Abstract
We developed “spoligoriftyping,” a 53-plex assay based on two preexisting methods, the spoligotyping and “rifoligotyping” assays, by combining them into a single assay. Spoligoriftyping allows simultaneous spoligotyping (i.e., clustered regularly interspaced short palindromic repeat [CRISPR]-based genotyping) and characterization of the main rifampin drug resistance mutations on the rpoB hot spot region in a few hours. This test partly uses the dual-priming-oligonucleotide (DPO) principle, which allows simultaneous efficient amplifications of rpoB and the CRISPR locus in the same sample. We tested this method on a set of 114 previously phenotypically and genotypically characterized multidrug-resistant (MDR) Mycobacterium tuberculosis or drug-susceptible M. tuberculosis DNA extracted from clinical isolates obtained from patients from Bulgaria, Nigeria, and Germany. We showed that our method is 100% concordant with rpoB sequencing results and 99.95% (3,911/3,913 spoligotype data points) correlated with classical spoligotyping results. The sensitivity and specificity of our assay were 99 and 100%, respectively, compared to those of phenotypic drug susceptibility testing. Such assays pave the way to the implementation of locally and specifically adapted methods of performing in a single tube both drug resistance mutation detection and genotyping in a few hours.
INTRODUCTION
New-generation high-throughput multiplexing instruments constitute a technological breakthrough that allows a change in biological assay design to miniaturization and point-of-care assays (3, 3a, 11, 13, 37, 39). Instead of separate molecular epidemiological investigations (either genotyping or clinical microbiology diagnostic assays) being performed either for molecular epidemiological investigations or to improve patient treatment, integrated methods, with both objectives in one tube, can now be developed. In tuberculosis (TB) control, the infectious disease caused by Mycobacterium tuberculosis complex (MTC), early diagnosis of multidrug-resistant TB (MDR-TB) is predicted to save the U.S. health care system $250,000 per case (6). It may also allow a reduction in the length of time that MDR-TB cases are infectious by as much as 6 weeks, an important feature in limiting MDR-TB spreading (6). The gold standard is the sequencing of drug resistance genes, e.g., rpoB for rifampin resistance. Current commercial molecular resistance identification tests include INNO-LiPA Rif.TB (Innogenetics, Ghent, Belgium), GenoType MTBDRplus (Hain Life Science GmbH, Nehren, Germany) (19, 20), and the new molecular beacon test GeneXpert MTB/Rif (Cepheid, Sunnyvale, CA); these tests allow direct rifampin resistance identification in sputum (4, 17). A change of paradigm in TB control from a culture-based toward a specimen-based approach seems to be on its way, even though numerous issues still remain (14). We should not ignore the fact that the Clinical and Laboratory Standards Institute (CLSI, Wayne, PA), which provides the standards for MDR-TB drug susceptibility testing (DST), requires that genotypic testing not replace phenotypic DST for the time being, but it can be an adjunct. Among other problems, the price of such assays remains very high for those countries that really need them, even though new business models try to promote the fast spreading of these technologies.
For the development of any nucleic acid-based assay, multiplexing capacity and sensitivity are important issues that can be addressed in several ways. Multiplexing can be designed in silico to provide the best theoretical framework. The PrimerPlex (Premier Biosoft, Palo Alto, CA) and MP Primer softwares can be used for this purpose (30, 31). To improve sensitivity, new molecular biological principles aimed at using a single pair of primers and multiple hybridization tags can also be used. Multiplex-ligation-dependent probe amplification (MLPA) and the molecular inversion probe (MIP) assay are two methods of creating multiplexed assays (15, 28, 29). Another multiplexing principle is the dual-priming oligonucleotide (DPO) assay, which was recently used for respiratory virus detection and in a single nucleotide polymorphism (SNP) typing assay targeting the cytochrome oxidase CYP2C19 gene (8). The DPO principle relies on primers containing two separate priming regions joined by a polydeoxyinosine linker (8). The longer 5′ segment initiates stable priming; the shorter 3′ segment determines target-specific extension. In this way, priming efficiency is improved, thus allowing multiplexing reactions to be more easily designed and produced (8).
Tuberculosis and, in particular, MDR-TB are increasingly challenging issues for international public health control (40). Rapid diagnostic tests allowing both simultaneous identification of Mycobacterium tuberculosis and drug susceptibility identification are needed, especially in those countries in which initial resistance is high and the transmission of MDR isolates is known to occur (e.g., Peru, the Philippines, South Africa, the Baltic countries, and the republics of the former USSR) (24, 26). Rifampin resistance is a good surrogate marker of MDR-TB (resistant to rifampin and isoniazid), and 95% of all mutations are limited to the 81-bp rifampin resistance-determining region (RRDR) of the rpoB gene (16). The link between phenotypic (i.e., DST of culture) and genotypic associations for the most frequent point mutations in rpoB is excellent (27).
Spoligotyping detects 43 spacers in the CRISPR region of MTC strains and allows characterization of clinical isolates at a phylogeographical level.
In this study, our goal was to combine in a single assay spoligotyping with “rifoligotyping,” which characterizes rpoB hot spot mutations (21, 25). These two assays were developed in 1997 and in 2002, respectively, and were originally developed as reverse line blot hybridization assays (21, 25). These two methods were adapted to the microbead-based format and were later transitioned to the latest magnetic-bead-based new-generation Magpix, a device that has a charge-coupled device (CCD) imager and two channels (a reporter channel and a classification channel at, respectively, 532 and 635 nm) (23). We report a 53-plex development method and expect that this new and inexpensive high-throughput format assay (running on 96 wells) will ultimately facilitate tuberculosis control by its fast and easy implementation in specialized TB laboratories that possess either a Bio-Plex (Bio-Rad, Hercules, CA) or a Luminex 200 (Luminex Corp., Austin, TX). Resistance to other first-line drugs (by detection of mutations in inhA, katG, rrs, rpsl, embB, and pncA) and other second-line drug resistance genes will have to be included later.
MATERIALS AND METHODS
Suspension microspheres and instruments.
The high-throughput systems used in this study were a classical Bio-Plex (Bio-Rad, Hercules, CA) and a Luminex 200 (Luminex Corp, Austin, TX). The corresponding softwares were Bio-Plex Manager (version 5.0) and xPonent (version 3.1.871.0). In-house oligonucleotide-precoupled MicroPlex microspheres were used throughout this study.
Patient isolates and DNA extraction methods.
DNAs were extracted by the cetyltrimethylammonium bromide (CTAB) method or by the thermolyzate method (33, 36). One hundred fourteen DNAs corresponding to 114 M. tuberculosis clinical isolates were used. Ninety-seven MDR-TB DNAs with known drug resistance and patients' information were from Bulgaria (National Center of Infectious and Parasitic Diseases and National TB Laboratory, Sofia, Bulgaria). Phenotypic drug susceptibility testing (DST) was done using the Bactec MGIT960 TB system by following the recommendations provided by the manufacturer (Becton, Dickinson, Franklin Lakes, NJ). Membrane-based spoligotyping and VNTR analysis of 24 loci were performed on 94 isolates (21). Four MDR-TB DNAs with rpoB sequenced (with GTC at codon 516, GAC at codon 526, TAC at codon 526, and TTG at codon 531) were kindly provided by S. Feuerriegel and S. Niemann (National Reference Center for Tuberculosis, Molecular Mycobacteriology Group, Borstel, Germany) and used as rpoB mutant controls. Thirteen drug-susceptible DNAs were from Nigeria (DST was also performed on a Bactec MGIT960 TB system) and provided by Lovett Lawson, Zankli Medical Center, Abuja, Nigeria, and used as rpoB wild-type controls.
Primer and probe oligonucleotide sequences and PCR protocol.
Primers and probes came from Eurogentec (Liège, Belgium) or Integrated DNA Technologies (Coralville, IA) (IDT) (Table 1). Spoligotyping probe sequences are published (42), and high-throughput spoligotyping was done as reported previously (10, 42). DPO primers were designed according to the DPO principle (8) and were chosen to amplify a 181-bp fragment encompassing the rpoB rifampin resistance-determining region (RRDR; 81 bp). Probes were designed manually or with the PrimerPlex software (version 2.60). Fifteen trials were required to obtain the right probes for codon 526. The final PCR protocol was as follows. In a total volume of 25 μl, using DRa-DRb, rpoB-Dfw, and rpoB-Drv dual-priming oligonucleotide primers (Table 1), the reaction mixture contained 2 μl of a DNA sample (20 to 40 ng), 0.2 mM each deoxynucleoside triphosphate (dNTP), 1 μM each primer, PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl), and 1.0 U of Taq polymerase. The following PCR program was used: 3 min at 95°C, followed by 25 cycles of 30 s at 95°C, 30 s at 65°C, and 30 s at 72°C, with a final elongation step at 72°C for 5 min. Hybridization of 2 μl of the PCR products in 50 μl of tetramethylammonium chloride buffer (1× TMAC) was performed after denaturation for 10 min at 95°C and then 20 min at 50°C. After centrifugation at 4,000 rpm and replacement of 35 μl of supernatant by 1× TMAC, streptavidin-phycoerythrin solution (Interchim SA, Montluçon, France) prepared in 1× TMAC was added to a final concentration of 2 μg/ml to reach a final volume of 75 μl. We allowed 5 min of incubation in the system at 50°C before reading the samples.
Table 1.
DPO and classical primers, sequencing primers, and specific probes used in this study
| Primer or probea | Name | Primer sequence | Reference |
|---|---|---|---|
| 1 | biot-DRa | 5′ GGTTTTGGGTCTGACGAC 3′b | J. Kamerbeek et al. (21) |
| 2 | DRb | 5′ CCGAGAGGGGACGGAAAC 3′ | J. Kamerbeek et al. (21) |
| 3 | rpoB_Dfw | 5′ CGGTGGTCGCCGCGATCAAGGAIIIIITCGGCA 3′ | This study (181-bp RRDR) |
| 4 | rpoB_Drv | 5′ CCGTAGTGCGACGGGTGCACGTIIIIIACCTCC 3′b | This study (181-bp RRDR) |
| 5 | TR1 | 5′ TACGGTCGGCGAGCTGATCC3′A. | A. Telenti et al. (34) |
| 6 | TR2b | 5′ TACGGCGTTTCGATGAACC3′ | A. Telenti et al. (34) |
| 7 | Spa_wt1 | 5′ AGCCAGCTGAGCCAATTC 3′c | This study |
| 8 | rpoB_516 wt | 5′ AATTCATGGACCAGAACA 3′c | This study |
| 9 | rpoB_516 mutGTC | 5′ AATTCATGGTCCAGAACA 3′c | This study |
| 10 | Spa_wt2 | 5′ AGAACAACCCGCTGTCGG 3′c | This study |
| 11 | rpoB_526 wt | 5′ GGGTTGACCCACAAGCGCC 3′c | This study |
| 12 | rpoB_526 mutGAC | 5′ GGGTTGACCGACAAGCGCC 3′c | This study |
| 13 | rpoB_526 mutTAC | 5′ GGGTTGACCTACAAGCGCC 3′c | This study |
| 14 | rpoB_531 wt | 5′ CCGACTGTCGGCGCTGGG 3′c | This study |
| 15 | rpoB_531 mutTTG | 5′ CCGACTGTTGGCGCTGGG 3′c | This study |
| 16 | rpoB_531 mutTGG | 5′ CCGACTGTGGGCGCTGGG 3′c | This study |
Primers 1 to 4 are classical primers, primers 5 and 6 are sequencing primers, and numbers 7 to 16 are probes.
5′ biotinylated; all other spoligotyping probes are described in the work of Zhang et al. (42).
Amino C12 linker at the 5′ terminus.
Principle of the test.
Spoligoriftyping is based on a simultaneous analysis of the polymorphism in the clustered regularly interspersed short palindromic region (CRISPR) and those in the rpoB gene hot spot region, as shown in Fig. 1. Specifically, spacers 1 to 43 are amplified, as well as the most frequent SNPs, i.e., TTG at codon 531, TGG at codon 531, GAC at codon 526, TAC at codon 526, and GTC at codon 516, with specific capture probes used for detection (note that the nomenclature for the rpoB gene codons is the Escherichia coli rpoB gene nomenclature [see also reference 16]). The use of spanning capture probes (Spa_wt1 and Spa_wt2) and wild-type capture probes for codons 531, 526, and 516 allowed the detection of wild-type or other potential SNPs present in the RRDR. If there is an SNP in the region covered by the Spa_wt1 probe, Spa_wt1 will not hybridize and the signal obtained by analyzing the specimen's mean fluorescence intensity (MFI) in the Luminex will be low (the value will be inferior to the cutoff, i.e., negative). In contrast, the Spa_wt1 probe (like the wild-type sequence in that region) will hybridize and its signal (MFI) will be higher than the cutoff, i.e., positive. This principle is the same for the Spa_wt2 probe. For a strain that is wild type at position 516, the MFI of probe rpoB_516 wt will be positive and the MFI of the probe rpoB_516 mut GTC will be negative. In the case of a strain with the mutation GTC at position 516, the MFI signal of the rpoB_516 mutGTC probe will be positive and that of probe rpoB_516 wt will be negative. In the case of a strain with a mutation in codon 516 other than GTC, both signals of probes rpoB_516 wt and rpoB_516 mutGTC will be negative. The same algorithm is applied to codons 526 and 531.
Fig 1.
Schematic representation of the spoligoriftyping test principle. (A) The CRISPR region is amplified by a single pair of primers that hybridize within the repeat sequences and overlap the spacers. (B) The full span of the rpoB gene hot spot region is determined with capture probes targeting either drug-resistant mutant SNPs or drug-susceptible wild-type sequences. (The first line is the hot spot sequence, and line 2 to line 11 represent the different capture probes.) wt, wild type.
Hence, to predict a rifampin-susceptible or rifampin-resistant phenotype, the adopted algorithm is as follows. (i) A rifampin-susceptible clinical isolate must be wild type in the whole RRDR; i.e., all probes, namely, Spa_wt1, rpoB_516 wt, Spa_wt2, rpoB_526 wt, and rpoB_531 wt, must provide a positive MFI relative to the cutoff. (ii) A rifampin-resistant strain must have at least one SNP; i.e., at least one of the preceding wild-type probes must give a negative MFI compared to the cutoff.
Statistical analysis and performance of the method.
Statistical analysis was performed using R software (version 2.14.2; www.r-project.org/). To determine reproducibility (intraoperator and intermachine reproducibility), a Pearson correlation test was performed.
To determine the cutoffs, the receiver operating characteristic (ROC) curve approach was adapted to our aims, which means ease of automatization; to provide highly reliable results, we chose to identify two cutoffs instead of one for each marker (a cutoff for negative values and a cutoff for positive values). Several cutoffs based on the distribution of negative and positive values were explored: means ± 1, 2, or 3 standard deviations. Those identifying the narrowest gray zone and resulting in 100% sensitivity and 100% specificity were kept for each marker (spacers and mutation probes). Means and standard deviations of values for reference set samples (samples with reference method results) were calculated with R software. The script for computing the sensitivity and specificity method performances is available upon request. The reference methods were reverse-line-blot membrane-based spoligotyping, sequencing for detection of mutations, and phenotypic DST for drug resistance identification. All methods were performed in a blind, random order and by operators from different teams (we obtained a subcontract for the sequencing and membrane spoligotyping performed in our coinvestigators' laboratories).
Interpretation of the MFI values.
We used the defined cutoffs to interpret the quantitative MFI values obtained in a Luminex (see Tables S1 and S2 in the supplemental material) as qualitative values, positive values (indicating the presence of the target), negative values (indicating the absence of the target), or undetermined values (for which it was necessary for an expert to determine the result).
Sequencing, spoligotyping, VNTR typing, and data analysis.
We amplified and sequenced a 411-bp rpoB fragment overlapping the RRDRs from 110 strains out of 114 studied isolates (97 MDR strains, 13 drug-susceptible strains) and for which independent spoligoriftyping test results were obtained. The mutations were already known for the other 4 strains from Germany. PCR was prepared as published previously (33) and sent to Beckman Coulter Genomics (Takeley, United Kingdom) for Sanger sequencing. Sequencing results were aligned by MultAlign (multiple sequence alignment with hierarchical clustering) (9), and the presence/absence of mutations was analyzed. Classical spoligotyping of membranes (Isogen Bioscience BV, Maarssen, The Netherlands) and VNTR typing were done at the National Center of Infectious and Parasitic Diseases according to standard published procedures (21, 32). Dendrograms were built using BioNumerics (version 6.6; Applied Maths, Sint-Martens-Latem, Belgium).
Validation of spoligoriftyping.
We validated the two components of the spoligoriftyping method, namely, spoligotyping and Rif resistance SNP detection, by (i) comparing the spoligotype patterns obtained by our test to those previously obtained from membranes for the same isolates and (ii) comparing the RRDR SNPs detected by our test to SNPs found by sequencing of the same region for the same isolates.
RESULTS
Method development.
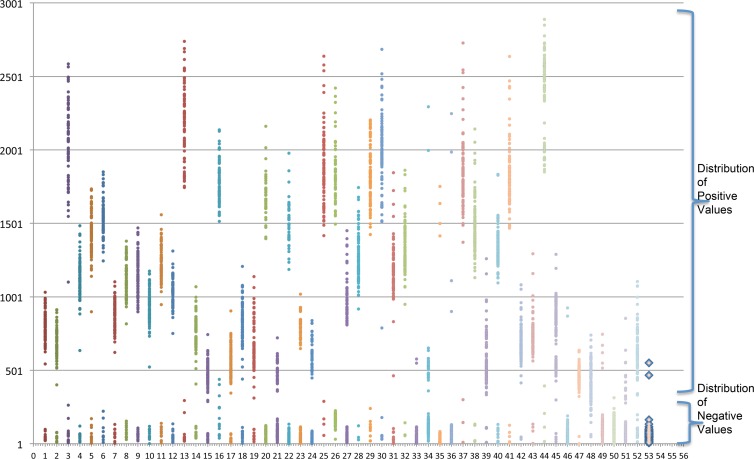
For all markers, intraoperator reproducibility was very good (Pearson correlation coefficient when the df is 2,382 [rdf = 2,382] = 0.993), as well as intermachine reproducibility (Pearson rdf = 3,008 = 0.923). The distribution of the MFI values was bimodal for all markers (Fig. 2)
Fig 2.
Distribution of mean fluorescence intensities (MFI) across samples. y axis, MFI results obtained by each capture probe (each data point represents an individual sample); x axis, the capture probes (probes 1 to 43 are direct repeat spacer capture probes, and probes 44 to 53 are rpoB SNP capture probes).
For the determination of the cutoff of each marker, we used two sets of samples. For one set (n = 88), spoligotype patterns in membranes were determined by both the reverse line blot assay and our new test, and for the second set (n = 74), results of both rpoB hot spot sequencing and our test were obtained. To allow automatization of MFI interpretation and to indirectly check DNA quality, we chose not to consider a single cutoff but instead two cutoffs separating 3 zones: one in which all samples are negative, a gray zone in which samples can be positive or negative, and a zone in which all samples are positive. To fix the cutoff between the negative and the gray zone, we compared the specificities of values derived from the distribution of negative results and kept the highest cutoff, ensuring the highest specificity. To fix the cutoff between results in the gray zone and the positive zone, we compared the sensitivities of cutoffs computed from the distribution of positive results and kept the highest cutoff, ensuring the highest sensitivity. For every spoligoriftyping marker except spacers 36 and 39, Spa_wt2, rpoB_526 wt, rpoB_526 mutGAC, rpoB_526 mutTAC, rpoB_531 wt, and rpoB_531 mutTTG, the cutoff for negative results was the mean of negative values plus 3 standard deviations and the cutoff for positive results was the mean of positive values minus 3 standard deviations. For spacers 36 and 39, Spa_wt2, rpoB_526 wt, rpoB_526 mutGAC, rpoB_526 mutTAC, rpoB_531 wt, and rpoB_531 mutTTG, the cutoffs were computed based on the mean ± 1 or the mean ± 2 standard deviations (for more details, see Table S2 in the supplemental material).
These pairs of cutoffs were applied in the subsequent parts of the study for the MFI interpretation for all strains. For the rare experimental MFI values located in the gray zone, an expert examination allowed us to interpret results as negative or positive. This may sometimes happen experimentally, since DNA quality/quantity is not optimal in many settings.
Spoligoriftyping results.
All samples (114 strains) except one provided a spoligoriftyping pattern. A spoligoriftyping pattern is a 53-character string with the spoligotype pattern (n = 43) plus the rpoB gene hot spot SNP pattern (n = 10). The full interpreted results are summarized in Table S1 in the supplemental material. Original raw Luminex MFI values can be obtained upon request.
Spoligotyping results.
This analysis allowed us to describe the Mycobacterium tuberculosis genetic diversity within Bulgaria's MDR Mycobacterium tuberculosis isolates. According to a classification into phylogeographical clades based on the spolDB4 database, 43% of the 95 isolates belonged to the Turkish (ST41) clade and 37% to the T1 clade (23% were in the ST53 cluster) (5). These two clades represented the majority of the isolates (76/95). The others clades were poorly prevalent (Table 2).
Table 2.
Lineage diversity of M. tuberculosis Bulgarian MDR clinical isolates
| Clade (cluster[s]) | No. of isolates | %a |
|---|---|---|
| Turkish (TUR, SIT41) | 41 | 43 |
| T1 (Euro-American) | 35 | 37 |
| T1 (SIT53) | 21 | 23 |
| T1 (SIT154) | 4 | 4 |
| East-Med1 (SIT284) | 4 | 4 |
| T1 (ST462) | 1 | 1 |
| T1 (SIT144) | 2 | 2 |
| T1 (new) | 2 | 2 |
| T1 (1280) | 1 | 1 |
| S-Bulgaria (SIT125) | 4 | 4 |
| T4 (SIT40) | 1 | 1 |
| X1 (SIT119) | 1 | 1 |
| Beijing (SIT1) | 3 | 3 |
| H3 (SIT50) | 1 | 1 |
| T2-T3 (SIT73) | 1 | 1 |
| S (SIT34) | 1 | 1 |
| U (SIT90) | 2 | 2 |
| U (NEW) | 3 | 3 |
| Ural (SIT262) | 1 | 1 |
| Ural (new) | 1 | 1 |
| Total | 95 | 100 |
Percentages are without decimals, and the total percentage is calculated by taking into account the total number of the isolates.
rpoB gene hot spot sequencing results.
One hundred DNAs were successfully sequenced; 10 DNAs could not be sequenced due to low levels of PCR products (in the majority of cases, these were thermolyzate DNAs). All the SNPs found in the rpoB hot spot by sequencing were correctly identified by our test.
Rifampin-typing results.
The 100 DNA samples that had been successfully sequenced served as references for verifying the performance of the rifampin-typing part of our test. Ninety-five clinical isolates were from Bulgaria (MDR strains), and the other five were from Nigeria (drug-susceptible clinical isolates). The most frequent rifampin mutation within the Bulgarian samples was the TTG SNP at codon 531. Eighty-two percent of samples (78/95) were found to have a mutation in codon 531 (76 isolates had the SNP TTG, and 2 had the SNP TGG), 8 strains harbored an SNP in codon 526 (6 had the SNP GAC, and 2 had the SNP TAC), and 2 strains had the SNP GTC at codon 516. All SNPs identified by sequencing were correctly detected by the spoligoriftyping test, including those in the region of sequence covered by the capture probe Spa_wt1 (n = 2) and by the capture probe Spa_wt2 (n = 1). Double mutations were also detected; one isolate had mutations both in codon 526 and in the region covered by the capture probe Spa_wt1, and one isolate had mutations in codon 516 and in the region covered by the capture probe Spa_wt1. One isolate classified as resistant by DST was found to be susceptible by the spoligoriftyping method. The 4 isolates from Germany with known SNPs were correctly detected by the new spoligoriftyping method, and their spoligotype pattern was obtained. Similarly, all 13 drug-susceptible isolates from Nigeria were identified as drug-susceptible TB clinical isolates and their correct spoligotype patterns obtained by the new assay (see Table S1 in the supplemental material for interpreted spoligoriftyping results).
Sensitivity and specificity of the spoligoriftyping test (method performances).
We used both sequencing and DST as gold standard tests to evaluate the new spoligoriftyping test's sensitivity and specificity for SNP detection and MDR strain prediction. We used the membrane spoligotyping results for 91 isolates from Bulgaria to evaluate the quality of our new assay for spoligotype pattern characterization. For these 91 isolates, the results of both methods were concordant for 89 strains. We observed discrepant spoligotype data points for 2 (of 3,913) strains. These two discrepancies seem to be due to an error in the interpretation of the membrane, as clear-cut MFI values were obtained twice with our test.
Both the sensitivity and specificity of spoligoriftyping were 100% when results were compared to those of the rpoB sequencing method, a gold standard for rifampin resistance mutation detection. Using phenotypic DST as the gold standard, the sensitivity and specificity of our test were, respectively, 99% (95% confidence interval = 0.97 to 1) and 100%.
Cluster analysis of VNTR-spoligoriftyping results and MDR-TB transmission.
Out of 94 clinical isolates for which a 24-locus VNTR profile was available, 71 isolates were determined to be in 16 clusters by cluster analysis, and only 57 remaining isolates were in 11 clusters when a combined spoligoriftyping-VNTR typing dendrogram was built (Fig. 3). Five MDR-TB clusters were found within the Turkish clade (SIT41), and MDR-TB transmission could not be ruled out for these clusters, representing 35 patients. Three SIT53 clusters and three others (SIT125, SIT144, SIT154) were also suggestive of MDR-TB transmission.
Fig 3.
Cluster analysis of 94 clinical isolates for which both results from VNTR analysis of 24 loci and spoligoriftyping results were available. From left to right, dendrogram of results of the unweighted-pair group method using mathematical averages (UPGMA) built based on a composite VNTR-spoligoriftyping similarity matrix, VNTR results (24 loci), spoligotyping results (columns 1 to 43), and rifampin-typing results (Spa_wt1, rif_516 wt [wt3], Spa_wt2, rif_526 wt [wt4], rif_531 wt [wt5], rif_516 mutGTC [mut1], rif_526 mutGAC [mut2], rif_526 mutTAC [mut3], rif_531 mutTTG [mut4], and rif_531 mutTGG [mut5], where “rif” means rpoB). Some prevalent clades are shaded (Turkish clade [clusters SIT41 and SIT125], SIT1-Beijing, and SIT284-East-Med1). Turq, Turkey.
DISCUSSION
We developed a high-throughput 53-plex technique that runs on the microbead-based multianalyte Luminex 200 device. We designated this test “spoligoriftyping,” based on two previous methods on which it was based, spoligotyping and rifoligotyping (21, 25). The spoligoriftyping test allowed us to perfectly perform spoligotyping simultaneously with rpoB hot spot mutation detection for rifampin resistance and prospective MDR-TB genetic testing. Compared to sequencing (spoligoriftyping versus sequencing), both the sensitivity and specificity of spoligoriftyping were 100% for the prediction of rifampin-resistant strains. The sensitivity decreased to 99% when phenotypic DST was used as the gold standard because 1 MDR clinical isolate from the Bulgarian collection was identified as drug susceptible by the spoligoriftyping test. Sequencing showed that this MDR strain had no SNP in the rpoB RRDR region (cluster I), which corroborated our test results. A thorough assessment of the specific history of this patient and of this clinical isolate did not allow us to understand this discrepancy between phenotypic and genotypic results.
Mutations outside the rpoB hot spot regions are not targeted by our test. It is well known in the literature that the majority of rifampin-resistant strains harbor SNPs in the rpoB RRDR, and with high frequency they are in codon 531, and our results corroborated this scientific evidence. However, a few strains harbor SNPs in other regions, like the so-called cluster II and III regions (16); in the cases of phenotypically rifampin-resistant strains that would be undetected by our test, sequencing of the entire rpoB gene should be performed.
Between tests, spoligoriftyping was also successfully used to rapidly characterize rpoB resistance-associated mutations in MTC strains from Pakistan (M. Yasmin, personal communication). The comparison of spoligotyping results obtained with membranes (the classical spoligotyping method) for 91 strains from Bulgaria to those obtained by spoligoriftyping of the same isolate collection showed concordance for 89 strain spoligotype patterns. Our team provided guidelines to improve spoligotyping on membranes based on the use of the microbead-based format, which allows technicians to achieve results with high sensitivity and a more robust and reliable interpretation (1). All of the spoligotype patterns of the 13 strains from Nigeria were also concordant to those obtained by microbead-based spoligotyping (22).
Hence, spoligoriftyping is a new test that may contribute efficiently to tuberculosis control at the local and global levels and especially in areas with high MDR-TB rates and/or with a high burden of HIV infection. Areas like KwaZulu-Natal (in South Africa), with about 10 million people, are an important increasing source of MDR-TB cases (there has been a 10-fold increase in the number of MDR-TB cases from 2001 [216 cases] to 2007 [2,799 cases]) (38). Other settings in Europe, like the Baltic countries, and the republics of the former USSR could also benefit from this test. For future use in developing countries, we are now transferring our assay to the Magpix system, with the aims of potentially increasing portability to point-of-care settings and reducing costs.
The current worldwide increase in MDR-TB prevalence makes it necessary to find methods for rapid identification of resistance genes. Several new molecular methods detecting drug resistance SNPs have already been set up (2, 3, 7, 18, 35, 37, 41). Like GenoType MTBDRplus or GeneXpert MTB/Rif (Cepheid), the spoligoriftyping test allows rpoB gene RRDR mutation detection with high sensitivity and specificity, with the additional advantage of allowing lineage or sublineage identification in a single test and in just a few hours. We are launching a worldwide multicenter study to validate the method's robustness. Ongoing improvements will be (i) the addition of more targets to detect isoniazid (katG, inhA), quinolone (gyrA, gyrB), streptomycin (rrs, rpsl), kanamycin (rrs, eis), or ethambutol (embB) resistance in a single test and (ii) the adaptation of these tests to portable, field-adapted, and new-generation multianalyte systems. Last but not least, compared to GeneXpert (estimated unitary cost, $17 U.S.), the estimated cost of spoligoriftyping could fall below $10, i.e., allowing worldwide implementation in specialized TB labs for an effective MDR-TB control strategy. Together with other tests, such as the MLPA-TB assay (3a, 29), high-throughput nucleic acid-based tests may become interesting alternative methods to phenotypic DST, especially if they become applicable not only to DNA extracted from culture but also to clinical samples (11).
In conclusion, we developed a new multiplex high-throughput genotyping and multidrug resistance molecular detection test for TB control that allows us to both perform molecular epidemiological studies and provide patient-relevant information.
Supplementary Material
ACKNOWLEDGMENTS
M.K.G. is a Ph.D. student supported by a joint grant from the Centre National de la Recherche Scientifique (CNRS) and the Fondation Mérieux.
We are especially grateful to Guy Vernet (Fondation Mérieux, Lyon). Stefan Niemann and Silke Feuerriegel from the Borstel Research Centre (NRL Laboratory for Germany) are also acknowledged for supplying the 4 MDR-TB DNAs described in this study. Christine Pourcel and Gilles Vergnaud, GPMS Team, UMR8621, are acknowledged for providing MDR-TB DNA from clinical isolates during preliminary experiments for rifampin SNP typing. M. François Topin (Luminex BV, Oosterhout, The Netherlands) is acknowledged for technical support.
Footnotes
Published ahead of print 18 July 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Abadia E, et al. 2011. The use of microbead-based spoligotyping for Mycobacterium tuberculosis complex to evaluate the quality of the conventional membrane-based method: providing guidelines for quality control issues when working on membranes. BMC Infect. Dis. 11:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnold C, et al. 2005. Single-nucleotide polymorphism-based differentiation and drug resistance detection in Mycobacterium tuberculosis from isolates or directly from sputum. Clin. Microbiol. Infect. 11:122–130 [DOI] [PubMed] [Google Scholar]
- 3. Bergval IL, et al. 2008. Development of multiplex assay for rapid characterization of Mycobacterium tuberculosis. J. Clin. Microbiol. 46:689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a. Bergval I, et al. Combined species identification genotyping and drug resistance detection of Mycobacterium tuberculosis cultures by MLPA on a bead-based array. PLoS One, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boehme CC, et al. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brudey K, et al. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics, and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. CDC 2011. Report of expert consultations on rapid molecular testing to detect drug-resistant tuberculosis in the United States. CDC, Atlanta, GA: http://www.cdc.gov/tb/topic/laboratory/rapidmoleculartesting/default.htm [Google Scholar]
- 7. Choi GE, et al. 2010. High-resolution melting curve analysis for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 48:3893–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chun JY, et al. 2007. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 35:e40 doi:10.1093/nar/gkm051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cowan LS, Diem L, Brake MC, Crawford JT. 2004. Transfer of a Mycobacterium tuberculosis genotyping method, spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. J. Clin. Microbiol. 42:474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deshpande A, et al. 2009. A rapid multiplex assay for nucleic acid-based diagnostics. J. Microbiol. Methods 80:155–163 [DOI] [PubMed] [Google Scholar]
- 12. Dubois Cauwelaert N, Ramarokoto H, Ravololonandriana P, Richard V, Rasolofo V. 2011. DNA extracted from stained sputum smears can be used in the MTBDRplus assay. J. Clin. Microbiol. 49:3600–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dunbar SA. 2006. Applications of Luminex® xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta 363:71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Evans CA. 2011. GeneXpert—a game-changer for tuberculosis control? PLoS Med. 8:e1001064 doi:10.1371/journal.pmed.1001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hardenbol P, et al. 2003. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 21:673–678 [DOI] [PubMed] [Google Scholar]
- 16. Heep M, et al. 2001. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39:107–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Helb D, et al. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hernandez-Neuta I, et al. 2010. Rifampin-isoniazid oligonucleotide typing: an alternative format for rapid detection of multidrug-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 48:4386–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herrera L, Jimenez S, Valverde A, Garcia-Aranda MA, Saez-Nieto JA. 2003. Molecular analysis of rifampicin-resistant Mycobacterium tuberculosis isolated in Spain (1996–2001). Description of new mutations in the rpoB gene and review of the literature. Int. J. Antimicrob. Agents 21:403–408 [DOI] [PubMed] [Google Scholar]
- 20. Hillemann D, Rusch-Gerdes S, Richter E. 2007. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 45:2635–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamerbeek J, et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lawson L, et al. 2012. A molecular epidemiological and genetic diversity study of tuberculosis in Ibadan, Nnewi and Abuja, Nigeria. PLoS One 7:e38409 doi:10.1371/journal.pone.0038409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin A, et al. 2011. Rapid O serogroup identification of the ten most clinically relevant STECs by Luminex® microbead-based suspension array. J. Microbiol. Methods 87:105–110 [DOI] [PubMed] [Google Scholar]
- 24. Mlambo CK, et al. 2008. Genotypic diversity of extensively drug-resistant tuberculosis (XDR-TB) in South Africa. Int. J. Tuberc. Lung Dis. 12:99–104 [PubMed] [Google Scholar]
- 25. Morcillo N, et al. 2002. A low cost, home-made, reverse-line blot hybridisation assay for rapid detection of rifampicin resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 6:959–965 [PubMed] [Google Scholar]
- 26. Mori T. 2007. MDR-TB—its characteristics and control in Asia-Pacific rim symposium in USJCMSP 10th international conference on emerging infectious diseases in the Pacific rim. Tuberculosis (Edinb.) 87(Suppl 1):S5–S9 [DOI] [PubMed] [Google Scholar]
- 27. Nikolayevskyy V, et al. 2009. Performance of the Genotype MTBDRPlus assay in the diagnosis of tuberculosis and drug resistance in Samara, Russian Federation. BMC Clin. Pathol. 9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Novais RC, Borsuk S, Dellagostin OA, Thorstenson YR. 2008. Molecular inversion probes for sensitive detection of Mycobacterium tuberculosis. J. Microbiol. Methods 72:60–66 [DOI] [PubMed] [Google Scholar]
- 29. Schouten J, et al. 2002. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 30:e57 doi:10.1093/nar/gnf056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen Z, et al. 2010. MPprimer: a program for reliable multiplex PCR primer design. BMC Bioinformatics 11:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song Y, et al. 2010. A multiplex single nucleotide polymorphism typing assay for detecting mutations that result in decreased fluoroquinolone susceptibility in Salmonella enterica serovars Typhi and Paratyphi A. J. Antimicrob. Chemother. 65:1631–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Supply P, et al. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tafaj S, et al. 2009. First insight into genetic diversity of the Mycobacterium tuberculosis complex in Albania obtained by multilocus variable-number tandem-repeat analysis and spoligotyping reveals the presence of Beijing multidrug-resistant isolates. J. Clin. Microbiol. 47:1581–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Telenti A, et al. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647–650 [DOI] [PubMed] [Google Scholar]
- 35. Van Der Zanden AG, Te Koppele-Vije EM, Vijaya Bhanu N, Van Soolingen D, Schouls LM. 2003. Use of DNA extracts from Ziehl-Neelsen-stained slides for molecular detection of rifampin resistance and spoligotyping of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Soolingen D, Hermans PWM, de Haas PEW, Sool DR, Embden JDA. 1991. The occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wada T, et al. 2004. Dual-probe assay for rapid detection of drug-resistant Mycobacterium tuberculosis by real-time PCR. J. Clin. Microbiol. 42:5277–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wallengren K, et al. 2011. Drug-resistant tuberculosis, KwaZulu-Natal, South Africa, 2001–2007. Emerg. Infect. Dis. 17:1913–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weile J, Knabbe C. 2009. Current applications and future trends of molecular diagnostics in clinical bacteriology. Anal. Bioanal. Chem. 394:731–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wright A, et al. 2009. Epidemiology of antituberculosis drug resistance 2002–07: an updated analysis of the Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Lancet 373:1861–1873 [DOI] [PubMed] [Google Scholar]
- 41. Yao C, et al. 2010. Detection of rpoB, katG and inhA gene mutations in Mycobacterium tuberculosis clinical isolates from Chongqing as determined by microarray. Clin. Microbiol. Infect. 16:1639–1643 [DOI] [PubMed] [Google Scholar]
- 42. Zhang J, et al. 2010. Mycobacterium tuberculosis complex CRISPR genotyping: improving efficiency, throughput and discriminative power of ‘spoligotyping’ with new spacers and a microbead-based hybridization assay. J. Med. Microbiol. 59:285–294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.