Abstract
Lytic enzymes are the group of hydrolases that break down structural polymers of the cell walls of various microorganisms. In this work, we determined the nucleotide sequences of the Lysobacter sp. strain XL1 alpA and alpB genes, which code for, respectively, secreted lytic endopeptidases L1 (AlpA) and L5 (AlpB). In silico analysis of their amino acid sequences showed these endopeptidases to be homologous proteins synthesized as precursors similar in structural organization: the mature enzyme sequence is preceded by an N-terminal signal peptide and a pro region. On the basis of phylogenetic analysis, endopeptidases AlpA and AlpB were assigned to the S1E family [clan PA(S)] of serine peptidases. Expression of the alpA and alpB open reading frames (ORFs) in Escherichia coli confirmed that they code for functionally active lytic enzymes. Each ORF was predicted to have the Shine-Dalgarno sequence located at a canonical distance from the start codon and a potential Rho-independent transcription terminator immediately after the stop codon. The alpA and alpB mRNAs were experimentally found to be monocistronic; transcription start points were determined for both mRNAs. The synthesis of the alpA and alpB mRNAs was shown to occur predominantly in the late logarithmic growth phase. The amount of alpA mRNA in cells of Lysobacter sp. strain XL1 was much higher, which correlates with greater production of endopeptidase L1 than of L5.
INTRODUCTION
To suppress competing microorganisms, bacteria produce and secrete into the ambient medium a broad arsenal of antimicrobial factors such as porins, nucleases, bacteriocins similar to phage tails, peptide antibiotics, etc. (1, 10, 28, 35, 39). Some of the microbial antagonism factors are lytic enzymes secreted by bacteria. A target of bacteriolytic enzymes is peptidoglycan, the main structural component of the bacterial cell wall. Depending on which bonds in peptidoglycan they hydrolyze, bacteriolytic enzymes are classified into four groups (17, 21). Glucosaminidases and muramidases cleave different bonds in peptidoglycan glucan chains, amidases hydrolyze the amide bond between muramic acid and the peptide subunit, and peptidases cleave the peptide bonds in peptide subunits or interpeptide bridges. Unlike bacteriophage-encoded lysins, whose action is directed to one species or even a limited group of bacterial strains (16, 31), bacteriolytic enzymes of bacterial origin have a broad spectrum of antimicrobial activity (3, 53). Given that peptidoglycan has a conserved structure, the probability of the emergence of bacteria resistant to the action of lytic enzymes, especially ones with broad substrate specificity, is extremely low. Owing to these properties, the use of bacteriolytic enzymes in medicine as antimicrobial agents, especially against pathogenic microorganisms with multiple drug resistance, is promising.
The bacterium Lysobacter sp. strain XL1 secretes a variety of lytic enzymes into the ambient medium. The antimicrobial lysoamidase preparation obtained from the culture liquid of this bacterium is active against a broad range of microorganisms, in particularly bacteria of the genera Staphylococcus, Bacillus, Streptococcus, Micrococcus, Kocuria, Peptostreptococcus, Corynebacterium, Streptomyces, and Alcaligenes; some other bacteria; and yeasts of the genera Saccharomyces, Candida, and Pseudozyma (24). The preparation also prevents the germination of bacterial and fungal spores (6, 37). To date, five bacteriolytic enzymes, a metalloprotease active against yeasts, and a phosphatase have been isolated from the culture liquid of Lysobacter sp. strain XL1 and characterized to various degrees (7, 42–44, 46, 47, 51).
The bacteriolytic enzymes of the lysoamidase preparation are represented by three endopeptidases, L1, L4, and L5, as well as N-acetylmuramoyl-l-alanine amidase L2 and muramidase L3 (42, 45, 47, 51). It is obvious that the occurrence of lytic enzymes specific to different bonds in peptidoglycan determines a broad range of lysoamidase antibacterial activities. From inhibition data, endopeptidases L1, L4, and L5 have been assigned to the serine protease catalytic type (42, 51). Endopeptidase L1, a small protein with a molecular mass of about 22 kDa, has a broad substrate specificity and hydrolyzes the amide bonds in proteins, peptides, and microbial cell walls. With respect to bacterial peptidoglycans, this enzyme exhibits both amidase and endopeptidase activities, the latter of which is the more potent. It hydrolyzes the amide bond between muramic acid and alanine and the peptide bonds between diaminopimelic acid and alanine, as well as between glycines of the interpeptide bridge. Endopeptidase L5 has a molecular mass of about 26 kDa. It occurs in culture medium in much smaller amounts than L1, and its enzymatic properties have been little studied. L1 and L5 are thermostable enzymes with reaction optima of 70 and 80°C, respectively.
Endopeptidases L1 and L5 have different spectra of antimicrobial activity (18, 19). For instance, cells of Bacillus subtilis and Micrococcus luteus are lysed by both enzymes. At the same time, only L1 is capable of destroying Staphylococcus aureus and Bacillus cereus cells, whereas L5 (unlike L1) lyses Kocuria rosea and Alcaligenes faecalis cells.
It has been shown that endopeptidases L1 and L5 are secreted via different pathways. Endopeptidase L5 is secreted via outer membrane vesicles, whereas L1 is not found in them. Secretion via vesicles promotes the delivery of L5 to peptidoglycans of Gram-negative bacteria through the outer membrane, which significantly expands the antimicrobial action spectrum of L5 (51).
Combination of lytic enzymes with different substrate specificities would enable the development of novel antimicrobial preparations with a broad action range. Cloning of genes of Lysobacter sp. strain XL1 lytic enzymes and their characterization are required both for further research and for applied research and development, in particular, to create producer strains.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Lysobacter sp. strain XL1 was from the collection of the Laboratory of Microbial Cell Surface Biochemistry. Cells of this strain were grown at 28°C on Luria-Bertani (LB) medium for isolation of genomic DNA and on CY medium (30) for isolation of RNA.
For cloning and plasmid isolation, we used Escherichia coli strain DH5α (52).
Oligonucleotide primers.
The oligonucleotide primers used in this study are listed in Table S1 in the supplemental material.
Isolation of nucleic acids.
Genomic DNA was isolated by phenol extraction as described previously (38). Plasmid DNA was isolated using a QIAquick Plasmid Purification kit (Qiagen). Total RNA was isolated from cells in the mid- or late-logarithmic growth phase using an RNeasy Protect Mini kit (Qiagen).
Construction of recombinant plasmids.
Plasmids pALPI-29a and pALPII-29a bear the alpA and alpB open reading frames (ORFs), respectively. To construct these plasmids, Lysobacter sp. strain XL1 DNA fragments that contained the alpA or alpB ORF were amplified by PCR with primers 14 and 15 and primers 16 and 17, respectively, and inserted into the NdeI and XhoI sites of vector pET-29a (Novagen). All constructs were confirmed by sequencing.
Vector pUC18-lic is intended for ligation-independent cloning of PCR fragments by forming 15-nucleotide (nt) 5′ protruding ends (Y. S. Lapteva, unpublished data). For this, the vector was digested with restriction endonuclease SmaI and treated with T4 DNA polymerase in the presence of dATP. The vector was used for determination of the 5′ ends of the alpA and alpB mRNAs.
Nucleic acid hybridization.
Southern and Northern blot hybridizations were performed as described previously (38). Radioactive labeling of DNA probes was carried out using a DECAprime II DNA Labeling kit (Ambion Inc.).
DNA-DNA hybridization.
Five micrograms of Lysobacter sp. strain XL1 genomic DNA was digested by restriction endonucleases, separated in a 1% agarose gel, and transferred to a Hybond-N membrane (GE Healthcare). As the probe, we used a 32P-labeled, 426-bp-long DNA fragment of Lysobacter sp. strain XL1 coding for the mature part of endopeptidase L1. The fragment was amplified by PCR using primers 1 and 2.
RNA-DNA hybridization.
Thirty micrograms of Lysobacter sp. strain XL1 total RNA was separated in a 1.2% denaturing agarose gel and transferred to a Hybond-N membrane. DNA probes specific to the alpA and alpB genes were amplified by PCR with primers 3 and 4 and primers 5 and 6, respectively.
Determination of mRNA 5′ ends.
The 5′ ends of the alpA and alpB mRNAs were determined by the 5′ rapid amplification of cDNA ends (RACE) protocol. The first cDNA strand was obtained with 5 μg of total RNA and 10 nmol of primer 7 or 8 specific to the ORF of alpA or alpB, respectively, using a RevertAid First Strand cDNA Synthesis kit (Fermentas). RNA was removed by alkaline hydrolysis. The cDNA was cleaned of nucleotides and primers. One-third of the purified DNA was ligated with 5 ng of oligonucleotide 9 using T4 RNA ligase at room temperature for 48 h. Amplification of DNA was carried out by PCR with primers 12 (0.5 μM), 10 (0.025 μM), and 13 (0.5 μM) for alpA and 12 (0.5 μM), 11 (0.025 μM), and 13 (0.5 μM) for alpB. PCR fragments were purified from a 2% agarose gel using a QIAquick Gel Extraction kit (Qiagen), treated with T4 DNA polymerase in the presence of dTTP, cloned into plasmid vector pUC18-lic, and sequenced.
Phylogenetic analysis of proteins.
Protein sequences homologous to AlpA and AlpB were searched for in the GenBank database (http://www.ncbi.nlm.nih.gov/) by using the BLAST program. Amino acid sequences of proteins were aligned using the CLUSTAL X program (49). The tree was constructed by means of the TREECON package (50) by the neighbor-joining method. The statistical significance of branching was assessed by bootstrap analysis of 1,000 alternative trees using the respective function of the TREECON program. The evolutionary distance was expressed as the number of substitutions per 100 amino acids (aa).
Expression of alpA and alpB ORFs in E. coli.
Cells of E. coli BL21(DE3) (48) were transformed with plasmid pALPI-29a or pALPII-29a, plated on dishes with agarized LB medium with kanamycin (25 μg/ml), and grown overnight at 37°C. The grown colonies were replated on agarized Davis medium (12) containing disrupted cells of S. aureus 209P, 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and 25 μg/ml kanamycin. Cells were grown at 28°C for 48 to 72 h.
Nucleotide sequence accession number.
The final sequence of the 9,017-bp fragment of the Lysobacter sp. strain XL1 genome determined in this study has been deposited in GenBank under accession number GU188567.
RESULTS AND DISCUSSION
Search for and determination of the nucleotide sequence of the endopeptidase L1 gene.
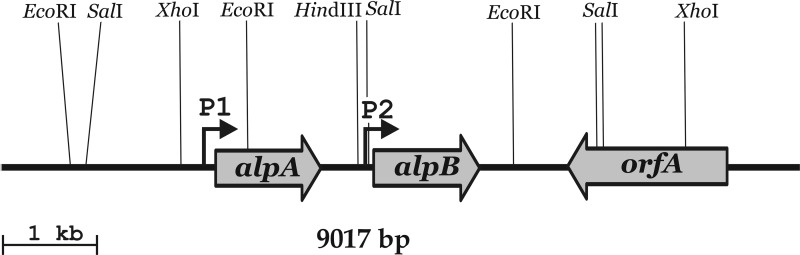
When cloning the gene for Lysobacter sp. strain XL1 endopeptidase L1, we based our approach on earlier data on the amino acid sequences of the N terminus of the mature enzyme and its proteolytic fragments (UniProtKB accession no. P85142). Considering the high similarity of the sequences of peptides of endopeptidase L1 and the α-lytic protease of Lysobacter enzymogenes (GenBank accession no. AAA74111) (14), which are 88 to 92% identical (29), we designed a pair of degenerate primers (1 and 2), amplified a Lysobacter sp. strain XL1 genome fragment 426 bp long with them by PCR, and determined its nucleotide sequence. Analysis of the in silico-translated sequence showed the fragment to contain a segment of the endopeptidase L1 gene. The fragment synthesized by PCR was used as a probe to identify the genomic-DNA restriction fragments bearing the endopeptidase L1 gene by hybridization (see Fig. S1 in the supplemental material). Then, by means of inverse PCR, we amplified EcoRI and SalI fragments (containing the alpA and alpB ORFs, Fig. 1) and established their nucleotide sequences. As we found, in addition to the endopeptidase L1 gene (alpA), this contig contained one more ORF that was named alpB. Its product, AlpB, exhibited a high degree of homology to AlpA (see below). Analysis showed that the putative protein AlpB contained a sequence corresponding to the experimentally established sequence of the N terminus of the mature region of Lysobacter sp. strain XL1 endopeptidase L5 (UniProtKB accession no. P85158). As the ORFs of endopeptidases L1 and L5 were found to be located next to each other, we assumed that the genes of lytic enzymes in the Lysobacter sp. strain XL1 genome could form a single cluster. For this reason, we determined the sequences of the genome segments flanking the alpA and alpB ORFs. As a result, a 9,017-bp contig was constructed in which no other lytic enzyme ORFs were found at distances of 2.4 kb upstream of alpA and 3.6 kb downstream of alpB (Fig. 1, GenBank accession no. GU188567). Downstream of the alpB gene in the sequenced fragment, there is an ORF designated orfA, oriented oppositely with respect to alpA and alpB. Analysis of the Pfam database (http://pfam.sanger.ac.uk) (15) showed that the 600-aa protein OrfA presumably belongs to the family of ABC transporters and contains a TMD (transmembrane domain; pf00664) and an ABC (ATP binding cassette; pf00005) domain characteristic of these proteins.
Fig 1.
Map of the Lysobacter sp. strain XL1 alpA, alpB, and orfA gene cluster (GenBank accession no. GU188567). Gray arrows indicate the arrangement and orientation of the ORFs. P1 and P2 are, respectively, transcription start points for the alpA and alpB genes. Locations of restriction endonuclease sites are shown.
AlpA and AlpB are secreted lytic peptidases.
The secreted proteases of microorganisms are synthesized, as a rule, in the form of inactive preproenzymes and contain, at the N end, a signal peptide required for translocation across the cytoplasmic membrane, a pro region, and a mature enzyme. The pro region, as has been shown for the α-lytic endopeptidase of L. enzymogenes (2, 4, 40), catalyzes the folding of the enzyme and, after autocatalytic cleavage, inhibits its activity. The latter function is important for preventing the digestion of peptidoglycan of the producing cell before the enzyme leaves the periplasmic space. We analyzed the amino acid sequences of AlpA and AlpB to establish the structural organization of these proteins. A search for signal peptides was carried out using the SignalP 3.0 program (13). Using this program, the signal peptides for both proteins were predicted to exist by the algorithm of the hidden Markov model with a probability of 1.000. According to the prediction, the positions of the processing sites are located, with a probability of 1.000, between aa 33 and 34 of AlpA and between aa 28 and 29 of AlpB. The boundaries between the pro regions and mature enzymes were determined on the basis of the data on the N-terminal sequences of mature enzymes L1 and L5 isolated from the culture liquid of Lysobacter sp. strain XL1. On the basis of this analysis, the proenzyme processing sites were found to be between aa 199 and 200 of AlpA and between aa 194 and 195 of AlpB. Summing up the data of the amino acid sequence analysis, it can be concluded that AlpA, with a length of 398 aa, has the following structure: signal peptide, 33 aa; pro region, 166 aa; mature enzyme, 199 aa. Close to it in size, AlpB, with a length of 399 aa, has a similar structural organization: signal peptide, 28 aa; pro region, 166 aa; mature enzyme, 205 aa.
The mature AlpA and AlpB enzymes are close in molecular mass, at 19.8 and 20.8 kDa, and in pI, at 9.63 and 9.44, respectively. Nevertheless, the electrophoretic mobilities of mature, naturally secreted AlpA and AlpB in denaturing PAGE (25) differ significantly and correspond to the mobilities of proteins with molecular masses of 22 and 26 kDa (51). The same electrophoretic mobilities were observed for mature recombinant endopeptidases AlpA and AlpB, preparations of which were obtained by in vitro renaturation of purified recombinant proenzymes and subsequent autocatalytic processing (O. R. Latypov, unpublished data). Besides, during the expression in E. coli of the alpA and alpB ORFs fused at the 3′ end with the sequence coding for 6×His, the mature endopeptidases secreted into the culture liquid contained a His tag at the C terminus and therewith preserved the difference in electrophoretic mobility (A. E. Kalinin, unpublished data). These data make it unlikely that there is posttranslational modification of AlpB or processing of AlpA at the C terminus. Apparently, an anomalous electrophoretic mobility of AlpB is due to features of the enzyme's structure, which is more resistant to the action of SDS. In favor of this assumption is the significantly greater temperature stability of AlpB than AlpA. It has been shown that the activity of AlpB drops almost 2-fold after incubation at 75°C for 15 min, whereas AlpA already loses half of its activity after incubation at 55°C for the same time (42). Besides, both endopeptidases are capable of preserving their activity in the presence of SDS (A. E. Kalinin, unpublished data).
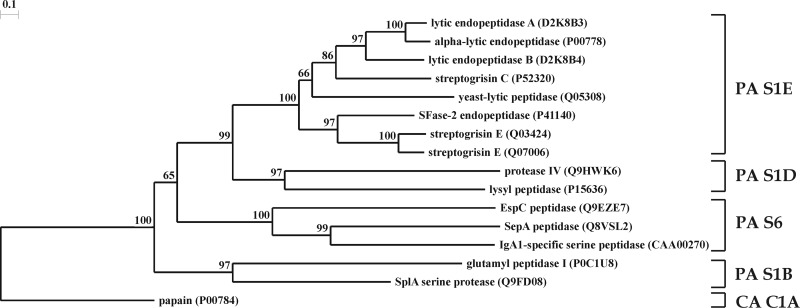
In the modern classification, peptidases are divided into clans, families, and subfamilies based on their evolutionary relationship, catalytic mechanism, and order in the polypeptide chain of amino acid residues involved in catalysis (5). A clan may contain enzymes that use various mechanisms of catalysis, whose relationship is exhibited only in the similarity of tertiary structures. The phylogenetic analysis of the sequences of mature endopeptidases AlpA and AlpB (Fig. 2) enabled us to assign these enzymes to clan PA, subclan S of serine peptidases, family S1, and subfamily S1E, the prototype of which is chymotrypsin A. Clan PA is a mixed clan and unites serine and cysteine peptidases. The sequences of AlpA and AlpB exhibit the highest similarity to the α-lytic protease of L. enzymogenes; the precursor of this enzyme is 78 and 58% identical to preproenzymes AlpA and AlpB, respectively. Alignment of the sequences of the L. enzymogenes α-lytic protease, AlpA, and AlpB (Fig. 3) revealed in the latter two the catalytic triad His (H235 for AlpA, H234 for AlpB), Asp (D262 for AlpA, D261 for AlpB), and Ser (S343 for AlpA and AlpB), characteristic of serine proteases of the subclan PA(S) (33). It should be noted that AlpA and AlpB, like all characterized representatives of the chymotrypsin family, are endopeptidases (7, 34) (MEROPS database; http://merops.sanger.ac.uk/).
Fig 2.
Phylogeny of endopeptidases AlpA and AlpB. This phylogenetic tree of bacterial serine proteases was constructed on the basis of homology between the amino acid sequences of mature enzymes. Accession numbers of sequences in the UniProt database are given. Enzymes from the following organisms are presented: P00778, L. enzymogenes; D2K8B3 and D2K8B4, Lysobacter sp. strain XL1; P52320 and Q07006, Streptomyces griseus; Q03424 and P41140, Streptomyces fradiae; Q05308, Rarobacter faecitabidus; P0C1U8 and Q9FD08, S. aureus; P15636, Achromobacter lyticus; Q9HWK6, Pseudomonas aeruginosa PAO1; Q8VSL2, Shigella flexneri; CAA00270 (GenBank accession number), Neisseria gonorrhoeae; Q9EZE7, E. coli; P00784, Carica papaya. Papain was selected as an outgroup. Protease clans (PA and CA) and families (S1E, S1B, S1D, and S6) are given at the right in accordance with the MEROPS peptidase database (34; http://merops.sanger.ac.uk/).
Fig 3.
Alignment of the amino acid sequences of Lysobacter sp. strain XL1 lytic endopeptidases AlpA and AlpB and the L. enzymogenes α-lytic protease (GenBank accession no. P00778). Sequences corresponding to signal peptides, pro regions, and mature enzymes are shown. The amino acid residues of the catalytic His-Asp-Ser triad are boxed.
The well-characterized α-lytic protease of L. enzymogenes has specificity toward peptide substrates with small hydrophobic side chains before a scissile bond. For this enzyme, it was shown that residues M343 and M363 (numbered according to Fig. 3), which line primary specificity pocket S1, as well as most of the amino acid residues of a surface loop consisting of aa 367 to 385 adjacent to the S1 pocket, are essential for the enzyme's specificity (26). Replacement of these residues with alanines broadened the specificity of the α-lytic protease significantly. In particular, the enzyme containing either an M343A or an M363A substitution is able to cleave peptides with large hydrophobic side chains before a scissile bond efficiently (9). The substrate specificity of AlpA and AlpB has not been studied yet. Sequence alignment of AlpA, AlpB, and the α-lytic protease of L. enzymogenes (Fig. 3) shows that the amino acid residues of AlpA and the α-lytic protease at the above sequence positions are identical, whereas those of AlpB differ significantly. In AlpB, the Met residues at positions 343 and 363 are replaced by Thr and there are 11 substitutions in the 19-aa surface loop. Therefore, it is feasible to suggest that the substrate specificity of AlpA is similar to that of the α-lytic protease, while AlpB has a different specificity. The different spectra of antibacterial activity of AlpA and AlpB count in favor of this suggestion. Nevertheless, it is not possible to definitively predict the substrate specificity of AlpB because of a lack of knowledge about the composition of the cell wall of the bacteria lysed by L5 and cleavage sites in interpeptide bridges.
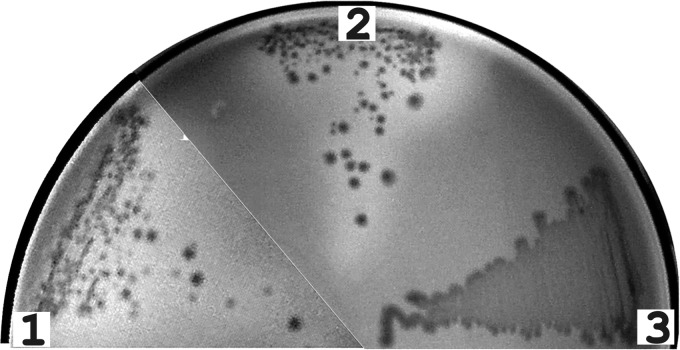
To check their functional activity, the alpA and alpB genes were individually cloned under the control of the T7lac promoter and expressed in E. coli. For this, E. coli BL21(DE3) cells transformed with corresponding plasmids (see Materials and Methods) were grown in the presence of the inducer on an agarized medium containing disrupted S. aureus cells. Lytic activity appeared as zones of lysis around colonies. As shown by the results of the assay depicted in Fig. 4, zones of lysis were observed only around colonies ofE. coli cells that expressed the alpA and alpB genes but were absent around colonies of host E. coli cells transformed with the plasmid vector. It should be noted that the alpA and alpB gene products proved toxic for E. coli cells, which was manifested in a significantly lower growth rate on Davis minimal medium in the presence of the inducer. It appears that the peptidoglycan of this bacterium is sensitive to the action of the AlpA and AlpB peptidases.
Fig 4.
Expression of the genes for Lysobacter sp. strain XL1 lytic endopeptidases AlpA and AlpB in E. coli. A lytic-activity test is shown. Cells of E. coli BL21(DE3) transformed with vector pET-29a (sector 3) or with plasmid pALPI-29a (sector 2) or pALPII-29 (sector 1) bearing the alpA or alpB gene, respectively, were grown on an agarized medium containing cell walls of S. aureus 209-P (see Materials and Methods). Lytic activity is seen as clarification zones around colonies.
On the basis of the analysis conducted, it can be concluded that the alpA and alpB genes code for secreted peptidases L1 and L5 of Lysobacter sp. strain XL1. It is unlikely that the putative ABC transporter OrfA, which we identified in this study, is involved in the secretion of AlpA or AlpB. ABC transporters carry diverse substrates, including proteins, across cellular membranes. Nevertheless, sequence analysis does not allow the prediction of their substrate specificity. In Gram-negative bacteria, an ABC transporter is a component of the type I secretion system that carries proteins that are synthesized without an N-terminal signal peptide (8), whereas proteins with an N-terminal signal peptide, such as AlpA and AlpB, are usually secreted by a type II or V secretion system.
The alpA and alpB ORFs are transcribed from their own promoters.
Lysobacter sp. strain XL1 releases ∼20 times as much endopeptidase L1 as L5 into its culture liquid (51). To explain the difference, we analyzed the translation and transcription signals for their ORFs.
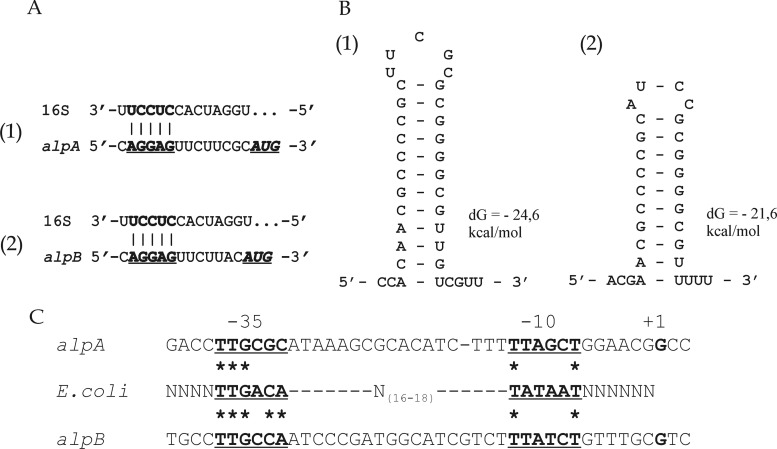
Analysis of the nucleotide sequences upstream of the alpA and alpB ORFs established the occurrence of a Shine-Dalgarno sequence before the initiating ATG codon at a distance of 8 nt for alpA and 7 nt for alpB. For both ORFs, it is a polypurine sequence, AGGAG, that is complementary to the sequence at the 3′ end of the 16S rRNA gene for various species of Lysobacter deposited in the GenBank database (Fig. 5A). The occurrence of a Shine-Dalgarno sequence at a canonical distance from the start codon, as well as the absence of stem-loop structures in this region (data not shown), does not make it possible to assume a significant difference in the efficiency of translation initiation between alpA and alpB.
Fig 5.
Analysis of translation and transcription signals for alpA (parts 1) and alpB (parts 2) of Lysobacter sp. strain XL1. (A) Fragments of the 5′ UTR sequences of the alpA (bp 2391 to 2407) and alpB (bp 4190 to 4205) mRNAs and the sequence at the 3′ end of the 16S rRNA of Lysobacter sp. strain Shinshu-th3 (bp 1530 to 1543; GenBank accession no. AB121774) are shown. The putative ribosome-binding sites in the mRNAs of alpA and alpB are in bold and underlined. The start codons are in bold italics and underlined. (B) Fragments of the 3′ UTR sequences of the alpA (bp 3628 to 3656) and alpB (bp 5430 to 5453) mRNAs capable of forming stem-loop structures are shown. The sequences were analyzed using the UNAFold package (27; http://mfold.rna.albany.edu/). Hairpin energy (in kcal/mol) is given on the right. Positions of the ribosome-binding sites and stem-loop structures are given in accordance with GenBank accession no. GU188567. (C) Analysis of alpA and alpB −35 and −10 regions compared with the corresponding consensus sequences of the E. coli σ70 promoter (bold and underlined). The transcription start point (+1) is in bold. Asterisks indicate identical bases.
Analysis of the nucleotide sequences located after the stop codons of the alpA and alpB ORFs made it possible to identify the occurrence of putative intrinsic transcription terminators (Fig. 5B) (36). Thus, at a distance of 26 nt from alpA ORF stop codon, there is a 29-member palindromic sequence capable of forming a hairpin with a perfect stem, rich in GC pairs (75%), of 12 bp and a central loop of 5 unpaired bases. So high a content of GC pairs is usually characteristic of Rho-independent terminators (11). At a distance of 27 nt from the alpB ORF stop codon, there is also a 24-member palindromic sequence that forms a stem-loop structure of 20 paired (80% GC) and 4 unpaired bases. Immediately after the hairpin, there is an oligo(dT) sequence.
The finding of potential transcription terminators, as well as the distance of 602 nt between these ORFs, suggested that the alpA and alpB genes have their own promoters. This suggestion was examined by Northern blot hybridization of the total RNA of Lysobacter sp. strain XL1 with DNA probes specific to the alpA and alpB ORFs (Fig. 6). As shown by the hybridization results, cells of Lysobacter sp. strain XL1 contain transcripts about 1.5 kb long that comprise the alpA or alpB ORF. Larger-size transcripts potentially containing both ORFs were not found. Besides, alpA mRNA is found in the mid-exponential growth phase and its level significantly increases by the late exponential phase-early stationary phase of growth, whereas the amount of alpB mRNA is significantly lower than that of alpA mRNA and the transcript is detected only by the late exponential growth phase.
Fig 6.
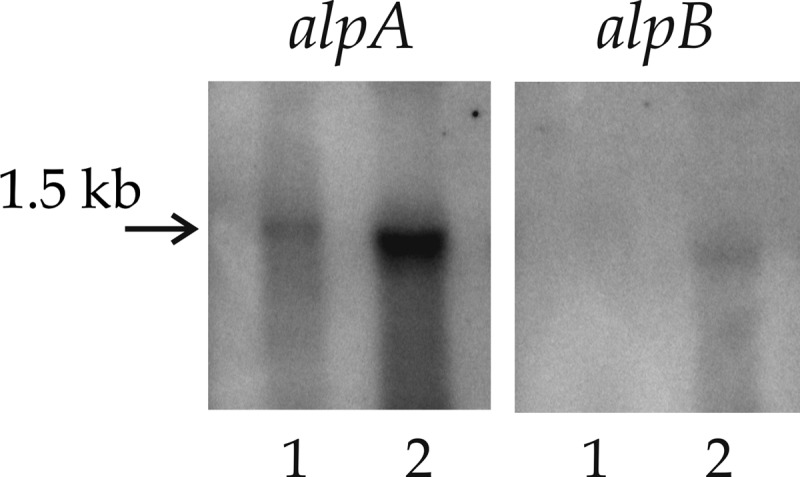
Transcription analysis of alpA and alpB expression. The total RNA of Lysobacter sp. strain XL1 cells in the mid-logarithmic (lanes 1) or late logarithmic (lanes 2) growth phase was hybridized with a 32P-labeled probe to alpA or alpB.
Using the 5′ RACE technique, we determined the 5′ ends of the alpA and alpB mRNAs (Table 1). In 8 of 11 clones of alpA and 7 of 9 clones of alpB, the positions of the transcription start points coincided. Three alpA cDNA clones and two alpB cDNA clones corresponded to a shorter 5′ untranslated region (UTR). We assume that this could be due to the premature termination of cDNA synthesis or a partial degradation of mRNA from the 5′ end. Thus, the length of the 5′ UTR of the alpA mRNA is 134 nt and that of the alpB mRNA 5′ UTR is 140 nt. In both mRNAs, the 5′-terminal nucleotide residue is G.
Table 1.
Analysis of 5′ ends of alpA and alpB mRNAs by the 5′ RACE protocol
| Gene and position of transcription start pointa | 5′ UTR length (nt) | No. of clones |
|---|---|---|
| alpA | ||
| 2271 | 134 | 8 |
| 2275 | 130 | 1 |
| 2276 | 129 | 1 |
| 2287 | 118 | 1 |
| alpB | ||
| 4063 | 140 | 7 |
| 4092 | 111 | 1 |
| 4111 | 92 | 1 |
In accordance with GenBank accession no. GU188567.
We analyzed the nucleotide sequences of the alpA and alpB genes in the −35 and −10 regions by aligning them with the corresponding consensus sequences of the E. coli σ70 promoter (Fig. 5C). This analysis showed that the putative promoter of alpB corresponds more closely to the E. coli sequence than that of alpA does. In the case of alpA, 3 out of 6 nt were found to match the E. coli consensus sequence in the −35 region and 2 nt were found to match it in the −10 region. In the case of alpB, there are five matches to the E. coli consensus sequence in the −35 region and two in the −10 region. In spite of the evolutionary conservation of σ70 (20, 22, 32), it is evident that the sequences of the promoters of Lysobacter bacteria in the −35 and −10 regions can differ significantly from the consensus sequences of the E. coli σ70 promoter. In particular, in Xanthomonas campestris, which, like Lysobacter sp. strain XL1, is a member of the Xanthomonadaceae family, an analysis of the frequency of occurrence of potentially strong gene promoters conducted on the basis of the E. coli σ70 consensus sequences has shown that such promoters are, in fact, absent from the genome (41). Besides, the activity of the alpA and alpB gene promoters can be regulated by transcription factors or else depend on alternative σ factors. In favor of this is the significant increase in the amount of the alpA and alpB mRNAs in cells in the late exponential growth phase. Involvement of transcription factors in the regulation of expression of lytic enzymes was shown for L. enzymogenes strain C3. In this strain, the production of lytic enzymes is regulated by the product of the gene clp, which has been assigned to the CRP family of global transcription regulators (23).
Thus, on the basis of the results of the analysis of the nucleotide sequences and cellular transcripts of alpA and alpB, it can be concluded that these genes are each transcribed from their own promoters. Nevertheless, these findings are not sufficient to explain the differences in the levels of alpA and alpB mRNAs, which can be due to differences in promoter strength, mRNA stability, or both.
The results obtained enable a more profound study of the regulation of alpA and alpB gene expression, as well as a study of the structural features of the encoded proteins responsible for the choice of secretion pathway and substrate specificities.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant 06-04-08240 from the Russian Foundation for Basic Research and the federal program Scientific and Scientific-Pedagogical Personnel of Innovative Russia 2009-2013, state contract P1045.
Footnotes
Published ahead of print 3 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abriouel H, Franz CM, Ben Omar N, Galvez A. 2011. Diversity and applications of Bacillus bacteriocins. FEMS Microbiol. Rev. 35: 201–232 [DOI] [PubMed] [Google Scholar]
- 2. Anderson DE, Peters RJ, Wilk B, Agard DA. 1999. Alpha-lytic protease precursor: characterization of a structured folding intermediate. Biochemistry 38: 4728–4735 [DOI] [PubMed] [Google Scholar]
- 3. Aratani H, Kawata S, Tsuruyama S, Yoshida N, Makisumi S. 1984. Purification and characterization of an aminopeptidase from bovine leukocytes. J. Biochem. 96: 107–115 [DOI] [PubMed] [Google Scholar]
- 4. Baker D, Silen JL, Agard DA. 1992. Protease pro region required for folding is a potent inhibitor of the mature enzyme. Proteins 12: 339–344 [DOI] [PubMed] [Google Scholar]
- 5. Barrett AJ, Tolle DP, Rawlings ND. 2003. Managing peptidases in the genomic era. Biol. Chem. 384: 873–882 [DOI] [PubMed] [Google Scholar]
- 6. Begunova EA, et al. 2006. Study of the effect of lysoamidase on the spore form of bacteria of the genus Bacillus. Dokl. Biochem. Biophys. 408: 152–154 [DOI] [PubMed] [Google Scholar]
- 7. Begunova EA, Stepnaya OA, Lysanskaya VY, Kulaev IS. 2003. Specificity of the action of lysoamidase on Staphylococcus aureus 209P cell walls. Biochemistry (Mosc.) 68: 735–739 [DOI] [PubMed] [Google Scholar]
- 8. Bleves S, et al. 2010. Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int. J. Med. Microbiol. 300: 534–543 [DOI] [PubMed] [Google Scholar]
- 9. Bone R, Silen JL, Agard DA. 1989. Structural plasticity broadens the specificity of an engineered protease. Nature 339: 191–195 [DOI] [PubMed] [Google Scholar]
- 10. Cascales E, et al. 2007. Colicin biology. Microbiol. Mol. Biol. Rev. 71: 158–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. d'Aubenton Carafa Y, Brody E, Thermes C. 1990. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J. Mol. Biol. 216: 835–858 [DOI] [PubMed] [Google Scholar]
- 12. Davis BD. 1949. The isolation of biochemically deficient mutants of bacteria by means of penicillin. Proc. Natl. Acad. Sci. U. S. A. 35: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- 14. Epstein DM, Wensink PC. 1988. The alpha-lytic protease gene of Lysobacter enzymogenes. The nucleotide sequence predicts a large prepro-peptide with homology to pro-peptides of other chymotrypsin-like enzymes. J. Biol. Chem. 263: 16586–16590 [PubMed] [Google Scholar]
- 15. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fischetti VA. 2008. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11: 393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghuysen JM. 1968. Use of bacteriolytic enzymes in determination of wall structure and their role in cell metabolism. Bacteriol. Rev. 32: 425–464 [PMC free article] [PubMed] [Google Scholar]
- 18. Granovsky IE, et al. 2010. Lytic protease AlpA of Lysobacter sp. XL1 bacteria, fragment of DNA coding lytic protease AlpA of Lysobacter sp. XL1 bacteria, method for preparation of lytic protease AlpA of Lysobacter sp. XL1 bacteria. Russian Federation patent 2407782. [Google Scholar]
- 19. Granovsky IE, et al. 2011. Lytic protease AlpB of Lysobacter sp. XL1 bacteria, fragment of DNA coding lytic protease AlpB of Lysobacter sp. XL1 bacteria, method for preparation of lytic protease AlpB of Lysobacter sp. XL1 bacteria. Russian Federation patent 2408725. [Google Scholar]
- 20. Gruber TM, Bryant DA. 1997. Molecular systematic studies of eubacteria, using sigma70-type sigma factors of group 1 and group 2. J. Bacteriol. 179: 1734–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Höltje JV, Mirelman D, Sharon N, Schwarz U. 1975. Novel type of murein transglycosylase in Escherichia coli. J. Bacteriol. 124: 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iyer LM, Koonin EV, Aravind L. 2004. Evolution of bacterial RNA polymerase: implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene 335: 73–88 [DOI] [PubMed] [Google Scholar]
- 23. Kobayashi DY, Reedy RM, Palumbo JD, Zhou JM, Yuen GY. 2005. A clp gene homologue belonging to the Crp gene family globally regulates lytic enzyme production, antimicrobial activity, and biological control activity expressed by Lysobacter enzymogenes strain C3. Appl. Environ. Microbiol. 71: 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kulaev IS, et al. 2006. Bacteriolytic complex, method for producing said complex and strain for carrying out said method. U.S. patent 7,150,985 B2. [Google Scholar]
- 25. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- 26. Mace JE, Wilk BJ, Agard DA. 1995. Functional linkage between the active site of alpha-lytic protease and distant regions of structure: scanning alanine mutagenesis of a surface loop affects activity and substrate specificity. J. Mol. Biol. 251: 116–134 [DOI] [PubMed] [Google Scholar]
- 27. Markham NR, Zuker M. 2008. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 453: 3–31 [DOI] [PubMed] [Google Scholar]
- 28. Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84: 499–510 [DOI] [PubMed] [Google Scholar]
- 29. Muranova TA, Krasovskaya LA, Tsfasman IM, Stepnaya OA, Kulaev IS. 2004. Structural investigations and identification of the extracellular bacteriolytic endopeptidase L1 from Lysobacter sp. XL1. Biochemistry (Mosc.) 69: 501–505 [DOI] [PubMed] [Google Scholar]
- 30. Novick RP, Brodsky R. 1972. Studies on plasmid replication. I. Plasmid incompatibility and establishment in Staphylococcus aureus. J. Mol. Biol. 68: 285–302 [DOI] [PubMed] [Google Scholar]
- 31. O'Flaherty S, Ross RP, Coffey A. 2009. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 33: 801–819 [DOI] [PubMed] [Google Scholar]
- 32. Paget MS, Helmann JD. 2003. The sigma70 family of sigma factors. Genome Biol. 4: 203 doi:10.1186/gb-2003-4-1-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rawlings ND, Barrett AJ. 1994. Families of serine peptidases. Methods Enzymol. 244: 19–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rawlings ND, Barrett AJ, Bateman A. 2012. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40: D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Riley MA, Wertz JE. 2002. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84: 357–364 [DOI] [PubMed] [Google Scholar]
- 36. Roberts JW, Shankar S, Filter JJ. 2008. RNA polymerase elongation factors. Annu. Rev. Microbiol. 62: 211–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ryazanova LP, Stepnaya OA, Suzina NE, Kulaev IS. 2005. Antifungal action of the lytic enzyme complex from Lysobacter sp. XL1. Process Biochem. 40: 557–564 [Google Scholar]
- 38. Sambrook J, Fritch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor NY [Google Scholar]
- 39. Severinov K, Semenova E, Kazakov A, Kazakov T, Gelfand MS. 2007. Low-molecular-weight post-translationally modified microcins. Mol. Microbiol. 65: 1380–1394 [DOI] [PubMed] [Google Scholar]
- 40. Silen JL, Frank D, Fujishige A, Bone R, Agard DA. 1989. Analysis of prepro-alpha-lytic protease expression in Escherichia coli reveals that the pro region is required for activity. J. Bacteriol. 171: 1320–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sinoquet C, Demey S, Braun F. 2008. Large-scale computational and statistical analyses of high transcription potentialities in 32 prokaryotic genomes. Nucleic Acids Res. 36: 3332–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stepanaia OA, Begunova EA, Tsfasman IM, Kulaev IS. 1996. Bacteriolytic enzyme preparation lysoamidase. Purification and some properties of bacteriolytic peptidase L1. Biokhimiia 61: 656–663. (In Russian.) [PubMed] [Google Scholar]
- 43. Stepanaia OA, et al. 1992. Some properties of bacteriolytic protease L2. Prikl. Biokhim. Mikrobiol. 28: 666–673. (In Russian.) [PubMed] [Google Scholar]
- 44. Stepnaia OA, Severin AI, Krupyanko VI, Kulaev IS. 1986. Purification and some properties of neutral phosphatase of the enzyme preparation of lysoamidase isolated from Pseudomonadaceae bacteria. Biokhimiia 51: 684–690. (In Russian.) [PubMed] [Google Scholar]
- 45. Stepnaia OA, Severin AI, Kulaev IS. 1986. Some physico-chemical properties of lytic proteinase L2 of the enzyme preparation lysoamidase isolated from bacteria of Pseudomonadaceae family. Biokhimiia 51: 909–915. (In Russian.) [PubMed] [Google Scholar]
- 46. Stepnaya OA, Tsfasman IM, Chaika IA, Muranova TA, Kulaev IS. 2008. Extracellular yeast-lytic enzyme of the bacterium Lysobacter sp. XL1. Biochemistry (Mosc.) 73: 310–314 [DOI] [PubMed] [Google Scholar]
- 47. Stepnaya OA, et al. 2005. Isolation and characterization of a new extracellular bacteriolytic endopeptidase of Lysobacter sp. XL1. Biochemistry (Mosc.) 70: 1031–1037 [DOI] [PubMed] [Google Scholar]
- 48. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189: 113–130 [DOI] [PubMed] [Google Scholar]
- 49. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Van de Peer Y, De Wachter R. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10: 569–570 [DOI] [PubMed] [Google Scholar]
- 51. Vasilyeva NV, Tsfasman IM, Suzina NE, Stepanaia OA, Kulaev IS. 2008. Secretion of bacteriolytic endopeptidase L5 of Lysobacter sp. XL1 into the medium by means of outer membrane vesicles. FEBS J. 275: 3827–3835 [DOI] [PubMed] [Google Scholar]
- 52. Woodcock DM, et al. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17: 3469–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yoshimoto T, Tsuru D. 1972. Studies on bacteriolytic enzymes. II. Purification and some properties of two types of staphylolytic enzymes from Streptomyces griseus. J. Biochem. 72: 379–390 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.