand the multiple roles of this virulence factor. Results from this study provide an important biological insight into NiV-host cell interaction and reveal the critical function of the nonstructural NiV C protein in the regulation of the chemokine-induced immune response and the control of virus pathogenicity, opening new avenues to better understand this emergent highly lethal infectious disease.
ACKNOWLEDGMENTS
The work was supported by INSERM and ANR-09-MIEN-018-01 and by a University of Malaya HIR grant (E0004-20001).
We thank H. Raoul and biosafety team members from INSERM biosafety level 4 (BSL4) Laboratory Jean Mérieux in Lyon for their assistance. We are also grateful to F. Wild, A. Sabine, M. Castelazzi, B. King, D. Gerlier, R. Buckland, and members of the group Immunobiology of Viral Infections, as well as N. Nazaret and A. Wierinckx (ProfileXpert, Lyon), for their help during the realization of the study. We acknowledge the contribution of the technical platforms Production et Analyze de Proteines (A. Chaboud) and Cellulonet (I. Grosjean) of SFR Bioscience Gerland-Lyon Sud (UMS344/US8).
Footnotes
Published ahead of print 25 July 2012
REFERENCES
- 1. Baron MD, Barrett T. 2000. Rinderpest viruses lacking the C and V proteins show specific defects in growth and transcription of viral RNAs. J. Virol. 74:2603–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Basler CF. 11 April 2012. Nipah and Hendra virus interactions with the innate immune system. Curr. Top. Microbiol. Immunol. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 3. Bossart KN, Wang LF, Eaton BT, Broder CC. 2001. Functional expression and membrane fusion tropism of the envelope glycoproteins of Hendra virus. Virology 290:121–135 [DOI] [PubMed] [Google Scholar]
- 4. Bromley SK, Mempel TR, Luster AD. 2008. Orchestrating the orchestrators: chemokines in control of T cell traffic. Nat. Immunol. 9:970–980 [DOI] [PubMed] [Google Scholar]
- 5. Bussolino F, et al. 1989. Granulocyte- and granulocyte-macrophage-colony stimulating factors induce human endothelial cells to migrate and proliferate. Nature 337:471–473 [DOI] [PubMed] [Google Scholar]
- 6. Chadha MS, et al. 2006. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 12:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan YP, Chua KB, Koh CL, Lim ME, Lam SK. 2001. Complete nucleotide sequences of Nipah virus isolates from Malaysia. J. Gen. Virol. 82:2151–2155 [DOI] [PubMed] [Google Scholar]
- 8. Childs K, et al. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190–200 [DOI] [PubMed] [Google Scholar]
- 9. Chua KB, et al. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432–1435 [DOI] [PubMed] [Google Scholar]
- 10. Chua KB, et al. 2002. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 4:145–151 [DOI] [PubMed] [Google Scholar]
- 11. Chua KB, et al. 2000. High mortality in Nipah encephalitis is associated with presence of virus in cerebrospinal fluid. Ann. Neurol. 48:802–805 [PubMed] [Google Scholar]
- 12. Ciancanelli MJ, Volchkova VA, Shaw ML, Volchkov VE, Basler CF. 2009. Nipah virus sequesters inactive STAT1 in the nucleus via a P gene-encoded mechanism. J. Virol. 83:7828–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Devaux P, Cattaneo R. 2004. Measles virus phosphoprotein gene products: conformational flexibility of the P/V protein amino-terminal domain and C protein infectivity factor function. J. Virol. 78:11632–11640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Devaux P, Hodge G, McChesney MB, Cattaneo R. 2008. Attenuation of V- or C-defective measles viruses: infection control by the inflammatory and interferon responses of rhesus monkeys. J. Virol. 82:5359–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eaton BT, Broder CC, Middleton D, Wang LF. 2006. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 4:23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Escoffier C, et al. 1999. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J. Virol. 73:1695–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garcin D, Marq JB, Strahle L, le Mercier P, Kolakofsky D. 2002. All four Sendai Virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295:256–265 [DOI] [PubMed] [Google Scholar]
- 18. Glass WG, Rosenberg HF, Murphy PM. 2003. Chemokine regulation of inflammation during acute viral infection. Curr. Opin. Allergy Clin. Immunol. 3:467–473 [DOI] [PubMed] [Google Scholar]
- 19. Goodbourn S, Randall RE. 2009. The regulation of type I interferon production by paramyxoviruses. J. Interferon Cytokine Res. 29:539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gotoh B, Komatsu T, Takeuchi K, Yokoo J. 2001. Paramyxovirus accessory proteins as interferon antagonists. Microbiol. Immunol. 45:787–800 [DOI] [PubMed] [Google Scholar]
- 21. Gotoh B, Komatsu T, Takeuchi K, Yokoo J. 2002. Paramyxovirus strategies for evading the interferon response. Rev. Med. Virol. 12:337–357 [DOI] [PubMed] [Google Scholar]
- 22. Guillaume V, et al. 2009. Acute Hendra virus infection: Analysis of the pathogenesis and passive antibody protection in the hamster model. Virology 387:459–465 [DOI] [PubMed] [Google Scholar]
- 23. Gurley E, et al. 2007. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg. Infect. Dis. 13:1031–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harcourt BH, et al. 2005. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg. Infect. Dis. 11:1594–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harcourt BH, et al. 2001. Molecular characterization of the polymerase gene and genomic termini of Nipah virus. Virology 287:192–201 [DOI] [PubMed] [Google Scholar]
- 26. Harcourt BH, et al. 2000. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271:334–349 [DOI] [PubMed] [Google Scholar]
- 27. Hasan MK, et al. 2000. Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J. Virol. 74:5619–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hermoso MA, Cidlowski JA. 2003. Putting the brake on inflammatory responses: the role of glucocorticoids. IUBMB Life 55:497–504 [DOI] [PubMed] [Google Scholar]
- 29. Horikami SM, Hector RE, Smallwood S, Moyer SA. 1997. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology 235:261–270 [DOI] [PubMed] [Google Scholar]
- 30. Horvath CM. 2004. Silencing STATs: lessons from paramyxovirus interferon evasion. Cytokine Growth Factor Rev. 15:117–127 [DOI] [PubMed] [Google Scholar]
- 31. Hossain MJ, et al. 2008. Clinical presentation of nipah virus infection in Bangladesh. Clin. Infect. Dis. 46:977–984 [DOI] [PubMed] [Google Scholar]
- 32. Hsu VP, et al. 2004. Nipah virus encephalitis reemergence, Bangladesh. Emerg. Infect. Dis. 10:2082–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jaffe EA, Nachman RL, Becker CG, Minick CR. 1973. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J. Clin. Invest. 52:2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kato A, et al. 2007. Importance of the anti-interferon capacity of Sendai virus C protein for pathogenicity in mice. J. Virol. 81:3264–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Komatsu T, Takeuchi K, Yokoo J, Gotoh B. 2004. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology 325:137–148 [DOI] [PubMed] [Google Scholar]
- 36. Leroy EM, et al. 2000. Human asymptomatic Ebola infection and strong inflammatory response. Lancet 355:2210–2215 [DOI] [PubMed] [Google Scholar]
- 37. Lo MK, et al. 2010. Characterization of the antiviral and inflammatory responses against Nipah virus in endothelial cells and neurons. Virology 404:78–88 [DOI] [PubMed] [Google Scholar]
- 38. Marianneau P, et al. 2010. Experimental infection of squirrel monkeys with nipah virus. Emerg. Infect. Dis. 16:507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mathieu C, et al. 2012. Lethal Nipah virus infection induces rapid overexpression of CXCL10. PLoS One 7:e32157 doi:10.1371/journal.pone.0032157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mathieu C, et al. 2011. Nipah virus uses leukocytes for efficient dissemination within a host. J. Virol. 85:7863–7871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mayo MA. 2002. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 147:1655–1663 [DOI] [PubMed] [Google Scholar]
- 42. Nakatsu Y, Takeda M, Ohno S, Koga R, Yanagi Y. 2006. Translational inhibition and increased interferon induction in cells infected with C protein-deficient measles virus. J. Virol. 80:11861–11867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Park MS, et al. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patterson JB, Thomas D, Lewicki H, Billeter MA, Oldstone MB. 2000. V and C proteins of measles virus function as virulence factors in vivo. Virology 267:80–89 [DOI] [PubMed] [Google Scholar]
- 45. Piqueras B, Connolly J, Freitas H, Palucka AK, Banchereau J. 2006. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood 107:2613–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reynes JM, et al. 2005. Nipah virus in Lyle's flying foxes, Cambodia. Emerg. Infect. Dis. 11:1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rockx B, et al. 2011. Clinical outcome of henipavirus infection in hamsters is determined by the route and dose of infection. J. Virol. 85:7658–7671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rodriguez JJ, Parisien JP, Horvath CM. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476–11483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shaw G, Morse S, Ararat M, Graham FL. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16:869–871 [DOI] [PubMed] [Google Scholar]
- 50. Shaw ML, Cardenas WB, Zamarin D, Palese P, Basler CF. 2005. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J. Virol. 79:6078–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shaw ML, Garcia-Sastre A, Palese P, Basler CF. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78:5633–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sleeman K, et al. 2008. The C, V and W proteins of Nipah virus inhibit minigenome replication. J. Gen. Virol. 89:1300–1308 [DOI] [PubMed] [Google Scholar]
- 53. Stone R. 2011. Epidemiology. Breaking the chain in Bangladesh. Science 331:1128–1131 [DOI] [PubMed] [Google Scholar]
- 54. Strahle L, Garcin D, Kolakofsky D. 2006. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 351:101–111 [DOI] [PubMed] [Google Scholar]
- 55. Strahle L, Garcin D, Le Mercier P, Schlaak JF, Kolakofsky D. 2003. Sendai virus targets inflammatory responses, as well as the interferon-induced antiviral state, in a multifaceted manner. J. Virol. 77:7903–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Takeuchi K, et al. 2005. Stringent requirement for the C protein of wild-type measles virus for growth both in vitro and in macaques. J. Virol. 79:7838–7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Valsamakis A, et al. 1998. Recombinant measles viruses with mutations in the C, V, or F gene have altered growth phenotypes in vivo. J. Virol. 72:7754–7761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Virtue ER, Marsh GA, Baker ML, Wang LF. 2011. Interferon production and signaling pathways are antagonized during henipavirus infection of fruit bat cell lines. PLoS One 6:e22488 doi:10.1371/journal.pone.0022488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Virtue ER, Marsh GA, Wang LF. 2011. Interferon signaling remains functional during henipavirus infection of human cell lines. J. Virol. 85:4031–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wong KT, et al. 2003. A golden hamster model for human acute Nipah virus infection. Am. J. Pathol. 163:2127–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wong KT, et al. 2002. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am. J. Pathol. 161:2153–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yoneda M, et al. 2006. Establishment of a Nipah virus rescue system. Proc. Natl. Acad. Sci. U. S. A. 103:16508–16513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoneda M, et al. 2010. The nonstructural proteins of Nipah virus play a key role in pathogenicity in experimentally infected animals. PLoS One 5:e12709 doi:10.1371/journal.pone.0012709 [DOI] [PMC free article] [PubMed] [Google Scholar]