Abstract
Microbial dysbiosis has been suggested to be involved in the pathogenesis of Crohn's disease (CD); however, many studies of gut microbial communities have been confounded by environmental and patient-related factors. In this study, the microbial flora of fecal samples from 19 children newly diagnosed with CD and 21 age-matched controls were analyzed using high-throughput sequencing to determine differences in the microbial composition between CD patients and controls. Analysis of the microbial composition of specific bacterial groups revealed that Firmicutes percentages were significantly lower in CD patients than in controls and that this was due largely to changes in the class Clostridia. Bacteroidetes and Proteobacteria percentages were higher and significantly higher in CD patients than in controls, respectively. Both the detection frequencies of Bacteroidetes and Firmicutes correlated (positively and negatively, respectively) with the calculated pediatric Crohn's disease activity index scores of patients. Upon further analysis, differences in the microbial compositions of patients with mild disease and moderate to severe disease were identified. Our findings indicate that a combination of different bacterial species or a dynamic interplay between individual species is important for disease and is consistent with the dysbiosis hypothesis of CD.
INTRODUCTION
Crohn's disease (CD) is a chronic relapsing idiopathic disease. CD and ulcerative colitis are the two most common forms of inflammatory bowel diseases (IBD) (41). CD is a multifactorial disease with unknown etiology. However, it is currently hypothesized that intestinal microorganisms, in association with a disruption of the gastrointestinal epithelium, stimulate and subsequently drive a dysregulated immune response in predisposed individuals (47).
In recent literature, dysbiosis, a breakdown in the balance between commensal and pathogenic intestinal bacteria, has been suggested to be involved in the pathogenesis of CD. A consistent finding across studies investigating dysbiosis in CD is that in patients the abundance of members of the Firmicutes is decreased, whereas the abundance of members of the Proteobacteria (Escherichia coli in particular)is increased compared to that in controls (41). These findings are supported by observations in mouse models of CD where prolific colonization by commensal bacteria, such as Enterobacteriaceae, drives the depletion of other groups, such as Firmicutes (39).
The results observed for CD-related changes in groups such as the Bacteroidetes are, however, more inconsistent. For example, while Frank et al. (21) and Ott et al. (45) reported Bacteroidetes to be significantly depleted in CD patients compared to in controls, Rehman et al. (46) and Swidsinski et al. (53) showed Bacteroidetes to be more prevalent in CD patients than in controls. The latter findings are supported by studies examining the intestinal microbiota in the TRUC (T-bet, RAG, ulcerative colitis) mouse model of spontaneous colitis, where bacteria belonging to the order Bacteroidales (phylum Bacteroidetes) were reported to be present in high numbers (23). Furthermore, Rehman et al. have reported the transcriptional activity of Bacteroidetes to be significantly increased in patients with CD compared with that in controls (46).
While the current literature on dysbiosis and CD provides significant insights into microbial changes associated with CD, the data have not been consistent. Possible factors for these inconsistencies include differences in the techniques employed to survey the intestinal microbiota, study design, stage of disease and its location, and control populations used. To avoid potential confounding factors associated with previous studies in adults, including previous treatment with antibiotics or anti-inflammatory therapies, differences in stage of disease, smoking, and alcohol intake, we conducted the current study in children newly diagnosed with CD who had not undergone prior antibiotic or anti-inflammatory therapy for CD and age-matched controls.
MATERIALS AND METHODS
Patients.
Twenty-two symptomatic children undergoing diagnostic colonoscopy and upper endoscopy at the Sydney Children's Hospital Randwick (Sydney, Australia) were included in the study.
Based upon standard endoscopic, histologic, and radiologic investigations (24), 19/22 children (12 male; mean age of 11.6 ± 2.5 years) were newly diagnosed with ileocolonic CD with upper-gut involvement. The pediatric Crohn's disease activity index (PCDAI) ranged from 7.5 to 70 (Table 1). Of the remaining three children, one was diagnosed with reflux esophagitis (female, 12.2 years), one was diagnosed with duodenal ulcer disease (male, 13.1 years), and one was diagnosed with a functional bowel disorder (male, 11.7 years).
Table 1.
Characteristics of children with Crohn's disease included in the study, the location of their disease, and their PCDAI at diagnosis
| Patient | Age (yrs) | Gender | Disease location | PCDAI |
|---|---|---|---|---|
| CD1 | 9 | F | L3 + L4 (ileocolon) | 7.5 |
| CD2 | 12 | F | L3 + L4 (ileocolon) | 57.5 |
| CD3 | 14 | M | L3 + L4 (ileocolon) | 45 |
| CD4 | 13 | M | L3 + L4 (ileocolon) | 22.5 |
| CD5 | 14 | M | L3 + L4 (ileocolon) | 52.5 |
| CD6 | 15 | F | L3 + L4 (ileocolon) | 52 |
| CD7 | 14 | F | L3 + L4 (ileocolon) | 25 |
| CD8 | 9 | M | L3 + L4 (ileocolon) | 35 |
| CD9 | 10 | M | L3 + L4 (ileocolon) | 20 |
| CD10 | 13 | M | L3 + L4 (ileocolon) | 20 |
| CD11 | 14 | M | L3 + L4 (ileocolon) | 27.5 |
| CD12 | 15 | M | L3 + L4 (ileocolon) | 37.5 |
| CD13 | 9 | M | L3 + L4 (ileocolon) | 27.5 |
| CD14 | 8.6 | M | L3 + L4 (ileocolon) | 25 |
| CD15 | 11.4 | F | L3 + L4 (ileocolon) | 37.5 |
| CD16 | 13 | F | L3 + L4 (ileocolon) | 70 |
| CD17 | 8.7 | M | L3 + L4 (ileocolon) | 10 |
| CD18 | 7.2 | M | L3 + L4 (ileocolon) | 42.5 |
| CD19 | 11 | F | L3 + L4 (ileocolon) | 52.5 |
Along with the three children who underwent colonoscopy and who were not diagnosed with IBD, 18 healthy children were also included in the study as controls. These 21 control children with no histological features of IBD comprised 13 males and had a mean age of 9.5 ± 4.2 years (P = 0.07).
No child involved in this study had undergone prior antibiotic or anti-inflammatory therapy in the previous 4 weeks. Informed consent was obtained from all children (or their parent/guardian for younger children) to be included in the study. This study was approved by the Research Ethics Committees of the University of New South Wales and the South East Sydney Area Health Service—Eastern Section, Sydney (ethics no. 03/163, 03/165, and 06/164). Fecal samples were collected from each child prior to colonoscopic examination.
DNA extraction and microbial community sequencing.
DNA extraction was performed using the ISOLATE fecal DNA kit (Bioline) according to the manufacturer's instructions. The concentration and quality of DNA was measured using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE).
The microbial community was assessed by high-throughput sequencing of the 16S rRNA gene. Tag-encoded FLX amplicon pyrosequencing (bTEFAP) was performed as described previously using the primers Gray28F (5′TTTGATCNTGGCTCAG) and Gray519r (5′ GTNTTACNGCGGCKGCTG) (2, 3, 20, 26) with the primers numbered in relation to E. coli 16S rRNA (variable regions 1 to 3). The sequence of the primers is not complementary to human DNA, thus preventing coamplification of human sequences. Moreover, the primers span the variable region of the rRNA gene so that discrimination between closely related taxa can be performed. At the same time, the positioning of the primers allows for amplification of a large proportion of known 16S rRNA sequences. Generation of the sequencing library utilized a one-step PCR with a total of 30 cycles, a mixture of Hot Start and HotStar high-fidelity Taq polymerases, and amplicons originating and sequencing extending from the 28F with an average read length of 400 bp. Tag-encoded FLX amplicon pyrosequencing analyses utilized a Roche 454 FLX instrument with Titanium reagents. This bTEFAP process was performed at the Molecular Research laboratory (MR DNA, Shallowater, TX) based upon established and validated protocols.
Data analysis.
The sequence data derived from the high-throughput sequencing process were analyzed, employing a pipeline developed at Molecular Research LP. Sequences are first depleted of barcodes and primers, and then short sequences of <200 bp, sequences with ambiguous base calls, and sequences with homopolymer runs exceeding 6 bp are all removed. Sequences were then denoised and chimeras were removed (Black Box Chimera Check software B2C2; n = 2,129). Operational taxonomic units (OTU) were defined after removal of singleton sequences (sequences appearing only once in the whole data set) with clustering set at 3% divergence (97% similarity) (9, 14–17, 19, 51). OTU were then taxonomically classified using BLASTn against a curated GreenGenes database (12) and compiled into each taxonomic level. Taxonomy was defined based on the following percentages: >97%, species; between 97% and 95%, unclassified species; between 95% and 90%, unclassified genus; between 90% and 85%, unclassified family; between 85% and 80%, unclassified order; between 80% and 77%, unclassified phylum; <77%, unclassified. Statistical analyses based on relative abundances of bacterial groups were performed using Primer-E. Permutational multivariate analysis of variance (PERMANOVA) (1), which overcomes multiple hypothesis testing, was performed to detect overall group differences between CD patients and controls. The question of the test was, “Are there differences in Bray-Curtis similarities between bacterial communities of CD patients and controls?” In order to explore for key variables, differences in relative abundances of bacterial taxa between CD patients and controls were tested using standard t tests. Upon stratification of CD patients into mild and moderate/severe groups, a one-way analysis of variance (ANOVA) with a post hoc Tukey's correction was used to correct for multiple testing. Although examining the multivariate significance of the data provides some protection against increased chance of type I error, it is still likely that comparisons may achieve statistical significance by chance.
RESULTS AND DISCUSSION
Evidence to support the role of microorganisms in the pathogenesis of CD has been demonstrated in both humans and animals (41). However, the specific microorganism or group of microorganisms responsible for the initiation of CD is yet to be determined. Thus, in an attempt to gain a better understanding of microbial involvement in CD, we examined the microbial flora in fecal samples of children newly diagnosed with CD and age-matched controls, as this population is relatively free of confounding factors.
The mean number of sequences per sample obtained for the 40 subjects in this study following data analysis was 2,609 ± 138 sequences. To check for sampling bias arising from grouping the subjects into CD patients and controls, the number of sequences per sample within each group was analyzed. The mean number of sequences per sample for CD patients and controls were 2,716 ± 220 and 2,513 ± 174 sequences, respectively, and this difference was not statistically significant (P = 0.47), indicating that no sampling bias due to the grouping existed.
Differences in microbial compositions of the control subjects.
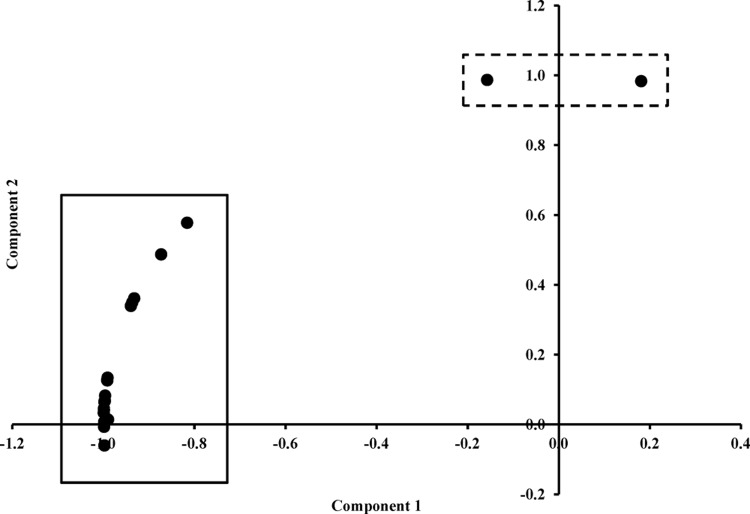
Principal component analysis was performed on the detection frequencies obtained for the control subjects in order to identify any outliers that could influence downstream analyses. Two subjects were shown to be outliers with respect to the other controls (Fig. 1). Interestingly, one of these outliers was the control subject diagnosed with a functional bowel disorder. Upon more specific analysis of the detection frequencies in this child, it was found that they had a high Actinobacteria count (17.1%), which was due to a putative Mycobacterium peregrinum infection (14.8%; 0% in all other subjects). Although M. peregrinum has yet to be clearly associated with human disease, this bacterium has been related to surgical site infections and catheter-related infections (44). Interestingly, infection with M. peregrinum was reported to be the cause of an outbreak of mycobacteriosis in an aviary containing Gouldian finches (Erythrura gouldiae) where affected birds developed granulomatous lesions of the liver and intestine (54). Another study reported that exposure of zebrafish to M. peregrinum led to clinical signs of mycobacteriosis (27). The second outlier (Fig. 1; see Fig. S1 in the supplemental material) was a healthy control that was found to have an unusually high level of Bacteroidetes (63.8%) that was shown to result from high levels of Prevotella copri (55.2%). Although P. copri has been previously isolated from human feces (28), no studies have associated this bacterium with any specific disease outcome. It is unknown why this healthy control had such high levels of P. copri within its fecal microbial composition. Given these anomalies, these two outliers were excluded from further multivariate and univariate comparisons with CD patients. Thus, for the final fecal microbiota comparisons, 19 control children (12 males) and 19 children with CD (12 males) were included.
Fig 1.
Principal component analysis of the bacterial community compositions of the control subjects.
Variations in microbial compositions between patients with Crohn's disease and controls.
Analysis of the microbial composition of CD patients and controls revealed that as a whole, error values within the detection frequencies of CD patients were relatively higher than that within the controls (percentage standard errors on the three major phyla Firmicutes, Bacteroidetes, and Proteobacteria were 11.6%, 26.7%, and 38.2% for CD patients and 3.0%, 21.8%, and 31.2% for controls, respectively), indicating that higher variations existed among individual CD patients than among controls. Despite this, PERMANOVA analysis showed that differences between CD patients and the remaining 19 controls were significant (P = 0.015), and several taxa were found to be significantly different in abundance in CD patients compared with controls using univariate analyses.
Firmicutes.
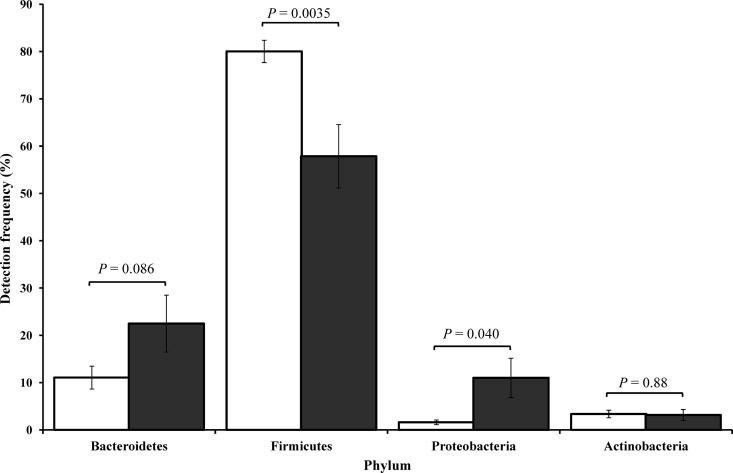
Analysis of the microbial composition for specific bacterial groups revealed that the abundance of Firmicutes was significantly lower in CD patients than in controls (P = 0.0035) (Table 2, Fig. 2). This shift in Firmicutes was found to be due largely to changes in the detection of class Clostridia (and order Clostridiales) that was significantly lower in CD patients than in controls (P = 0.0009) (Table 2). The decrease in relative abundance of members of the Firmicutes in the children with CD is consistent with recent findings on microbial composition changes in CD patients (41). For example, Rehman et al. showed the abundance of Firmicutes to be significantly lower in patients with CD (52.7%) than in healthy controls (78.6%; P < 0.01) (46).
Table 2.
Differences in detection frequencies of bacterial groups between CD patients and controls
| Phylum | Class | Order/family | Genus | Species | CD (%) | Controls (%) | P valuea |
|---|---|---|---|---|---|---|---|
| Firmicutes | 57.9 ± 6.7 | 80.0 ± 2.4 | 0.0035* | ||||
| Clostridia | 49.6 ± 7.3 | 77.4 ± 2.4 | 0.0009* | ||||
| Clostridiaceae | 10.3 ± 4.1 | 5.7 ± 1.4 | 0.29 | ||||
| Clostridium | 14.4 ± 3.9 | 8.8 ± 1.1 | 0.21 | ||||
| Lachnospiraceae | 18.0 ± 5.7 | 30.8 ± 4.6 | 0.11 | ||||
| Coprococcus | 0.93 ± 0.26 | 5.1 ± 0.8 | 4.4 × 10−5* | ||||
| Roseburia | 4.4 ± 1.8 | 10.4 ± 2.0 | 0.031* | ||||
| Roseburia faecis | 1.5 ± 0.7 | 4.5 ± 0.9 | 0.016* | ||||
| Coprococcus eutactus | 0.025 ± 0.017 | 0.95 ± 0.29 | 0.0045* | ||||
| Coprococcus “Clostridium sp. SS2/1” | 0.33 ± 0.13 | 2.4 ± 0.7 | 0.012* | ||||
| Ruminococcaceae | 15.0 ± 3.0 | 27.4 ± 2.2 | 0.0033* | ||||
| Oscillospira | 0.26 ± 0.16 | 1.8 ± 0.6 | 0.033* | ||||
| Subdoligranulum | 0.68 ± 0.45 | 3.5 ± 0.7 | 0.0029* | ||||
| Faecalibacterium prausnitzii | 9.9 ± 2.4 | 14.1 ± 2.2 | 0.20 | ||||
| Bacteroidetes | 22.5 ± 6.0 | 11.1 ± 2.4 | 0.086 | ||||
| Bacteroides | 19.6 ± 5.4 | 8.4 ± 2.3 | 0.065 | ||||
| Actinobacteria | 3.2 ± 1.2 | 3.4 ± 0.8 | 0.88 | ||||
| Actinomycetales | 0.24 ± 0.12 | 0.069 ± 0.031 | 0.19 | ||||
| Proteobacteria | 11.0 ± 4.2 | 1.6 ± 0.5 | 0.040* | ||||
| Gammaproteobacteria | 9.6 ± 4.1 | 1.1 ± 0.5 | 0.056 | ||||
| Enterobacteriales | 9.2 ± 4.3 | 0.71 ± 0.36 | 0.058 | ||||
| Escherichia | 3.2 ± 1.4 | 0.29 ± 0.16 | 0.062 | ||||
| Enterobacter | 2.3 ± 1.0 | 0.13 ± 0.05 | 0.061 | ||||
| Enterobacter hormaechei | 2.3 ± 1.0 | 0.11 ± 0.05 | 0.059 | ||||
| Alphaproteobacteria | 0.34 ± 0.13 | 0.10 ± 0.08 | 0.13 | ||||
| Rhizobiales | 0.10 ± 0.03 | 0 | 0.0033* | ||||
| Bradyrhizobiaceae | 0.036 ± 0.016 | 0 | 0.045* | ||||
| Betaproteobacteria | 1.0 ± 0.3 | 0.43 ± 0.19 | 0.12 | ||||
| Burkholderiales | 0.91 ± 0.28 | 0.42 ± 0.19 | 0.11 |
*, P < 0.05.
Fig 2.
Bacterial diversity observed in Crohn's disease patients and age-matched controls. Differences in detection frequencies of bacterial phyla between CD patients (gray) and controls (white). Firmicutes (CD patients, 57.9 ± 6.7%; controls, 80.0 ± 2.4%; P = 0.0035); Proteobacteria (CD patients, 11.0 ± 4.2%; controls, 1.6 ± 0.5%; P = 0.040); Bacteroidetes (CD patients, 22.5 ± 6.0%; controls, 11.1 ± 2.4%; P = 0.086); Actinobacteria (CD patients, 3.2 ± 1.2%; controls, 3.4 ± 0.8%; P = 0.88). Error bars are the standard errors of the means.
Three families (Clostridiaceae, Lachnospiraceae, and Ruminococcaceae) were found to have various relative abundances when the two subject groups were compared. Clostridiaceae, specifically members of the Clostridium genus, which comprises several important pathogens, were numerically higher in CD patients than in controls (Table 2), although this was not significant (P = 0.29). In contrast, Lachnospiraceae counts were higher in controls than in CD patients (Table 2); however, again this did not reach significance (P = 0.11). Closer analysis of this family showed that two genera, Coprococcus and Roseburia (P = 4.4 × 10−5 and P = 0.031, respectively), were driving the results (Table 2). Species of interest (those with significantly different relative abundances between CD patients and controls) were Roseburia faecis, Coprococcus eutactus, and a Coprococcus strain that was initially named Clostridium sp. strain SS2/1 (Coprococcus “Clostridium sp. SS2/1”) (Table 2).
Ruminococcaceae numbers were found to be significantly higher in controls than in CD patients (P = 0.0033). These results were partly due to significant differences in Oscillospira (P = 0.033) and Subdoligranulum (P = 0.0029). Unlike previous observations in adult patients (41, 48), where significantly lower abundances of Faecalibacterium prausnitzii have been reported in CD patients than in controls, no significant difference in Faecalibacterium prausnitzii was observed between the CD children and controls (P = 0.20); however, the frequency in CD children was lower than that in controls (Table 2). Interestingly, F. prausnitzii is believed to be an anti-inflammatory commensal bacterium, which when decreased in abundance increases the risk of CD recurrence (49). Our contrasting findings may reflect differences in the immunomodulatory roles of the gut microflora in adults and children and/or the fact that our patients were newly diagnosed with the disease.
One CD patient (CD9) who had a relatively high Firmicutes count (92.2%) compared with that of other CD patients (average, 57.9 ± 6.7%) was found to have unusually high counts of Streptococcus spp., including S. parasanguinis (30.9%), S. thermophilus (30.3%), and S. oralis (8.3%). Streptococcus spp. have been previously associated with CD, with streptococcal DNA being identified in CD patients but not in healthy controls (25).
Bacteroidetes.
In the current study, numerically higher percentages of Bacteroidetes were observed in CD patients than in controls (P = 0.086) (Table 2, Fig. 2). The drop in Firmicutes detection levels correlated with the increase in Bacteroidetes levels (CD patients, Pearson's correlation coefficient [r17] = −0.71, P = 0.0006; controls, r17 = −0.82, P < 0.0001). Similar correlations were observed when comparing the detection frequencies of Bacteroidia, Bacteroidales, and Bacteroidaceae in CD patients and controls. The only genus within Bacteroidaceae showing consistent results in its detection frequency between CD patients and controls that was close to significance was Bacteroides (P = 0.065) (Table 2). While the difference between the CD patients and controls improved statistically from the phylum to genus level (P = 0.086 to 0.065), it still did not reach significance (discussed further below). As reported previously by Swidsinski et al. (52), the Bacteroides within each patient were not represented by a single species but by multiple species. The change in the level of bacteria from the phylum Bacteroidetes was of particular interest given the study by Bloom et al. (5), who showed common commensal Bacteroides species, such as Bacteroides vulgatus and Bacteroides thetaiotaomicron, to colonize IBD-susceptible and -nonsusceptible mice equivalently but to induce disease exclusively in susceptible animals.
Fusobacteria.
In the current study, Fusobacteria were detected in 8/19 CD patients (patient CD2, having a high detection frequency of 29.2%) and 1/19 controls, with the one positive-control patient being the control diagnosed with duodenal ulcer disease (Fusobacterium canifelinum). None of the healthy controls had Fusobacteria. The species detected in our CD patients were Fusobacterium equinum (n = 2), Fusobacterium canifelinum (n = 3), Fusobacterium nucleatum (n = 1), Fusobacterium periodonticum (n = 1), and the misclassified (10) Clostridium rectum (n = 1). Fusobacterium nucleatum, which was detected in patient CD14, has been recently associated with colorectal carcinoma (34). Moreover, Strauss et al. (50) isolated Fusobacterium spp. from a significantly higher number of IBD (CD, n = 17; ulcerative colitis, n = 4; IBD unclassified, n = 1) patients (15/22, 68.2%) than non-IBD controls (9/34, 26.5%). Interestingly, these authors found that Fusobacterium strains originating from inflamed biopsy tissue of IBD patients were significantly more invasive than strains isolated from healthy tissue from either IBD patients or control patients (50).
Actinobacteria.
The frequencies of members of the phylum Actinobacteria were similar in both the CD and control subjects (P = 0.88) (Table 2, Fig. 2). However, in one CD patient (CD6), a very high Actinobacteria count (22.2%) was noted, which was shown to be due to the presence of two species, Collinsella aerofaciens (8.2%) and Bifidobacterium longum (9.9%). Interestingly, Swidsinski et al. (52) have previously reported a preferential increase in the frequency of C. aerofaciens in IBD patients. Moreover, the frequency of members of the order Actinomycetales was higher in CD patients than in controls (Table 2), and these bacteria were detected in 13/19 CD patients in comparison to 5/19 controls. However, these differences were not significant (P = 0.19).
Proteobacteria.
In the current study, Proteobacteria percentages were significantly higher in CD patients than in controls (P = 0.040) (Table 2, Fig. 2). Further analysis revealed that the detection rates of Proteobacteria were mostly driven by Gammaproteobacteria detection (P = 0.056) (Table 2).
The increased detection of Gammaproteobacteria in CD patients resulted from the detection of Enterobacteriales (P = 0.058), specifically Escherichia (P = 0.062) and Enterobacter (P = 0.061) (Table 2). The specific species detected within the Escherichia genus based on our BLAST sequence analyses of the pyrosequencing reads were Escherichia coli, Escherichia fergusonii, and Escherichia albertii. Interestingly, previous studies have reported adherent and invasive E. coli (AIEC) to be associated with ileal CD, with one study reporting the isolation of AIEC in 22% of ileal biopsy samples from CD patients compared with 6.2% of controls (11). Moreover, a further study has reported the isolation of AIEC in 29% of CD patients compared with 9% of controls (42). While E. coli was detected in our CD patients, whether this is an AIEC strain cannot be ascertained. Within the Enterobacter genus, only one species, Enterobacter hormaechei, was detected in our CD patients. While the detection rate in CD patients was higher than in controls (Table 2), this did not quite reach significance (P = 0.059).
One CD patient (CD11) who was found to have very high Proteobacteria counts (77.4%) was found to possibly have an unclassified Citrobacter infection (46.2%). This is of some interest given that in mice Citrobacter rodentium infection has been shown to elicit a highly polarized Th1 response, including upregulation of interleukin-12, gamma interferon, and tumor necrosis factor alpha, and a pathology very similar to that found in mouse models of inflammatory bowel disease (29). More recently, Th17 cells have been implicated in C. rodentium infections (37), a finding which is of interest considering the Th17 response has also been implicated in the development of Crohn's disease (6).
The Klebsiella granulomatis and Klebsiella variicola were detected in 4 and 5 CD patients, respectively, and 0 controls, although in one control Klebsiella pneumoniae was detected. In 2009, Gutierrez et al. reported Klebsiella DNA to be present in the blood of one CD patient but not in any healthy controls (25). Moreover, the presence of K. pneumoniae has been correlated with colitis in a genetically deficient mouse model (23), although this is not supported by our findings.
In 2008, Wagner et al. used a genus-specific bacterial 16S PCR as well as nested PCR to investigate the prevalence and diversity of Pseudomonas species in ileal biopsy specimens of 32 children with CD at initial endoscopic examination, as well as in control children with noninflammatory bowel disease (non-IBD) (55). This showed that 58% of CD patients demonstrated positive results for Pseudomonas spp., which was significantly higher than for non-IBD controls (33%). In the current study, members of the Pseudomonadaceae were detected in 5 CD patients (26.3%) and in no controls. Further analysis revealed that Pseudomonas balearica was the species detected only in the CD patients.
While a higher abundance of Alphaproteobacteria was detected in CD patients than in controls (Table 2), this difference was not significant (P = 0.13). Comparison of the order Rhizobiales (detected in 10/19 CD patients versus 0/19 controls) showed a significantly higher abundance in CD patients than in controls (P = 0.0033) (Table 2). Bradyrhizobiaceae (detected in 6/19 CD patients versus 0/19 controls) within the Rhizobiales also showed a significantly higher abundance in CD patients than in controls (P = 0.045) (Table 2). The species identified were an unclassified Balneimonas (n = 3) and Afipia broomae (n = 3). Other Alphaproteobacteria species identified were Methylobacterium adhaesivum (n = 2) and Methylobacterium hispanicum (n = 2) (within Methylobacteriaceae), Aurantimonas coralicida (n = 2) (within Aurantimonadaceae), Rhodoplanes cryptolactis (n = 1) and Gemmiger formicilis (n = 1) (within Hyphomicrobiaceae), and Rhizobium spp. IRBG 74 (n = 1) (within Rhizobiaceae). While it appears that A. broomae may be an opportunistic pathogen in elderly people (7), the higher abundance of Alphaproteobacteria in CD patients is more likely to have resulted from changes in the gut environment of patients (e.g., higher methane production and higher reactive nitrogen species production), as these bacteria tend to be environmental rather than host related.
Betaproteobacteria were also higher in abundance in CD patients than in controls, the predominant order detected being Burkholderiales (Table 2). Three species of interest were detected within this order, Cupriavidus gilardii (4 CD patients versus 0 controls), Massilia timonae (5 CD patients versus 0 controls), and Paucimonas lemoignei (4 CD patients versus 0 controls). C. gilardii is not known for its pathogenicity; however, it has been reported to infect humans, in one case causing fatal sepsis following the dissemination of an infection with intestinal focus (33). M. timonae has also been reported to infect humans, being isolated from blood, cerebrospinal fluid, and bone samples (36). Interestingly, Neisseriales were detected in 6 CD patients and in 1 control, the species detected being Eikenella corrodens (3 CD patients versus 0 controls) and an unclassified Vogesella species (3 CD patients versus 1 control).
Deltaproteobacteria, which have been associated with IBD (38), were detected in four CD patients (CD3, CD5, CD6, and CD12) and no controls. The bacteria were identified as an unclassified Entotheonella (belonging to Entotheonellales) (CD3 and CD5), Bilophila wadsworthia (belonging to Desulfovibrionales) (CD6 and CD12), and an unclassified Haliangium (belonging to Myxococcales) (CD6). Both Entotheonella spp. and Haliangium spp. appear to be environmental bacteria with no association with human disease (8, 22). In contrast, B. wadsworthia has been associated with human infections, including appendicitis and cholecystitis (4).
Bacteria from the class Epsilonproteobacteria were detected in two patients (CD1 and CD8) and no controls. The bacterium was identified as Campylobacter concisus, a species recently associated with IBD (32, 41). Interestingly, C. concisus strains isolated from chronic intestinal diseases such as CD have been shown to be more invasive than strains isolated from acute intestinal disease and healthy subjects (31, 40).
Possible bacterial species associated with initiating and driving CD in the 19 CD patients in this study are detailed in Table 3. These bacteria were chosen based on their previous association with CD in other studies, their pathogenic potential in humans, or their relative abundance in CD patients versus controls in this study.
Table 3.
Possible bacterial species associated with initiating and driving Crohn's disease in the CD patients in this studya
| Patient | Possible agents associated with disease |
|---|---|
| CD1 | Campylobacter concisus; Pseudomonas balearica; Cupriavidus gilardii |
| CD2 | Fusobacterium equinum (27.4%); Klebsiella spp. |
| CD3 | Cupriavidus gilardii; Pseudomonas balearica; Massilia timonae; Fusobacterium equinum |
| CD4 | Ruminococcus torques (21.1%) |
| CD5 | Klebsiella variicola; Escherichia fergusonii; Bacteroides spp. (73.7%); Fusobacterium canifelinum |
| CD6 | Massilia timonae; Escherichia fergusonii; Bilophila wadsworthia; Collinsella aerofaciens (8.2%); Fusobacterium canifelinum |
| CD7 | Cupriavidus gilardii; Pseudomonas balearica; Massilia timonae; Escherichia fergusonii; Clostridium difficile (6.5%) |
| CD8 | Campylobacter concisus; Clostridium difficile (24.7%); Cupriavidus gilardii; Pseudomonas balearica; Massilia timonae; Klebsiella spp.; Escherichia fergusonii |
| CD9 | Streptococcus spp. (74.0%) |
| CD10 | Klebsiella spp.; Escherichia fergusonii; Bacteroides spp. (50.2%); Fusobacterium canifelinum |
| CD11 | Citrobacter unclassified (46.2%); Escherichia fergusonii; Klebsiella spp.; Pseudomonas balearica; Massilia timonae |
| CD12 | Bilophila wadsworthia; Fusobacterium canifelinum |
| CD13 | Clostridium difficile (2.2%) |
| CD14 | Escherichia coli; Shigella sonnei; Fusobacterium nucleatum; Fusobacterium periodonticum; Peptostreptococcus anaerobius (13.9%) |
| CD15 | Unusual Alphaproteobacteria detected |
| CD16 | Escherichia coli; Shigella sonnei; Bacteroides spp. (56.3%) |
| CD17 | Escherichia coli; Shigella sonnei |
| CD18 | Bacteroides spp. (30.0%) |
| CD19 | Clostridium rectum (Fusobacteria); Bacteroides spp. (57.1%); Parabacteroides spp. (19.1%) |
These bacteria were chosen based on their previous association with CD in other studies, their pathogenic potential in humans, or their relative abundance in CD patients versus controls in this study.
Effect of age and gender on microbial composition.
While age has been reported to affect the intestinal microbial composition in infants or old people (18), in the current study no correlation between microbial composition and age was observed, and this may relate to the fact that the children were not infants and had a fairly narrow age range. While Lay et al. (35) and Dicksved et al. (13) have reported no significant correlation between microbial composition and age, Mueller et al. (43) reported both Enterobacteria and Bacteroides-Prevotella detection levels to be influenced by the subject's age. In addition to age, a number of previous studies have also investigated the effect of gender on the fecal microbial composition of subjects. In the current study, the detection frequencies of Firmicutes, Bacteroidetes, and Proteobacteria were compared in females and males in both the CD patients and controls. This showed that although Bacteroidetes counts appeared to be higher in females (both CD patients and controls) and Proteobacteria counts appeared to be higher in males (both CD patients and controls), no statistical difference was observed between females and males within either subject subgroup (Table 4).
Table 4.
Effect of gender on the detection frequencies of three bacterial phyla
| Subjects | Gender | Bacterial detection frequencies (%) |
|||||
|---|---|---|---|---|---|---|---|
| Firmicutes | P value | Bacteroidetes | P value | Proteobacteria | P value | ||
| CD patients | Female | 53.9 | 0.66 | 26.6 | 0.61 | 6.8 | 0.47 |
| Male | 60.2 | 20.1 | 13.5 | ||||
| Controls | Female | 80.2 | 0.95 | 13.5 | 0.45 | 0.47 | 0.085 |
| Male | 79.9 | 9.6 | 2.3 | ||||
Associations between the inflammatory index of Crohn's disease patients.
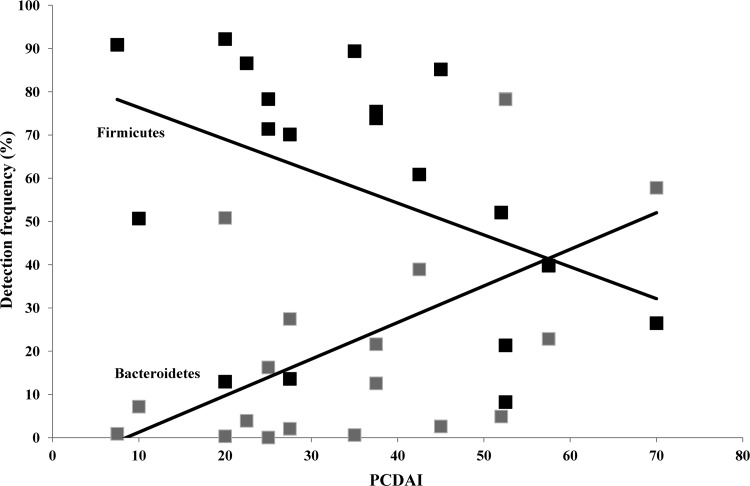
As one of the key hallmarks of CD is chronic inflammation, the microbial composition of patients was assessed with respect to their calculated PCDAI at diagnosis. Correlation analysis revealed that there existed a strong positive correlation between the detection frequencies of Bacteroidetes and the calculated PCDAI scores of each patient (r17 = 0.544, P = 0.016) (Fig. 3). In contrast, a negative correlation was identified between the detection frequencies of Firmicutes and PCDAI (r17 = −0.425, P = 0.070) (Fig. 3); however, this did not reach significance.
Fig 3.
Correlations between detection frequencies of Firmicutes and Bacteroidetes and patient PCDAI. Firmicutes, black (r17 = −0.425, P = 0.070); Bacteroidetes, gray (r17 = 0.544, P = 0.016).
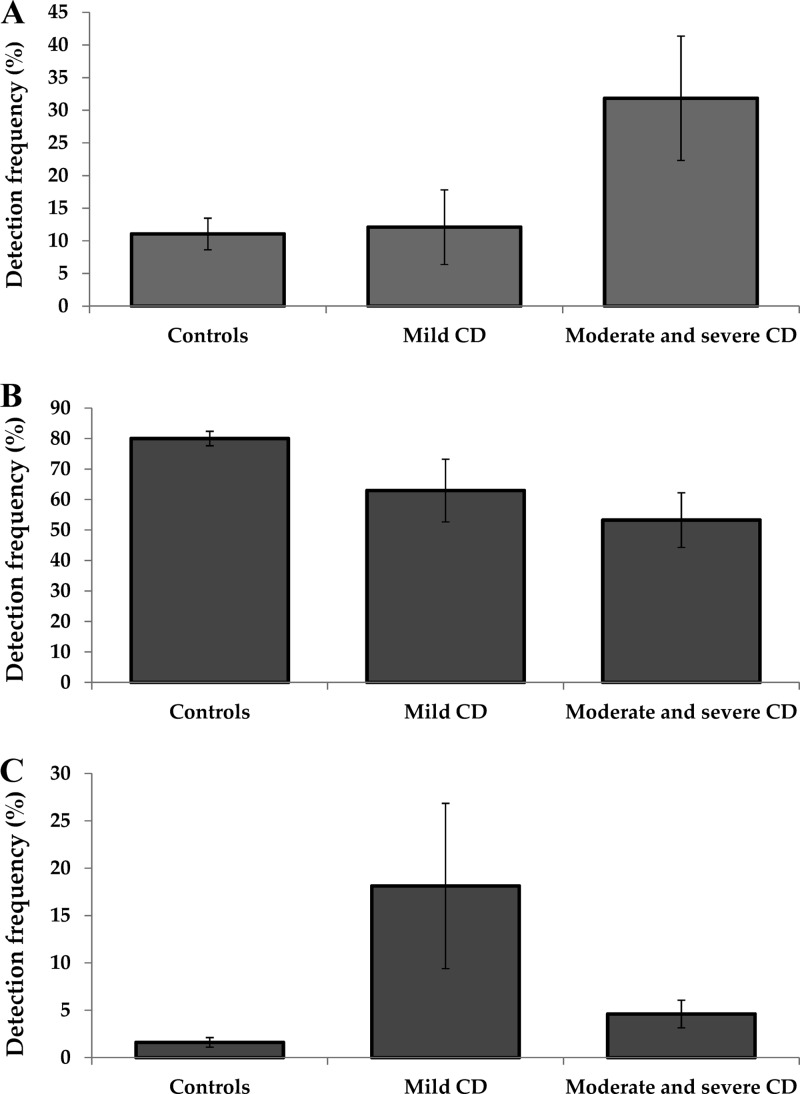
Due to the observed correlation between microbial composition and PCDAI, patients were stratified into two groups consisting of mild disease (PCDAI < 30) and moderate to severe disease (PCDAI ≥ 30) (30). This categorization of CD patients based on disease activity revealed that Bacteroidetes were present at significantly higher levels in moderate to severe disease (31.8 ± 9.5%) than in controls (11.1 ± 2.4%, P = 0.021), whereas patients with mild disease (12.1 ± 5.7%) had levels similar to those of controls (P = 0.99) (Fig. 4). The finding that Bacteroidetes were similar in patients with mild disease and controls suggests that their abundance is either as a result of inflammation or is driving inflammation. Interestingly, Firmicutes were present at numerically lower levels in mild (63.0 ± 10.3%, P = 0.15) and significantly lower levels in moderate to severe disease (53.3 ± 9.0%, P = 0.0097) than in controls (80.0 ± 2.4%) (Fig. 4), suggesting that inflammation may have a negative effect on Firmicutes or that changes in the Bacteroidetes and Proteobacteria (below) may affect their abundance. Of particular interest was the finding that Proteobacteria were present at higher levels in mild (18.1 ± 8.7%) than in moderate to severe (4.6 ± 1.5%, P = 0.070) disease and controls (1.6 ± 0.5%, P = 0.0085) (Fig. 4). Given the higher levels of Proteobacteria in patients with mild disease than in patients with moderate/severe disease, this may suggest that this group could play a role in the initiation of the disease.
Fig 4.
Microbial composition of patients with mild CD and moderate/severe CD and in controls. (A) Bacteroidetes; (B) Firmicutes; (C) Proteobacteria.
Conclusions.
In conclusion, significant differences in the microbial composition of patients with CD and controls were detected, highlighting specific bacterial groups that may be associated with disease outcome. Furthermore, the observed correlation between specific bacterial groups and patient PCDAI emphasizes the involvement of these groups in initiating or driving inflammation and possibly the disease.
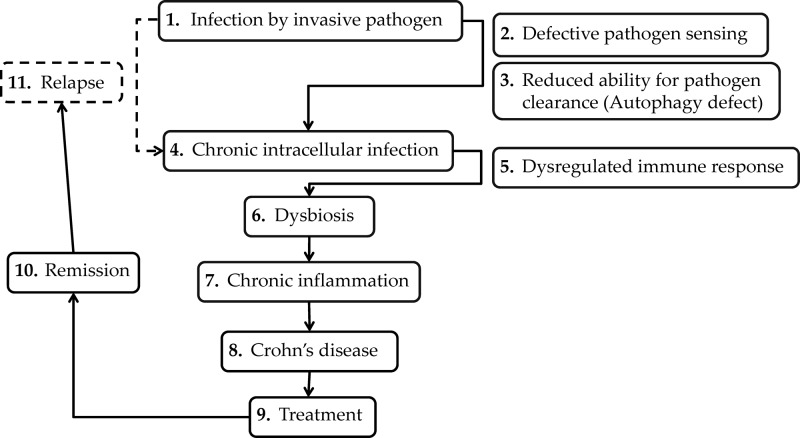
We propose the hypothesis (Fig. 5) whereby an invasive pathogen (possibly a member of the Proteobacteria) capable of surviving intracellularly (as evidenced by the association between autophagy and CD) establishes an infection in patients. Defective pathogen sensing and reduced ability for pathogen clearance by the host (47) result in a chronic intracellular infection. As a result of a dysregulated immune response, chronic inflammation occurs, which is driven by the dysbiotic intestinal microflora, and in particular Bacteroides species. Following treatment, disease remission occurs; however, reinfection leads to relapse of the disease.
Fig 5.
Proposed mechanism of initiation and recurrence of Crohn's disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was made possible by the support of the National Health and Medical Research Council, Australia, and The University of New South Wales Goldstar award. N.O.K. is supported by an Early Career fellowship from the National Health and Medical Research Council, Australia.
No conflicts of interest exist.
Footnotes
Published ahead of print 25 July 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Australia Ecol. 26:32–46 [Google Scholar]
- 2. Andreotti R, et al. 2011. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. 11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey MT, et al. 2010. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect. Immun. 78:1509–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baron EJ. 1997. Bilophila wadsworthia: a unique Gram-negative anaerobic rod. Anaerobe 3:83–86 [DOI] [PubMed] [Google Scholar]
- 5. Bloom SM, et al. 2011. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe 9:390–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brand S. 2009. Crohn's disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn's disease. Gut 58:1152–1167 [DOI] [PubMed] [Google Scholar]
- 7. Brenner DJ, et al. 1991. Proposal of Afipia gen. nov., with Afipia felis sp. nov. (formerly the cat scratch disease bacillus), Afipia clevelandensis sp. nov. (formerly the Cleveland Clinic Foundation strain), Afipia broomeae sp. nov., and three unnamed genospecies. J. Clin. Microbiol. 29:2450–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bruck WM, Sennett SH, Pomponi SA, Willenz P, McCarthy PJ. 2008. Identification of the bacterial symbiont Entotheonella sp. in the mesohyl of the marine sponge Discodermia sp. ISME J. 2:335–339 [DOI] [PubMed] [Google Scholar]
- 9. Capone KA, Dowd SE, Stamatas GN, Nikolovski J. 2011. Diversity of the human skin microbiome early in life. J. Investig. Dermatol. 131:2026–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collins MD, et al. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812–826 [DOI] [PubMed] [Google Scholar]
- 11. Darfeuille-Michaud A, et al. 2004. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127:412–421 [DOI] [PubMed] [Google Scholar]
- 12. DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dicksved J, et al. 2007. Molecular fingerprinting of the fecal microbiota of children raised according to different lifestyles. Appl. Environ. Microbiol. 73:2284–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dowd SE, et al. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dowd SE, et al. 2011. Survey of fungi and yeast in polymicrobial infections in chronic wounds. J. Wound Care 20:40–47 [DOI] [PubMed] [Google Scholar]
- 16. Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA. 2008. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog. Dis. 5:459–472 [DOI] [PubMed] [Google Scholar]
- 17. Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461 [DOI] [PubMed] [Google Scholar]
- 18. Egert M, de Graaf AA, Smidt H, de Vos WM, Venema K. 2006. Beyond diversity: functional microbiomics of the human colon. Trends Microbiol. 14:86–91 [DOI] [PubMed] [Google Scholar]
- 19. Eren AM, et al. 2011. Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PLoS One 6:e26732 doi:10.1371/journal.pone.0026732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Finegold SM, et al. 2010. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 16:444–453 [DOI] [PubMed] [Google Scholar]
- 21. Frank DN, et al. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104:13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fudou R, Jojima Y, Iizuka T, Yamanaka S. 2002. Haliangium ochraceum gen. nov., sp. nov. and Haliangium tepidum sp. nov.: novel moderately halophilic myxobacteria isolated from coastal saline environments. J. Gen. Appl. Microbiol. 48:109–116 [DOI] [PubMed] [Google Scholar]
- 23. Garrett WS, et al. 2010. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8:292–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Griffiths AM, et al. 2004. Inflammatory bowel disease, p 789–825 In Walker A, Goulet OJ, Kleinman RE, Sherman PM, Shneider BL, Sanderson I. (ed), Pediatric gastrointestinal disease, 4th ed, vol 1 BC Decker Inc., Hamilton, Ontario, Canada [Google Scholar]
- 25. Gutierrez A, et al. 2009. Cytokine association with bacterial DNA in serum of patients with inflammatory bowel disease. Inflamm. Bowel Dis. 15:508–514 [DOI] [PubMed] [Google Scholar]
- 26. Handl S, Dowd SE, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS. 2011. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol. Ecol. 76:301–310 [DOI] [PubMed] [Google Scholar]
- 27. Harriff MJ, Bermudez LE, Kent ML. 2007. Experimental exposure of zebrafish, Danio rerio (Hamilton), to Mycobacterium marinum and Mycobacterium peregrinum reveals the gastrointestinal tract as the primary route of infection: a potential model for environmental mycobacterial infection. J. Fish Dis. 30:587–600 [DOI] [PubMed] [Google Scholar]
- 28. Hayashi H, Shibata K, Sakamoto M, Tomita S, Benno Y. 2007. Prevotella copri sp. nov. and Prevotella stercorea sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 57:941–946 [DOI] [PubMed] [Google Scholar]
- 29. Higgins LM, Frankel G, Douce G, Dougan G, MacDonald TT. 1999. Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect. Immun. 67:3031–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hyams J, et al. 2005. Evaluation of the pediatric Crohn disease activity index: a prospective multicenter experience. J. Pediatr. Gastroenterol. Nutr. 41:416–421 [DOI] [PubMed] [Google Scholar]
- 31. Kaakoush NO, et al. 2011. The pathogenic potential of Campylobacter concisus strains associated with chronic intestinal diseases. PLoS One 6:e29045 doi:10.1371/journal.pone.0029045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaakoush NO, Mitchell HM. 2012. Campylobacter concisus—a new player in intestinal disease. Front. Cell. Infect. Microbiol. 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karafin M, et al. 2010. Fatal infection caused by Cupriavidus gilardii in a child with aplastic anemia. J. Clin. Microbiol. 48:1005–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kostic AD, et al. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22:292–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lay C, et al. 2005. Colonic microbiota signatures across five northern European countries. Appl. Environ. Microbiol. 71:4153–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lindquist D, et al. 2003. Characteristics of Massilia timonae and Massilia timonae-like isolates from human patients, with an emended description of the species. J. Clin. Microbiol. 41:192–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu JZ, Pezeshki M, Raffatellu M. 2009. Th17 cytokines and host-pathogen interactions at the mucosa: dichotomies of help and harm. Cytokine 48:156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loubinoux J, Bronowicki JP, Pereira IA, Mougenel JL, Faou AE. 2002. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol. Ecol. 40:107–112 [DOI] [PubMed] [Google Scholar]
- 39. Lupp C, et al. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:119–129 [DOI] [PubMed] [Google Scholar]
- 40. Man SM, et al. 2010. Host attachment, invasion, and stimulation of proinflammatory cytokines by Campylobacter concisus and other non-Campylobacter jejuni Campylobacter species. J. Infect. Dis. 202:1855–1865 [DOI] [PubMed] [Google Scholar]
- 41. Man SM, Kaakoush NO, Mitchell HM. 2011. The role of bacteria and pattern-recognition receptors in Crohn's disease. Nat. Rev. Gastroenterol. Hepatol. 8:152–168 [DOI] [PubMed] [Google Scholar]
- 42. Martin HM, et al. 2004. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology 127:80–93 [DOI] [PubMed] [Google Scholar]
- 43. Mueller S, et al. 2006. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl. Environ. Microbiol. 72:1027–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nagao M, et al. 2009. Surgical site infection due to Mycobacterium peregrinum: a case report and literature review. Int. J. Infect. Dis. 13:209–211 [DOI] [PubMed] [Google Scholar]
- 45. Ott SJ, et al. 2004. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53:685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rehman A, et al. 2010. Transcriptional activity of the dominant gut mucosal microbiota in chronic inflammatory bowel disease patients. J. Med. Microbiol. 59:1114–1122 [DOI] [PubMed] [Google Scholar]
- 47. Sartor RB. 1997. Enteric microflora in IBD: pathogens or commensals? Inflamm. Bowel Dis. 3:230–235 [PubMed] [Google Scholar]
- 48. Sokol H, Lay C, Seksik P, Tannock GW. 2008. Analysis of bacterial bowel communities of IBD patients: what has it revealed? Inflamm. Bowel Dis. 14:858–867 [DOI] [PubMed] [Google Scholar]
- 49. Sokol H, et al. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105:16731–16736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Strauss J, et al. 2011. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 17:1971–1978 [DOI] [PubMed] [Google Scholar]
- 51. Swanson KS, et al. 2011. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 5:639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Swidsinski A, et al. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44–54 [DOI] [PubMed] [Google Scholar]
- 53. Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. 2005. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43:3380–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vitali SD, Eden PA, Payne KL, Vaughan RJ. 2006. An outbreak of mycobacteriosis in Gouldian finches caused by Mycobacterium peregrinum. Vet. Clin. North Am. Exot. Anim. Pract. 9:519–522 [DOI] [PubMed] [Google Scholar]
- 55. Wagner J, et al. 2008. Identification and characterisation of Pseudomonas 16S ribosomal DNA from ileal biopsies of children with Crohn's disease. PLoS One 3:e3578 doi:10.1371/journal.pone.0003578 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.